Plasmodium, the Apicomplexa Outlier When It Comes to Protein Synthesis
Abstract
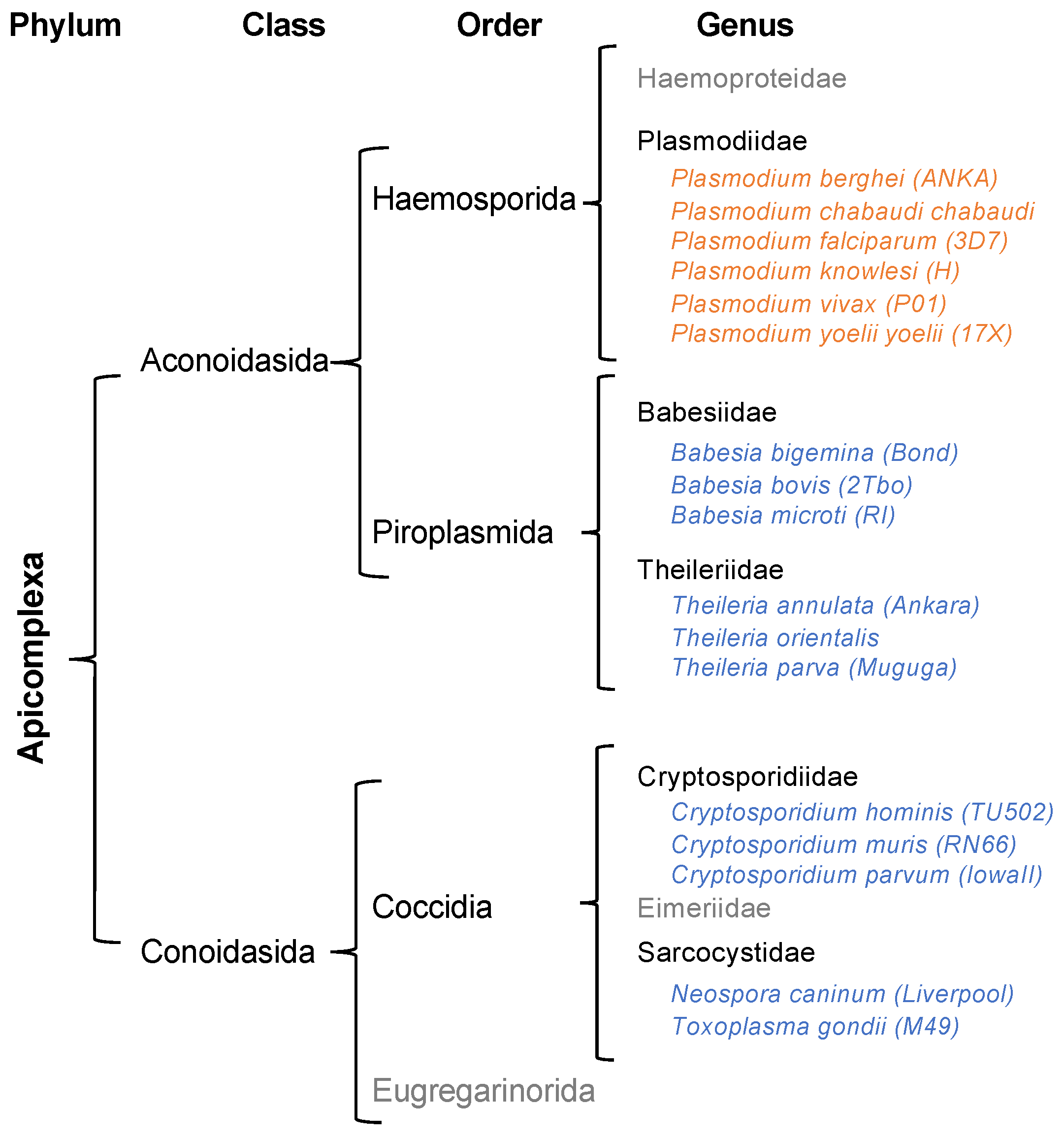
:1. Introduction
2. Material and Methods
2.1. Sequence Retrieval and Analysis
2.2. Complex Modeling with AlphaFold2
3. Results
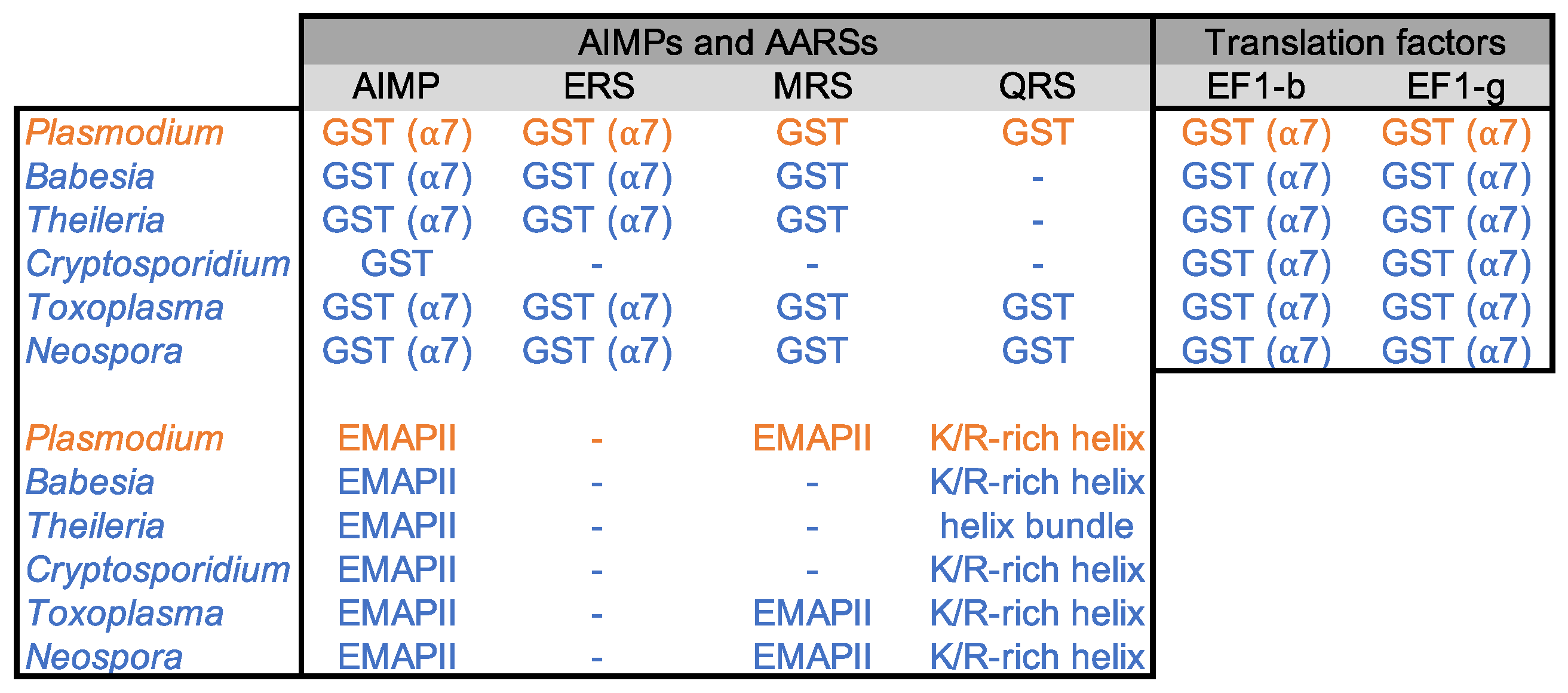
3.1. Identification of GST-like Domains
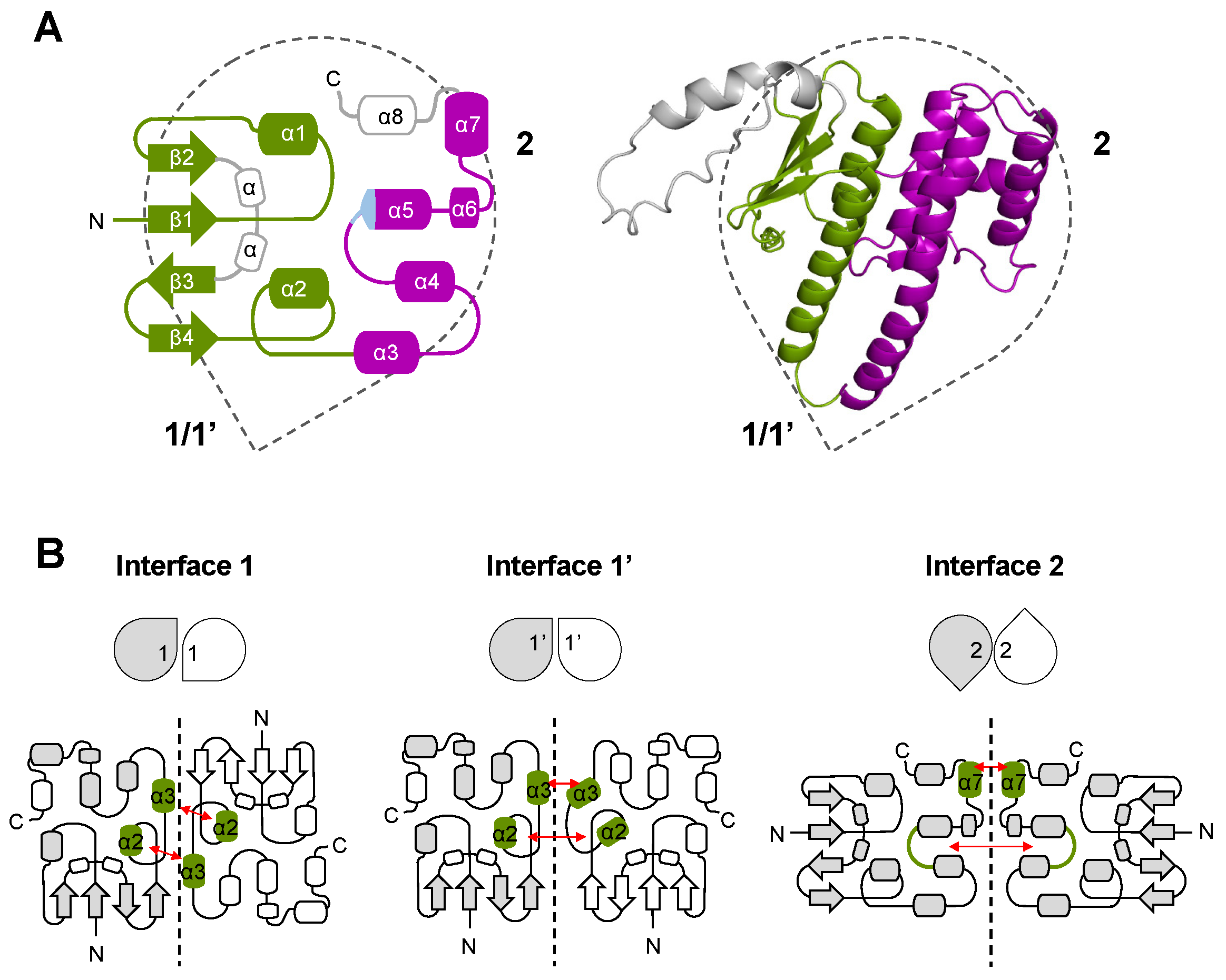
3.2. Description of GST-like Interactions
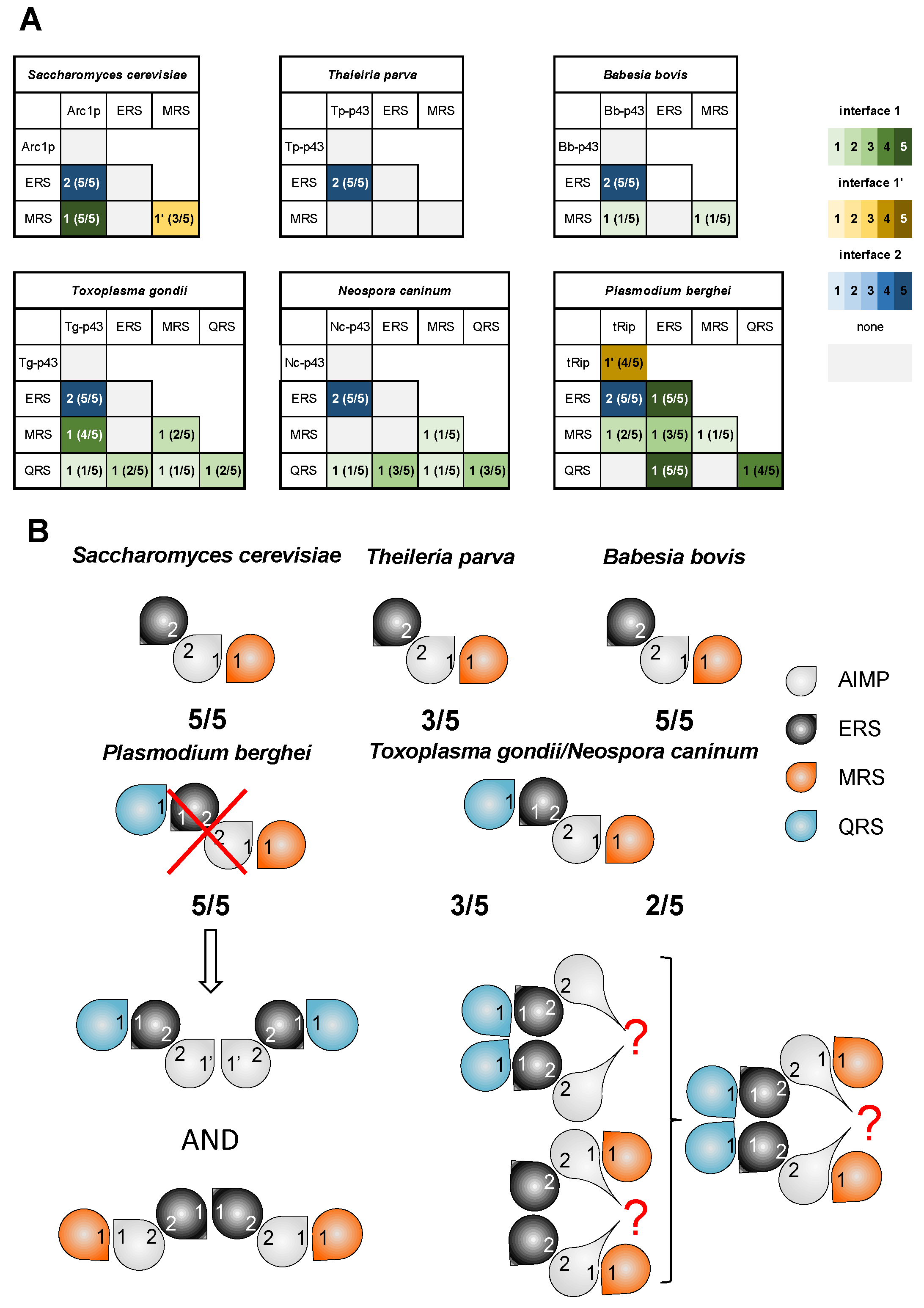
3.3. Pre-Tests for MSC Modeling
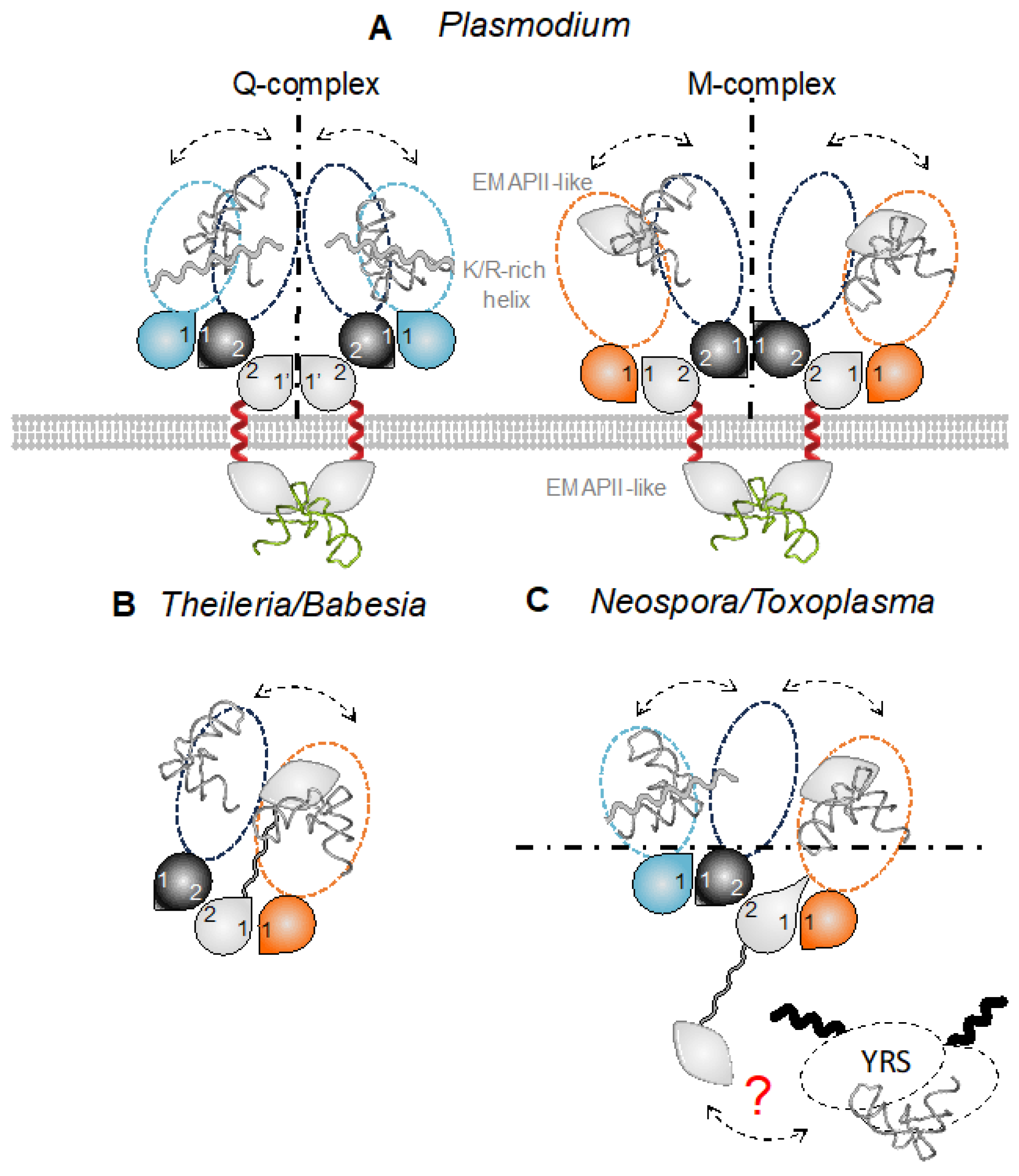
3.4. ColabFold Predicts the Same Interaction Network in All Apicomplexa MSCs
3.5. The Possibility of Homodimerization of Tg-p43 and Modeling of Two T. gondii MSCs
3.6. Potential Consequences of tRNA Binding by MSCs
3.7. What Makes Plasmodium Translation So Different That It Requires tRNA Import via Membrane-Associated MSCs?
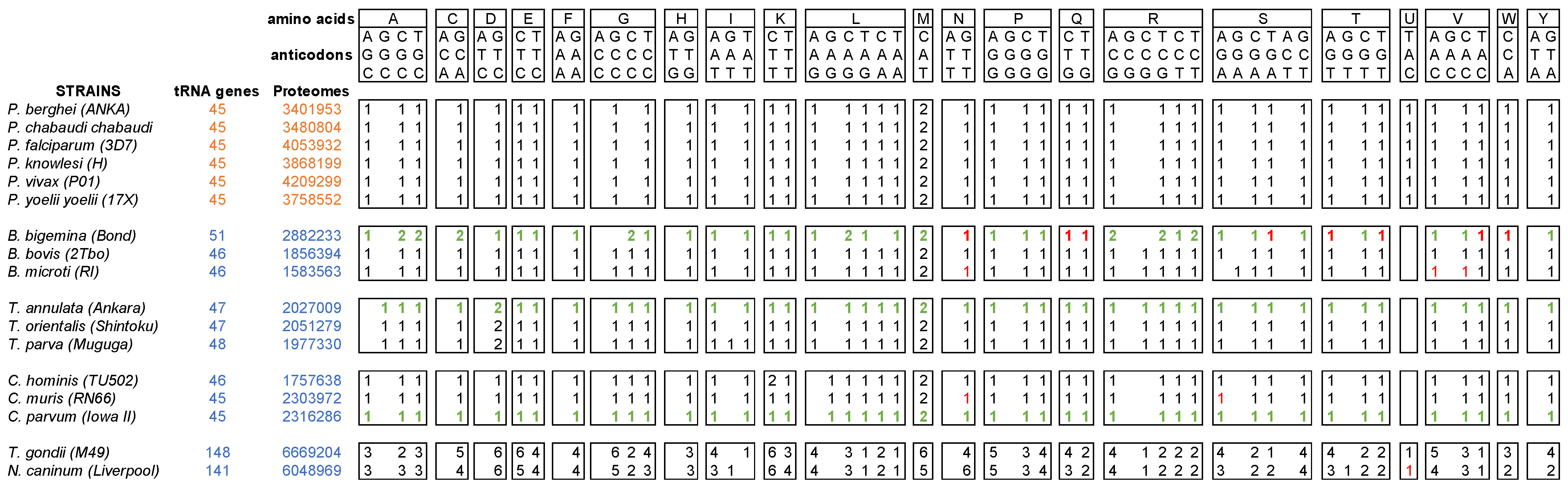
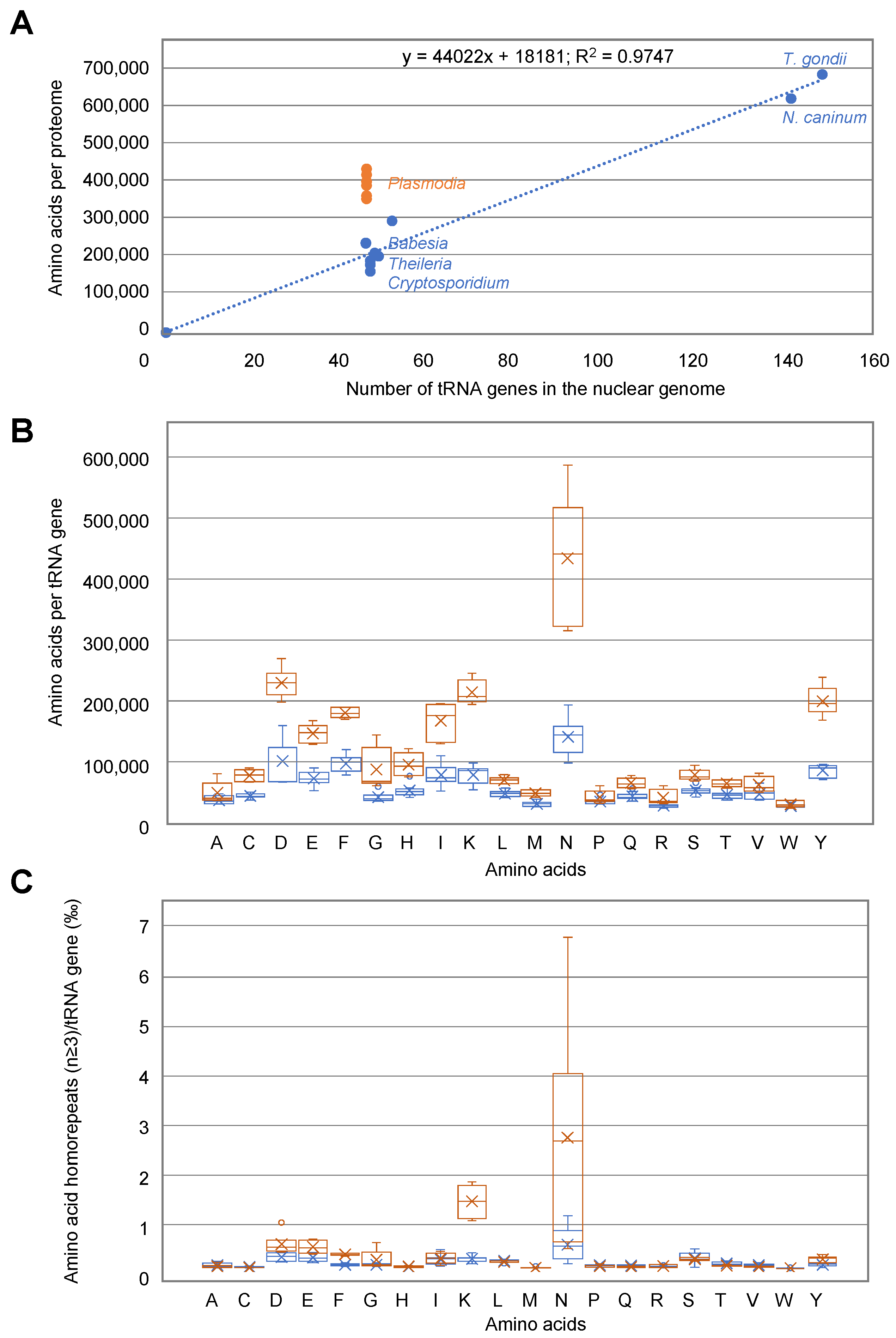
3.8. Correlation between Amino Acid Usage and tRNA Availability in Apicomplexa Parasites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Janouskovec, J.; Paskerova, G.G.; Miroliubova, T.S.; Mikhailov, K.V.; Birley, T.; Aleoshin, V.V.; Simdyanov, T.G. Apicomplexan-like Parasites Are Polyphyletic and Widely but Selectively Dependent on Cryptic Plastid Organelles. eLife 2019, 8, e49662. [Google Scholar] [CrossRef] [PubMed]
