Apoptosis Inhibitor 5: A Multifaceted Regulator of Cell Fate
Abstract
:1. Introduction
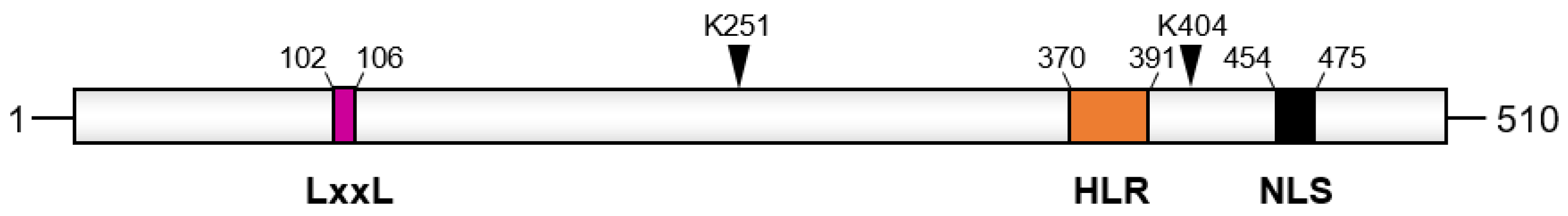
2. Structure and Binding Partners of API5
3. Physiological Functions of API5
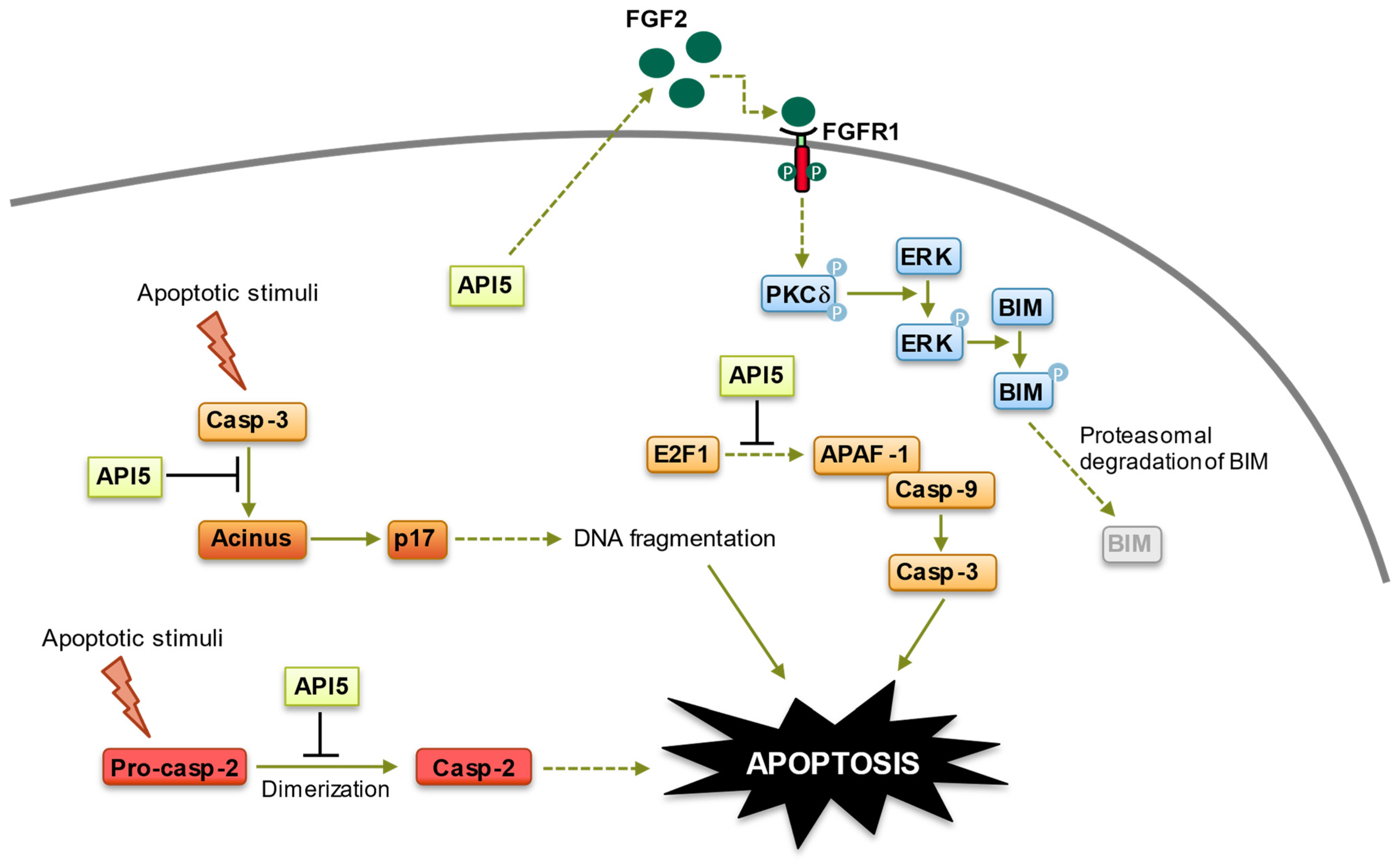
3.1. Anti-Apoptotic Functions of API5
3.1.1. Inhibition of E2F1 (E2F Transcription Factor 1)-Induced Apoptosis
3.1.2. Inhibition of Acinus (Apoptotic Chromatin Condensation Inducer in the Nucleus)-Induced Apoptotic DNA Fragmentation
3.1.3. Inhibition of Caspase-2 Activation
3.1.4. Fibroblast Growth Factor Receptor 1 (FGFR1)/Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Signaling-Mediated Degradation of BIM
3.2. Cell Cycle Regulation Functions of API5
3.3. mRNA Export Functions of API5
3.4. Modulation of TLR4 Signaling in Dendritic Cells and Adjuvant Effect of API5
3.5. Paneth Cell Protective Action of API5 as a Putative Therapeutic Target for Crohn’s Disease
3.6. Enhancement of Viral Replication by API5
4. API5 and Cancer
4.1. API5 Expression and Prognosis Value
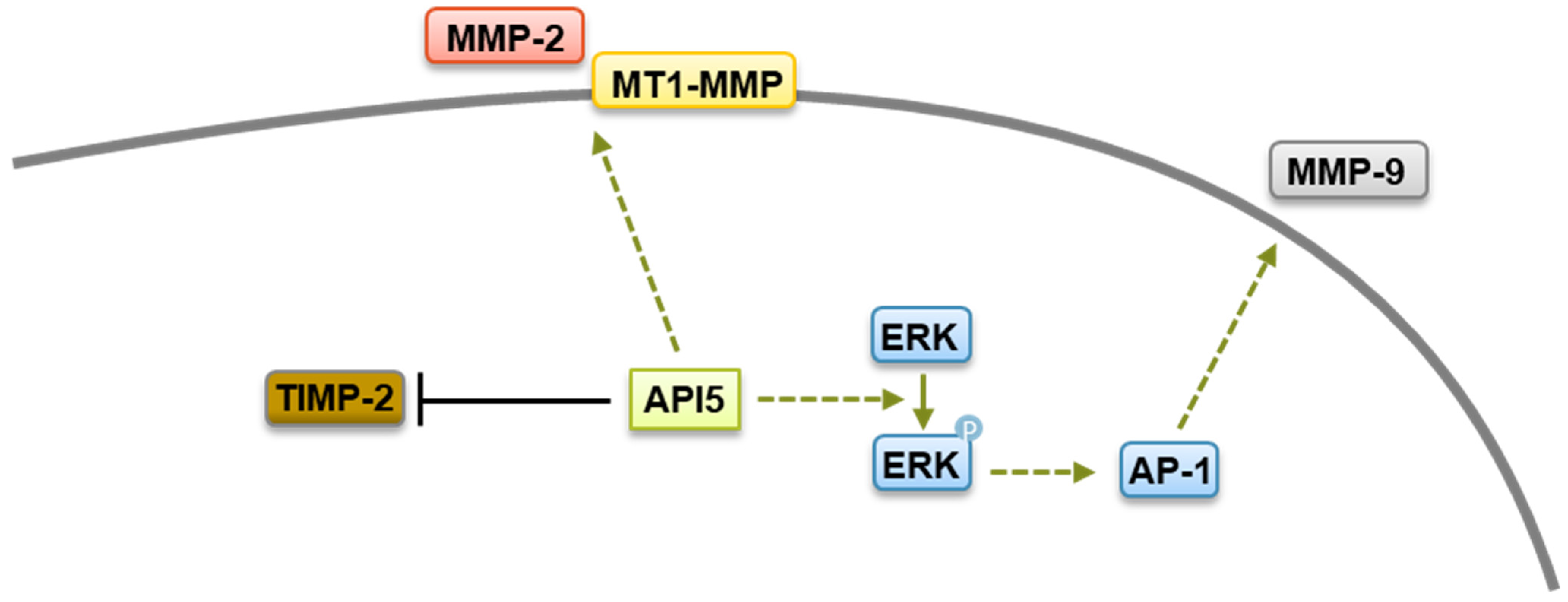
4.2. API5’s Role on Cancer Metastasis, Immune Response, and Survival
4.3. Targeting API5 as a Therapeutic Approach
5. Conclusions and Future Directions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vermeulen, K.; Van Bockstaele, D.R.; Berneman, Z.N. Apoptosis: Mechanisms and relevance in cancer. Ann. Hematol. 2005, 84, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, Y.; Steller, H. Live to die another way: Modes of programmed cell death and the signals emanating from dying cells. Nat. Rev. Mol. Cell Biol. 2015, 16, 329–344. [Google Scholar] [CrossRef]
- Geske, F.J.; Gerschenson, L.E. The biology of apoptosis. Hum. Pathol. 2001, 32, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Meier, P.; Vousden, K.H. Lucifer’s labyrinth—Ten years of path finding in cell death. Mol. Cell 2007, 28, 746–754. [Google Scholar] [CrossRef]
- Igney, F.H.; Krammer, P.H. Death and anti-death: Tumour resistance to apoptosis. Nat. Rev. Cancer 2002, 2, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.S. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011, 30, 87. [Google Scholar] [CrossRef]
- Pfeffer, C.M.; Singh, A.T.K. Apoptosis: A Target for Anticancer Therapy. Int. J. Mol. Sci. 2018, 19, 448. [Google Scholar] [CrossRef]
- Tuveson, D.; Hanahan, D. Translational medicine: Cancer lessons from mice to humans. Nature 2011, 471, 316–317. [Google Scholar] [CrossRef]
- Green, D.R. Caspases and Their Substrates. Cold Spring Harb. Perspect. Biol. 2022, 14, a041012. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Festjens, N.; Declercq, W.; Berghe, T.V.; Vandenabeele, P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007, 14, 44–55. [Google Scholar] [CrossRef]
- Thornberry, N.A.; Lazebnik, Y. Caspases: Enemies within. Science 1998, 281, 1312–1316. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yuan, J. Caspases in apoptosis and beyond. Oncogene 2008, 27, 6194–6206. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 2002, 9, 459–470. [Google Scholar] [CrossRef]
- Krueger, A.; Schmitz, I.; Baumann, S.; Krammer, P.H.; Kirchhoff, S. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J. Biol. Chem. 2001, 276, 20633–20640. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Figueiredo-Pereira, C.; Oudot, C.; Vieira, H.L.A.; Brenner, C. Mitochondrion: A Common Organelle for Distinct Cell Deaths? Int. Rev. Cell Mol. Biol. 2017, 331, 245–287. [Google Scholar] [PubMed]
- Bao, Q.; Shi, Y. Apoptosome: A platform for the activation of initiator caspases. Cell Death Differ. 2007, 14, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Sinha, S.; Kroemer, G. Bcl-2 family members: Dual regulators of apoptosis and autophagy. Autophagy 2008, 4, 600–606. [Google Scholar] [CrossRef]
- Kontomanolis, E.N.; Koutras, A.; Syllaios, A.; Schizas, D.; Mastoraki, A.; Garmpis, N.; Diakosavvas, M.; Angelou, K.; Tsatsaris, G.; Pagkalos, A.; et al. Role of Oncogenes and Tumor-suppressor Genes in Carcinogenesis: A Review. Anticancer. Res. 2020, 40, 6009–6015. [Google Scholar] [CrossRef]
- Plati, J.; Bucur, O.; Khosravi-Far, R. Apoptotic cell signaling in cancer progression and therapy. Integr. Biol. 2011, 3, 279–296. [Google Scholar] [CrossRef]
- Reed, J.C. Apoptosis-targeted therapies for cancer. Cancer Cell 2003, 3, 17–22. [Google Scholar] [CrossRef]
- Souers, A.J.; Leverson, J.D.; Boghaert, E.R.; Ackler, S.L.; Catron, N.D.; Chen, J.; Dayton, B.D.; Ding, H.; Enschede, S.H.; Fairbrother, W.J.; et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013, 19, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Kotschy, A.; Szlavik, Z.; Murray, J.; Davidson, J.; Maragno, A.L.; Le Toumelin-Braizat, G.; Chanrion, M.; Kelly, G.L.; Gong, J.-N.; Moujalled, D.M.; et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 2016, 538, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Caenepeel, S.; Brown, S.P.; Belmontes, B.; Moody, G.; Keegan, K.S.; Chui, D.; Whittington, D.A.; Huang, X.; Poppe, L.; Cheng, A.C.; et al. AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies. Cancer Discov. 2018, 8, 1582–1597. [Google Scholar] [CrossRef]
- Tron, A.E.; Belmonte, M.A.; Adam, A.; Aquila, B.M.; Boise, L.H.; Chiarparin, E.; Cidado, J.; Embrey, K.J.; Gangl, E.; Gibbons, F.D.; et al. Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia. Nat. Commun. 2018, 9, 5341. [Google Scholar] [CrossRef] [PubMed]
- Tewari, M.; Yu, M.; Ross, B.; Dean, C.; Giordano, A.; Rubin, R. AAC-11, a novel cDNA that inhibits apoptosis after growth factor withdrawal. Cancer Res. 1997, 57, 4063–4069. [Google Scholar]
- Basset, C.; Bonnet-Magnaval, F.; Navarro, M.G.-J.; Touriol, C.; Courtade, M.; Prats, H.; Garmy-Susini, B.; Lacazette, E. Api5 a new cofactor of estrogen receptor alpha involved in breast cancer outcome. Oncotarget 2017, 8, 52511–52526. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, G.; Feugeas, J.-P.; Gu, Y.; Leboeuf, C.; El Bouchtaoui, M.; Lu, H.; Espié, M.; Janin, A.; Di Benedetto, M. High expression of apoptosis protein (Api-5) in chemoresistant triple-negative breast cancers: An innovative target. Oncotarget 2019, 10, 6577–6588. [Google Scholar] [CrossRef]
- Cho, H.; Chung, J.-Y.; Song, K.-H.; Noh, K.H.; Kim, B.W.; Chung, E.J.; Ylaya, K.; Kim, J.H.; Kim, T.W.; Hewitt, S.M.; et al. Apoptosis inhibitor-5 overexpression is associated with tumor progression and poor prognosis in patients with cervical cancer. BMC Cancer 2014, 14, 545. [Google Scholar] [CrossRef]
- Kim, J.W.; Cho, H.S.; Kim, J.H.; Hur, S.Y.; Kim, T.E.; Lee, J.M.; Kim, I.K.; Namkoong, S.E. AAC-11 overexpression induces invasion and protects cervical cancer cells from apoptosis. Lab. Investig. 2000, 80, 587–594. [Google Scholar] [CrossRef]
- Krejci, P.; Pejchalova, K.; Rosenbloom, B.E.; Rosenfelt, F.P.; Tran, E.L.; Laurell, H.; Wilcox, W.R. The antiapoptotic protein Api5 and its partner, high molecular weight FGF2, are up-regulated in B cell chronic lymphoid leukemia. J. Leukoc. Biol. 2007, 82, 1363–1364. [Google Scholar] [CrossRef]
- Noh, K.H.; Kim, S.-H.; Kim, J.H.; Song, K.-H.; Lee, Y.-H.; Kang, T.H.; Han, H.D.; Sood, A.K.; Ng, J.; Kim, K.; et al. API5 confers tumoral immune escape through FGF2-dependent cell survival pathway. Cancer Res. 2014, 74, 3556–3566. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Moriyama, S.; Yukiue, H.; Kobayashi, Y.; Nakashima, Y.; Kaji, M.; Fukai, I.; Kiriyama, M.; Yamakawa, Y.; Fujii, Y. Expression of the antiapoptosis gene, AAC-11, as a prognosis marker in non-small cell lung cancer. Lung Cancer 2001, 34, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H.; Cho, H.; Lee, H.J.; Oh, S.J.; Woo, S.R.; Hong, S.O.; Jang, H.S.; Noh, K.H.; Choi, C.H.; Chung, J.Y.; et al. API5 confers cancer stem cell-like properties through the FGF2-NANOG axis. Oncogenesis 2017, 6, e285. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H.; Kim, S.H.; Noh, K.H.; Bae, H.C.; Kim, J.H.; Lee, H.J.; Song, J.; Kang, T.H.; Kim, D.W.; Oh, S.J.; et al. Apoptosis Inhibitor 5 Increases Metastasis via Erk-mediated MMP expression. BMB Rep. 2015, 48, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, H.; Liu, B.; Ma, W.; Xue, X.; Chen, J.; Zhou, Q. Gene expression levels of CSNK1A1 and AAC-11, but not NME1, in tumor tissues as prognostic factors in NSCLC patients. Med. Sci. Monit. 2010, 16, CR357-64. [Google Scholar] [PubMed]