- Chandley, P.; Ranjan, R.; Kumar, S.; Rohatgi, S. Host-Parasite Interactions during Plasmodium Infection: Implications for Immunotherapies. Front. Immunol. 2023, 13, 1091961. [Google Scholar] [CrossRef] [PubMed]
- Cowman, A.F.; Healer, J.; Marapana, D.; Marsh, K. Malaria: Biology and Disease. Cell 2016, 167, 610–624. [Google Scholar] [CrossRef] [PubMed]
- Attias, M.; Teixeira, D.E.; Benchimol, M.; Vommaro, R.C.; Crepaldi, P.H.; De Souza, W. The Life-Cycle of Toxoplasma Gondii Reviewed Using Animations. Parasites Vectors 2020, 13, 588. [Google Scholar] [CrossRef] [PubMed]
- Matta, S.K.; Rinkenberger, N.; Dunay, I.R.; Sibley, L.D. Toxoplasma gondii Infection and Its Implications within the Central Nervous System. Nat. Rev. Microbiol. 2021, 19, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Guérin, A.; Striepen, B. The Biology of the Intestinal Intracellular Parasite Cryptosporidium. Cell Host Microbe 2020, 28, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Dhal, A.K.; Panda, C.; Yun, S.-I.; Mahapatra, R.K. An Update on Cryptosporidium Biology and Therapeutic Avenues. J. Parasit. Dis. 2022, 46, 923–939. [Google Scholar] [CrossRef] [PubMed]
- Plattner, F.; Soldati-Favre, D. Hijacking of Host Cellular Functions by the Apicomplexa. Annu. Rev. Microbiol. 2008, 62, 471–487. [Google Scholar] [CrossRef]