- Li, X.; Gao, X.; Wei, Y.; Deng, L.; Ouyang, Y.; Chen, G.; Li, X.; Zhang, Q.; Wu, C. Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell 2011, 23, 1416–1434. [Google Scholar] [CrossRef]
- Klim, J.; Gładki, A.; Kucharczyk, R.; Zielenkiewicz, U.; Kaczanowski, S. Ancestral State Reconstruction of the Apoptosis Machinery in the Common Ancestor of Eukaryotes. G3 2018, 8, 2121–2134. [Google Scholar] [CrossRef]
- Chen, M.; Wu, W.; Liu, D.; Lv, Y.; Deng, H.; Gao, S.; Gu, Y.; Huang, M.; Guo, X.; Liu, B.; et al. Evolution and Structure of API5 and Its Roles in Anti-Apoptosis. Protein Pept. Lett. 2021, 28, 612–622. [Google Scholar] [CrossRef]
- Morris, E.J.; Michaud, W.A.; Ji, J.-Y.; Moon, N.-S.; Rocco, J.W.; Dyson, N.J. Functional identification of Api5 as a suppressor of E2F-dependent apoptosis in vivo. PLoS Genet. 2006, 2, e196. [Google Scholar] [CrossRef]
- Han, B.G.; Kim, K.H.; Lee, S.J.; Jeong, K.C.; Cho, J.W.; Noh, K.H.; Kim, T.W.; Kim, S.J.; Yoon, H.J.; Suh, S.W.; et al. Helical repeat structure of apoptosis inhibitor 5 reveals protein-protein interaction modules. J. Biol. Chem. 2012, 287, 10727–10737. [Google Scholar] [CrossRef]
- Chothia, C.; Gough, J.; Vogel, C.; Teichmann, S.A. Evolution of the protein repertoire. Science 2003, 300, 1701–1703. [Google Scholar] [CrossRef] [PubMed]
- Apic, G.; Gough, J.; Teichmann, S.A. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 2001, 310, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Caetano-Anolles, G. The evolutionary mechanics of domain organization in proteomes and the rise of modularity in the protein world. Structure 2009, 17, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Kobe, B.; Deisenhofer, J. The leucine-rich repeat: A versatile binding motif. Trends Biochem. Sci. 1994, 19, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Andrade, M.A.; Perez-Iratxeta, C.; Ponting, C.P. Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 2001, 134, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Tu, D.; Li, W.; Ye, Y.; Brunger, A.T. Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc. Natl. Acad. Sci. USA 2007, 104, 15599–15606. [Google Scholar] [CrossRef]
- Bong, S.M.; Bae, S.-H.; Song, B.; Gwak, H.; Yang, S.-W.; Kim, S.; Nam, S.; Rajalingam, K.; Oh, S.J.; Kim, T.W.; et al. Regulation of mRNA export through API5 and nuclear FGF2 interaction. Nucleic Acids Res. 2020, 48, 6340–6352. [Google Scholar] [CrossRef]
- Van den Berghe, L.; Laurell, H.; Huez, I.; Zanibellato, C.; Prats, H.; Bugler, B. FIF [fibroblast growth factor-2 (FGF-2)-interacting-factor], a nuclear putatively antiapoptotic factor, interacts specifically with FGF-2. Mol. Endocrinol. 2000, 14, 1709–1724. [Google Scholar] [CrossRef]
- Ahel, D.; Hořejší, Z.; Wiechens, N.; Polo, S.E.; Garcia-Wilson, E.; Ahel, I.; Flynn, H.; Skehel, M.; West, S.C.; Jackson, S.P.; et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 2009, 325, 1240–1243. [Google Scholar] [CrossRef]
- Mayank, A.K.; Sharma, S.; Nailwal, H.; Lal, S.K. Nucleoprotein of influenza A virus negatively impacts antiapoptotic protein API5 to enhance E2F1-dependent apoptosis and virus replication. Cell Death Dis. 2015, 6, e2018. [Google Scholar] [CrossRef]
- Imre, G.; Berthelet, J.; Heering, J.; Kehrloesser, S.; Melzer, I.M.; Lee, B.I.; Thiede, B.; Dötsch, V.; Rajalingam, K. Apoptosis inhibitor 5 is an endogenous inhibitor of caspase-2. EMBO Rep. 2017, 18, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Park, H.J.; Park, J.H.; Hong, E.J.; Jang, G.-Y.; Jung, I.D.; Han, H.D.; Lee, S.-H.; Vo, M.-C.; Lee, J.-J.; et al. A novel function of API5 (apoptosis inhibitor 5), TLR4-dependent activation of antigen presenting cells. OncoImmunology 2018, 7, e1472187. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; Lahiri, M. Interplay between p300 and HDAC1 regulate acetylation and stability of Api5 to regulate cell proliferation. Sci. Rep. 2021, 11, 16427. [Google Scholar] [CrossRef] [PubMed]