- Rubio Gomez, M.A.; Ibba, M. Aminoacyl-tRNA Synthetases. RNA 2020, 26, 910–936. [Google Scholar] [CrossRef]
- Guo, M.; Yang, X.-L. Architecture and Metamorphosis. In Aminoacyl-tRNA Synthetases in Biology and Medicine; Kim, S., Ed.; Topics in Current Chemistry; Springer: Dordrecht, The Netherlands, 2013; Volume 344, pp. 89–118. ISBN 978-94-017-8700-0. [Google Scholar]
- Laporte, D.; Huot, J.L.; Bader, G.; Enkler, L.; Senger, B.; Becker, H.D. Exploring the Evolutionary Diversity and Assembly Modes of Multi-Aminoacyl-tRNA Synthetase Complexes: Lessons from Unicellular Organisms. FEBS Lett. 2014, 588, 4268–4278. [Google Scholar] [CrossRef]
- Havrylenko, S.; Mirande, M. Aminoacyl-tRNA Synthetase Complexes in Evolution. Int. J. Mol. Sci. 2015, 16, 6571–6594. [Google Scholar] [CrossRef]
- Jaramillo Ponce, J.R.; Théobald-Dietrich, A.; Bénas, P.; Paulus, C.; Sauter, C.; Frugier, M. Solution X-ray Scattering Highlights Discrepancies in Plasmodium Multi-Aminoacyl-tRNA Synthetase Complexes. Protein Sci. 2023, 32, e4564. [Google Scholar] [CrossRef] [PubMed]
- Bour, T.; Mahmoudi, N.; Kapps, D.; Thiberge, S.; Bargieri, D.; Ménard, R.; Frugier, M. Apicomplexa-Specific tRip Facilitates Import of Exogenous tRNAs into Malaria Parasites. Proc. Natl. Acad. Sci. USA 2016, 113, 4717–4722. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Chhibber-Goel, J.; Sharma, M.; Parvez, S.; Harlos, K.; Sharma, A.; Yogavel, M. Crystal Structures of the Two Domains That Constitute the Plasmodium vivax P43 Protein. Acta Crystallogr. D Struct. Biol. 2020, 76, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo Ponce, J.R.; Kapps, D.; Paulus, C.; Chicher, J.; Frugier, M. Discovery of Two Distinct Aminoacyl-tRNA Synthetase Complexes Anchored to the Plasmodium Surface tRNA Import Protein. J. Biol. Chem. 2022, 298, 101987. [Google Scholar] [CrossRef] [PubMed]
- Pitolli, M.; Cela, M.; Kapps, D.; Chicher, J.; Despons, L.; Frugier, M. Comparative Proteomics Uncovers Low Asparagine Insertion in Plasmodium tRip-KO Proteins. BioRxiv 2023. submitted. [Google Scholar] [CrossRef]
- Wasmuth, J.; Daub, J.; Peregrín-Alvarez, J.M.; Finney, C.A.M.; Parkinson, J. The Origins of Apicomplexan Sequence Innovation. Genome Res. 2009, 19, 1202–1213. [Google Scholar] [CrossRef] [PubMed]
- Warrenfeltz, S.; Basenko, E.Y.; Crouch, K.; Harb, O.S.; Kissinger, J.C.; Roos, D.S.; Shanmugasundram, A.; Silva-Franco, F. EuPathDB: The Eukaryotic Pathogen Genomics Database Resource. Methods Mol. Biol. 2018, 1757, 69–113. [Google Scholar] [CrossRef]
- Amos, B.; Aurrecoechea, C.; Barba, M.; Barreto, A.; Basenko, E.Y.; Bażant, W.; Belnap, R.; Blevins, A.S.; Böhme, U.; Brestelli, J.; et al. VEuPathDB: The Eukaryotic Pathogen, Vector and Host Bioinformatics Resource Center. Nucleic Acids Res. 2022, 50, D898–D911. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Moretti, S.; Armougom, F.; Wallace, I.M.; Higgins, D.G.; Jongeneel, C.V.; Notredame, C. The M-Coffee Web Server: A Meta-Method for Computing Multiple Sequence Alignments by Combining Alternative Alignment Methods. Nucleic Acids Res. 2007, 35, W645–W648. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An Expanded Database of Transfer RNA Genes Identified in Complete and Draft Genomes. Nucleic Acids Res. 2016, 44, D184–D189. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L.; DeLano, W. PyMOL 2020. Available online: http://www.pymol.org/pymol (accessed on 22 November 2023).
- Källberg, M.; Wang, H.; Wang, S.; Peng, J.; Wang, Z.; Lu, H.; Xu, J. Template-Based Protein Structure Modeling Using the RaptorX Web Server. Nat. Protoc. 2012, 7, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Dragani, B.; Stenberg, G.; Melino, S.; Petruzzelli, R.; Mannervik, B.; Aceto, A. The Conserved N-Capping Box in the Hydrophobic Core of Glutathione S-Transferase P1–1 Is Essential for Refolding. J. Biol. Chem. 1997, 272, 25518–25523. [Google Scholar] [CrossRef]
- van Rooyen, J.M.; Murat, J.-B.; Hammoudi, P.-M.; Kieffer-Jaquinod, S.; Coute, Y.; Sharma, A.; Pelloux, H.; Belrhali, H.; Hakimi, M.-A. Assembly of the Novel Five-Component Apicomplexan Multi-Aminoacyl-tRNA Synthetase Complex Is Driven by the Hybrid Scaffold Protein Tg-P43. PLoS ONE 2014, 9, e89487. [Google Scholar] [CrossRef] [PubMed]