- Habault, J.; Thonnart, N.; Pasquereau-Kotula, E.; Bagot, M.; Bensussan, A.; Villoutreix, B.O.; Marie-Cardine, A.; Poyet, J.L. PAK1-dependent anti-tumor effect of AAC-11-derived peptides on Sézary syndrome malignant CD4+ T lymphocytes. J. Investig. Dermatol. 2021, 141, 2261–2271. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Hu, B.; Wang, X.; Yan, Y.; Zhou, J.; Lin, L.; Xu, Y.; Zheng, X.; Zhou, J. DeSUMOylation of Apoptosis Inhibitor 5 by Avibirnavirus VP3 Supports Virus Replication. mBio 2021, 12, e0198521. [Google Scholar] [CrossRef] [PubMed]
- Chellappan, S.P.; Hiebert, S.; Mudryj, M.; Horowitz, J.M.; Nevins, J.R. The E2F transcription factor is a cellular target for the RB protein. Cell 1991, 65, 1053–1061. [Google Scholar] [CrossRef]
- Wells, J.; Graveel, C.R.; Bartley, S.M.; Madore, S.J.; Farnham, P.J. The identification of E2F1-specific target genes. Proc. Natl. Acad. Sci. USA 2002, 99, 3890–3895. [Google Scholar] [CrossRef]
- Muller, H.; Bracken, A.P.; Vernell, R.; Moroni, M.C.; Christians, F.; Grassilli, E.; Prosperini, E.; Vigo, E.; Oliner, J.D.; Helin, K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001, 15, 267–285. [Google Scholar] [CrossRef]
- Wells, J.; Boyd, K.E.; Fry, C.J.; Bartley, S.M.; Farnham, P.J. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 2000, 20, 5797–5807. [Google Scholar] [CrossRef]
- Lazzerini Denchi, E.; Helin, K. E2F1 is crucial for E2F-dependent apoptosis. EMBO Rep. 2005, 6, 661–668. [Google Scholar] [CrossRef]
- Qin, X.Q.; Livingston, D.M.; Kaelin, W.G.; Adams, P.D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc. Natl. Acad. Sci. USA 1994, 91, 10918–10922. [Google Scholar] [CrossRef]
- Shan, B.; Lee, W.H. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol. Cell. Biol. 1994, 14, 8166–8173. [Google Scholar] [PubMed]
- Field, S.J.; Tsai, F.Y.; Kuo, F.; Zubiaga, A.M.; Kaelin, W.G.; Livingston, D.M.; Orkin, S.H.; Greenberg, M.E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 1996, 85, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Bell, L.A.; Ryan, K.M. Life and death decisions by E2F-1. Cell Death Differ. 2003, 11, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Rigou, P.; Piddubnyak, V.; Faye, A.; Rain, J.-C.; Michel, L.; Calvo, F.; Poyet, J.-L. The antiapoptotic protein AAC-11 interacts with and regulates Acinus-mediated DNA fragmentation. EMBO J. 2009, 28, 1576–1588. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, Z.; Yang, S.J.; Ye, K. Acinus-provoked protein kinase C delta isoform activation is essential for apoptotic chromatin condensation. Cell Death Differ. 2007, 14, 2035–2046. [Google Scholar] [CrossRef]
- Joselin, A.P.; Schulze-Osthoff, K.; Schwerk, C. Loss of Acinus inhibits oligonucleosomal DNA fragmentation but not chromatin condensation during apoptosis. J. Biol. Chem. 2006, 281, 12475–12484. [Google Scholar] [CrossRef]
- Zermati, Y.; Garrido, C.; Amsellem, S.; Fishelson, S.; Bouscary, D.; Valensi, F.; Varet, B.; Solary, E.; Hermine, O. Caspase activation is required for terminal erythroid differentiation. J. Exp. Med. 2001, 193, 247–254. [Google Scholar] [CrossRef]
- Schwerk, C.; Prasad, J.; Degenhardt, K.; Erdjument-Bromage, H.; White, E.; Tempst, P.; Kidd, V.J.; Manley, J.L.; Lahti, J.M.; Reinberg, D. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol. Cell. Biol. 2003, 23, 2981–2990. [Google Scholar] [CrossRef]
- Zhou, Z.; Licklider, L.J.; Gygi, S.P.; Reed, R. Comprehensive proteomic analysis of the human spliceosome. Nature 2002, 419, 182–185. [Google Scholar] [CrossRef]
- Rappsilber, J.; Ryder, U.; Lamond, A.I.; Mann, M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002, 12, 1231–1245. [Google Scholar] [CrossRef]