- Simader, H.; Hothorn, M.; Köhler, C.; Basquin, J.; Simos, G.; Suck, D. Structural Basis of Yeast Aminoacyl-tRNA Synthetase Complex Formation Revealed by Crystal Structures of Two Binary Sub-Complexes. Nucleic Acids Res. 2006, 34, 3968–3979. [Google Scholar] [CrossRef]
- Karanasios, E.; Simader, H.; Panayotou, G.; Suck, D.; Simos, G. Molecular Determinants of the Yeast Arc1p–Aminoacyl-tRNA Synthetase Complex Assembly. J. Mol. Biol. 2007, 374, 1077–1090. [Google Scholar] [CrossRef]
- Simos, G.; Sauer, A.; Fasiolo, F.; Hurt, E.C. A Conserved Domain within Arc1p Delivers tRNA to Aminoacyl-tRNA Synthetases. Mol. Cell 1998, 1, 235–242. [Google Scholar] [CrossRef]
- Simos, G.; Segref, A.; Fasiolo, F.; Hellmuth, K.; Shevchenko, A.; Mann, M.; Hurt, E.C. The Yeast Protein Arc1p Binds to tRNA and Functions as a Cofactor for the Methionyl- and Glutamyl-tRNA Synthetases. EMBO J. 1996, 15, 5437–5448. [Google Scholar] [CrossRef]
- Gardner, M.J.; Hall, N.; Fung, E.; White, O.; Berriman, M.; Hyman, R.W.; Carlton, J.M.; Pain, A.; Nelson, K.E.; Bowman, S.; et al. Genome Sequence of the Human Malaria Parasite Plasmodium falciparum. Nature 2002, 419, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Jackson, K.E.; Habib, S.; Frugier, M.; Hoen, R.; Khan, S.; Pham, J.S.; Pouplana, L.R.D.; Royo, M.; Santos, M.A.S.; Sharma, A.; et al. Protein Translation in Plasmodium Parasites. Trends Parasitol. 