- Tange, T.O.; Shibuya, T.; Jurica, M.S.; Moore, M.J. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 2005, 11, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Baliga, B.C.; Read, S.H.; Kumar, S. The biochemical mechanism of caspase-2 activation. Cell Death Differ. 2004, 11, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Brown-Suedel, A.N.; Bouchier-Hayes, L. Caspase-2 Substrates: To Apoptosis, Cell Cycle Control, and Beyond. Front. Cell Dev. Biol. 2020, 8, 610022. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Garcia-Carbonell, R.; Yamachika, S.; Zhao, P.; Dhar, D.; Loomba, R.; Kaufman, R.J.; Saltiel, A.R.; Karin, M. ER Stress Drives Lipogenesis and Steatohepatitis via Caspase-2 Activation of S1P. Cell 2018, 175, 133–145.e15. [Google Scholar] [CrossRef] [PubMed]
- Dehkordi, M.H.; Munn, R.G.K.; Fearnhead, H.O. Non-Canonical Roles of Apoptotic Caspases in the Nervous System. Front. Cell Dev. Biol. 2022, 10, 840023. [Google Scholar] [CrossRef] [PubMed]
- Imre, G.; Heering, J.; Takeda, A.-N.; Husmann, M.; Thiede, B.; zu Heringdorf, D.M.; Green, D.R.; van der Goot, F.G.; Sinha, B.; Dötsch, V.; et al. Caspase-2 is an initiator caspase responsible for pore-forming toxin-mediated apoptosis. EMBO J. 2012, 31, 2615–2628. [Google Scholar] [CrossRef]
- Itoh, N.; Ornitz, D.M. Fibroblast growth factors: From molecular evolution to roles in development, metabolism and disease. J. Biochem. 2011, 149, 121–130. [Google Scholar] [CrossRef]
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef]
- Jang, H.S.; Woo, S.R.; Song, K.-H.; Cho, H.; Chay, D.B.; Hong, S.-O.; Lee, H.-J.; Oh, S.J.; Chung, J.-Y.; Kim, J.-H.; et al. API5 induces cisplatin resistance through FGFR signaling in human cancer cells. Exp. Mol. Med. 2017, 49, e374. [Google Scholar] [CrossRef]
- Luciano, F.; Jacquel, A.; Colosetti, P.; Herrant, M.; Cagnol, S.; Auberger, P. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene 2003, 22, 6785–6793. [Google Scholar] [CrossRef]
- Kohno, M.; Pouyssegur, J. Targeting the ERK signaling pathway in cancer therapy. Ann. Med. 2006, 38, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, T.; Dass, C.R.; Choong, P.F. Bim-targeted cancer therapy: A link between drug action and underlying molecular changes. Mol. Cancer Ther. 2009, 8, 3173–3180. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Jove Navarro, M.; Basset, C.; Arcondéguy, T.; Touriol, C.; Perez, G.; Prats, H.; Lacazette, E. Api5 contributes to E2F1 control of the G1/S cell cycle phase transition. PLoS ONE 2013, 8, e71443. [Google Scholar] [CrossRef] [PubMed]
- DeGregori, J. The genetics of the E2F family of transcription factors: Shared functions and unique roles. Biochim. Biophys. Acta 2002, 1602, 131–150. [Google Scholar] [CrossRef] [PubMed]
- Kuttanamkuzhi, A.; Panda, D.; Malaviya, R.; Gaidhani, G.; Lahiri, M. Altered expression of anti-apoptotic protein Api5 affects breast tumorigenesis. BMC Cancer 2023, 23, 374. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Hurt, E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007, 8, 761–773. [Google Scholar] [CrossRef]
- Wolyniak, M.J.; Cole, C.N. Harnessing genomics to explore the processes and evolution of mRNA export. RNA Biol. 2008, 5, 68–72. [Google Scholar] [CrossRef]
- Natalizio, B.J.; Wente, S.R. Postage for the messenger: Designating routes for nuclear mRNA export. Trends Cell Biol. 2013, 23, 365–373. [Google Scholar] [CrossRef]
- Borden, K.L.B. The Nuclear Pore Complex and mRNA Export in Cancer. Cancers 2020, 13, 42. [Google Scholar] [CrossRef]
- Hurt, J.A.; Silver, P.A. mRNA nuclear export and human disease. Dis. Model. Mech. 2008, 1, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Shen, H. UAP56—A key player with surprisingly diverse roles in pre-mRNA splicing and nuclear export. BMB Rep. 2009, 42, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Medzhitov, R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004, 5, 987–995. [Google Scholar] [CrossRef] [PubMed]
- McClure, R.; Massari, P. TLR-Dependent Human Mucosal Epithelial Cell Responses to Microbial Pathogens. Front. Immunol. 2014, 5, 386. [Google Scholar] [CrossRef] [PubMed]