2011, 27, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Small-Howard, A.L.; Berry, M.J. Unique Features of Selenocysteine Incorporation Function within the Context of General Eukaryotic Translational Processes. Biochem. Soc. Trans. 2005, 33, 1493–1497. [Google Scholar] [CrossRef] [PubMed]
- Deinert, K.; Fasiolo, F.; Hurt, E.C.; Simos, G. Arc1p Organizes the Yeast Aminoacyl-tRNA Synthetase Complex and Stabilizes Its Interaction with the Cognate tRNAs. J. Biol. Chem. 2001, 276, 6000–6008. [Google Scholar] [CrossRef] [PubMed]
- Kyriacou, S.V.; Deutscher, M.P. An Important Role for the Multienzyme Aminoacyl-tRNA Synthetase Complex in Mammalian Translation and Cell Growth. Mol. Cell 2008, 29, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.; Kwon, N.H.; Lee, J.Y.; Park, M.C.; Kang, E.; Kim, H.H.; Kang, T.J.; Kim, S. AIMP3/P18 Controls Translational Initiation by Mediating the Delivery of Charged Initiator tRNA to Initiation Complex. J. Mol. Biol. 2012, 423, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, S.; Agou, F.; Robinson, J.-C.; Mirande, M. The P43 Component of the Mammalian Multi-Synthetase Complex Is Likely To Be the Precursor of the Endothelial Monocyte-Activating Polypeptide II Cytokine. J. Biol. Chem. 1997, 272, 32573–32579. [Google Scholar] [CrossRef] [PubMed]