- Wallet, S.M.; Puri, V.; Gibson, F.C. Linkage of Infection to Adverse Systemic Complications: Periodontal Disease, Toll-Like Receptors, and Other Pattern Recognition Systems. Vaccines 2018, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, A.; Vandenabeele, P.; Krysko, D.V. Necroptosis: The release of damage-associated molecular patterns and its physiological relevance. Immunity 2013, 38, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Nace, G.; Evankovich, J.; Eid, R.; Tsung, A. Dendritic cells and damage-associated molecular patterns: Endogenous danger signals linking innate and adaptive immunity. J. Innate Immun. 2011, 4, 6–15. [Google Scholar] [CrossRef]
- Duan, T.; Du, Y.; Xing, C.; Wang, H.Y.; Wang, R.-F. Toll-Like Receptor Signaling and Its Role in Cell-Mediated Immunity. Front. Immunol. 2022, 13, 812774. [Google Scholar] [CrossRef]
- Halfvarson, J.; Bodin, L.; Tysk, C.; Lindberg, E.; Järnerot, G. Inflammatory bowel disease in a Swedish twin cohort: A long-term follow-up of concordance and clinical characteristics. Gastroenterology 2003, 124, 1767–1773. [Google Scholar] [CrossRef]
- Mu, C.; Zhao, Q.; Zhao, Q.; Yang, L.; Pang, X.; Liu, T.; Li, X.; Wang, B.; Fung, S.Y.; Cao, H. Multi-omics in Crohn’s disease: New insights from inside. Comput. Struct. Biotechnol. J. 2023, 21, 3054–3072. [Google Scholar] [CrossRef] [PubMed]
- Cadwell, K.; Liu, J.Y.; Brown, S.L.; Miyoshi, H.; Loh, J.; Lennerz, J.K.; Kishi, C.; Kc, W.; Carrero, J.A.; Hunt, S.; et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008, 456, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.C.; Bevins, C.L. Paneth cells: Maestros of the small intestinal crypts. Annu. Rev. Physiol. 2013, 75, 289–311. [Google Scholar] [CrossRef] [PubMed]
- Adolph, T.E.; Tomczak, M.F.; Niederreiter, L.; Ko, H.-J.; Böck, J.; Martinez-Naves, E.; Glickman, J.N.; Tschurtschenthaler, M.; Hartwig, J.; Hosomi, S.; et al. Paneth cells as a site of origin for intestinal inflammation. Nature 2013, 503, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Matsuzawa-Ishimoto, Y.; Shono, Y.; Gomez, L.E.; Hubbard-Lucey, V.M.; Cammer, M.; Neil, J.; Dewan, M.Z.; Lieberman, S.R.; Lazrak, A.; Marinis, J.M.; et al. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J. Exp. Med. 2017, 214, 3687–3705. [Google Scholar] [CrossRef] [PubMed]
- Matsuzawa-Ishimoto, Y.; Yao, X.; Koide, A.; Ueberheide, B.M.; Axelrad, J.E.; Reis, B.S.; Parsa, R.; Neil, J.A.; Devlin, J.C.; Rudensky, E.; et al. The gammadelta IEL effector API5 masks genetic susceptibility to Paneth cell death. Nature 2022, 610, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.T.; Cortens, J.P.; Du, Q.; Wilkins, J.A.; Coombs, K.M. Influenza virus induces apoptosis via BAD-mediated mitochondrial dysregulation. J. Virol. 2013, 87, 1049–1060. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Marques, J.T.; Yamashita, M.; Peters, K.L.; Smith, K.; Desai, A.; Williams, B.R.G.; Sen, G.C. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 2010, 29, 1762–1773. [Google Scholar] [CrossRef]
- Zamarin, D.; García-Sastre, A.; Xiao, X.; Wang, R.; Palese, P. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 2005, 1, e4. [Google Scholar] [CrossRef]
- Garcia-Sastre, A.; Biron, C.A. Type 1 interferons and the virus-host relationship: A lesson in detente. Science 2006, 312, 879–882. [Google Scholar] [CrossRef]
- Carvalho, R.; Milne, A.N.; Polak, M.; Offerhaus, G.J.; Weterman, M.A. A novel region of amplification at 11p12-13 in gastric cancer, revealed by representational difference analysis, is associated with overexpression of CD44v6, especially in early-onset gastric carcinomas. Genes Chromosomes Cancer 2006, 45, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Klingbeil, P.; Natrajan, R.; Everitt, G.; Vatcheva, R.; Marchio, C.; Palacios, J.; Buerger, H.; Reis-Filho, J.S.; Isacke, C.M. CD44 is overexpressed in basal-like breast cancers but is not a driver of 11p13 amplification. Breast Cancer Res. Treat. 2010, 120, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Jarvinen, A.K.; Autio, R.; Kilpinen, S.; Saarela, M.; Leivo, I.; Grénman, R.; Mäkitie, A.A.; Monni, O. High-resolution copy number and gene expression microarray analyses of head and neck squamous cell carcinoma cell lines of tongue and larynx. Genes Chromosomes Cancer 2008, 47, 500–509. [Google Scholar] [CrossRef]
- Fukuda, Y.; Kurihara, N.; Imoto, I.; Yasui, K.; Yoshida, M.; Yanagihara, K.; Park, J.-G.; Nakamura, Y.; Inazawa, J. CD44 is a potential target of amplification within the 11p13 amplicon detected in gastric cancer cell lines. Genes Chromosomes Cancer 2000, 29, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Krejci, P.; Koci, L.; Chlebova, K.; Hyzdalova, M.; Hofmanova, J.; Jira, M.; Kysela, P.; Kozubik, A.; Kala, Z. Apoptosis inhibitor 5 (API-5; AAC-11; FIF) is upregulated in human carcinomas in vivo. Oncol. Lett. 2012, 3, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Zhang, W.; Shi, Y.; Gong, J. Pim-2 activates API-5 to inhibit the apoptosis of hepatocellular carcinoma cells through NF-kappaB pathway. Pathol. Oncol. Res. 2010, 16, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Curran, S.; Murray, G.I. Matrix metalloproteinases: Molecular aspects of their roles in tumour invasion and metastasis. Eur. J. Cancer 2000, 36, 1621–1630. [Google Scholar] [CrossRef]
- Coussens, L.M.; Fingleton, B.; Matrisian, L.M. Matrix metalloproteinase inhibitors and cancer: Trials and tribulations. Science 2002, 295, 2387–2392. [Google Scholar] [CrossRef]
- Wu, B.; Crampton, S.P.; Hughes, C.C. Wnt signaling induces matrix metalloproteinase expression and regulates T cell transmigration. Immunity 2007, 26, 227–239. [Google Scholar] [CrossRef]
- Costa, D.B.; Halmos, B.; Kumar, A.; Schumer, S.T.; Huberman, M.S.; Boggon, T.J.; Tenen, D.G.; Kobayashi, S. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007, 4, 1669–1679, discussion 1680. [Google Scholar] [CrossRef] [PubMed]
- Hubner, A.; Barrett, T.; Flavell, R.A.; Davis, R.J. Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol. Cell 2008, 30, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Janin, Y.L. Peptides with anticancer use or potential. Amino Acids 2003, 25, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, L.; Li, M.; Budai, M.M.; Wang, J. Mitochondrion-Mediated Cell Death through Erk1-Alox5 Independent of Caspase-9 Signaling. Cells 2022, 11, 3053. [Google Scholar] [CrossRef]
- Chen, Q.; Song, S.; Wei, S.; Liu, B.; Honjo, S.; Scott, A.; Jin, J.; Ma, L.; Zhu, H.; Skinner, H.D.; et al. ABT-263 induces apoptosis and synergizes with chemotherapy by targeting stemness pathways in esophageal cancer. Oncotarget 2015, 6, 25883–25896. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.J.; Ko, H.-J.; Liao, T.-S.; Zheng, X.-R.; Chou, P.-H.; Wang, L.-T.; Lin, R.-W.; Chen, C.-H.; Wang, C. Severe cellular stress activates apoptosis independently of p53 in osteosarcoma. Cell Death Discov. 2021, 7, 275. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lee, A.T.C.; Ma, J.Z.I.; Wang, J.; Ren, J.; Yang, Y.; Tantoso, E.; Li, K.-B.; Ooi, L.L.P.J.; Tan, P.; et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J. Biol. Chem. 2008, 283, 13205–13215. [Google Scholar] [CrossRef]
- Chopra, M.; Dharmarajan, A.M.; Meiss, G.; Schrenk, D. Inhibition of UV-C light-induced apoptosis in liver cells by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 2009, 111, 49–63. [Google Scholar] [CrossRef]
- Yuan, J.; Liu, Z.; Liu, J.; Fan, R. Circ_0060055 Promotes the Growth, Invasion, and Radioresistance of Glioblastoma by Targeting MiR-197-3p/API5 Axis. Neurotox. Res. 2022, 40, 1292–1303. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Burnett, J.C.; Rossi, J.J. RNA-based therapeutics: Current progress and future prospects. Chem. Biol. 2012, 19, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.; Patil, S.; Zhao, X.; Miao, Z.; Qian, A. RNA Therapeutics—Research and Clinical Advancements. Front. Mol. Biosci. 2021, 8, 710738. [Google Scholar] [CrossRef] [PubMed]
- Attar, N.; Kurdistani, S.K. Exploitation of EP300 and CREBBP Lysine Acetyltransferases by Cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a026534. [Google Scholar] [CrossRef] [PubMed]
- Filippakopoulos, P.; Knapp, S. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 2014, 13, 337–356. [Google Scholar] [CrossRef] [PubMed]
- Habault, J.; Thonnart, N.; Ram-Wolff, C.; Bagot, M.; Bensussan, A.; Poyet, J.-L.; Marie-Cardine, A. Validation of AAC-11-Derived Peptide Anti-Tumor Activity in a Single Graft Sezary Patient-Derived Xenograft Mouse Model. Cells 2022, 11, 2933. [Google Scholar] [CrossRef] [PubMed]
- Jagot-Lacoussiere, L.; Kotula, E.; Villoutreix, B.O.; Bruzzoni-Giovanelli, H.; Poyet, J.L. A Cell-Penetrating Peptide Targeting AAC-11 Specifically Induces Cancer Cells Death. Cancer Res. 2016, 76, 5479–5490. [Google Scholar] [CrossRef] [PubMed]
- Habault, J.; Kaci, A.; Pasquereau-Kotula, E.; Fraser, C.; Chomienne, C.; Dombret, H.; Braun, T.; Pla, M.; Poyet, J.-L. Prophylactic and therapeutic antileukemic effects induced by the AAC-11-derived Peptide RT53. OncoImmunology 2020, 9, 1728871. [Google Scholar] [CrossRef] [PubMed]
- Habault, J.; Fraser, C.; Pasquereau-Kotula, E.; Born-Bony, M.; Marie-Cardine, A.; Poyet, J.-L. Efficient Therapeutic Delivery by a Novel Cell-Penetrating Peptide Derived from Acinus. Cancers 2020, 12, 1858. [Google Scholar] [CrossRef]
- Pasquereau-Kotula, E.; Habault, J.; Kroemer, G.; Poyet, J.-L. The anticancer peptide RT53 induces immunogenic cell death. PLoS ONE 2018, 13, e0201220. [Google Scholar] [CrossRef]
- Papo, N.; Shai, Y. Host defense peptides as new weapons in cancer treatment. Cell. Mol. Life Sci. 2005, 62, 784–790. [Google Scholar] [CrossRef]
- Rosell, M.; Fernández-Recio, J. Hot-spot analysis for drug discovery targeting protein-protein interactions. Expert Opin. Drug Discov. 2018, 13, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Milroy, L.G.; Grossmann, T.N.; Hennig, S.; Brunsveld, L.; Ottmann, C. Modulators of protein-protein interactions. Chem. Rev. 2014, 114, 4695–4748. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.W.; Kann, M.G. Chapter 4: Protein interactions and disease. PLoS Comput. Biol. 2012, 8, e1002819. [Google Scholar] [CrossRef]
- Lu, H.; Zhou, Q.; He, J.; Jiang, Z.; Peng, C.; Tong, R.; Shi, J. Recent advances in the development of protein-protein interactions modulators: Mechanisms and clinical trials. Signal Transduct. Target. Ther. 2020, 5, 213. [Google Scholar] [CrossRef] [PubMed]
- Mabonga, L.; Kappo, A.P. Protein-protein interaction modulators: Advances, successes and remaining challenges. Biophys. Rev. 2019, 11, 559–581. [Google Scholar] [CrossRef]

| Protein | Gene Name | Biological Impact | Reference |
|---|---|---|---|
| Fibroblast growth factor 2 | FGF2 | Modulation of mRNA nuclear export | [47,48] |
| Apoptotic chromatin condensation inducer in the nucleus | ACIN1 | Regulation of apoptotic DNA fragmentation | [48] |
| Amplified in liver cancer 1 | ALC1 | Not determined | [49] |
| Nucleoprotein of influenza A virus | NP | Stimulation of E2F1- mediated apoptosis | [50] |
| Caspase-2 | CASP2 | Inhibition of caspase-2 activation | [51] |
| Estrogen receptor α | ERα | Gene expression regulation | [26] |
| Toll-like receptor 4 | TLR4 | Modulation of TLR4 signaling (agonist effect) | [52] |
| Leucine-rich pentatricopeptide repeat containing | LRPPRC | Modulation of mRNA nuclear export | [47] |
| U2AF65-associated protein 56 | UAP56 | Modulation of mRNA nuclear export | [47] |
| P300 | P300 | Regulation of API5 stability | [53] |
| Histone deacetylase 1 | HDAC1 | Regulation of API5 stability | [53] |
| p21-activated kinase 1 | PAK1 | Not determined | [54] |
| VP3 protein of Avibirnavirus | VP3 | Regulation of API5 SUMOylation | [55] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbas, H.; Derkaoui, D.K.; Jeammet, L.; Adicéam, E.; Tiollier, J.; Sicard, H.; Braun, T.; Poyet, J.-L. Apoptosis Inhibitor 5: A Multifaceted Regulator of Cell Fate. Biomolecules 2024, 14, 136. https://doi.org/10.3390/biom14010136
Abbas H, Derkaoui DK, Jeammet L, Adicéam E, Tiollier J, Sicard H, Braun T, Poyet J-L. Apoptosis Inhibitor 5: A Multifaceted Regulator of Cell Fate. Biomolecules. 2024; 14(1):136. https://doi.org/10.3390/biom14010136
Chicago/Turabian StyleAbbas, Hafsia, Dalia Kheira Derkaoui, Louise Jeammet, Emilie Adicéam, Jérôme Tiollier, Hélène Sicard, Thorsten Braun, and Jean-Luc Poyet. 2024. "Apoptosis Inhibitor 5: A Multifaceted Regulator of Cell Fate" Biomolecules 14, no. 1: 136. https://doi.org/10.3390/biom14010136