- Arif, A.; Jia, J.; Mukhopadhyay, R.; Willard, B.; Kinter, M.; Fox, P.L. Two-Site Phosphorylation of EPRS Coordinates Multimodal Regulation of Noncanonical Translational Control Activity. Mol. Cell 2009, 35, 164–180. [Google Scholar] [CrossRef]
- Lee, S.W.; Cho, B.H.; Park, S.G.; Kim, S. Aminoacyl-tRNA Synthetase Complexes: Beyond Translation. J. Cell Sci. 2004, 117, 3725–3734. [Google Scholar] [CrossRef]
- Guo, M.; Schimmel, P. Essential Nontranslational Functions of tRNA Synthetases. Nat. Chem. Biol. 2013, 9, 145–153. [Google Scholar] [CrossRef]
- Cela, M.; Théobald-Dietrich, A.; Rudinger-Thirion, J.; Wolff, P.; Geslain, R.; Frugier, M. Identification of Host tRNAs Preferentially Recognized by the Plasmodium Surface Protein tRip. Nucleic Acids Res. 2021, 49, 10618–10629. [Google Scholar] [CrossRef]
- Bermudez-Santana, C.; Attolini, C.S.-O.; Kirsten, T.; Engelhardt, J.; Prohaska, S.J.; Steigele, S.; Stadler, P.F. Genomic Organization of Eukaryotic tRNAs. BMC Genom. 2010, 11, 270. [Google Scholar] [CrossRef] [PubMed]
- DePristo, M.A.; Zilversmit, M.M.; Hartl, D.L. On the Abundance, Amino Acid Composition, and Evolutionary Dynamics of Low-Complexity Regions in Proteins. Gene 2006, 378, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Frugier, M.; Bour, T.; Ayach, M.; Santos, M.A.; Rudinger-Thirion, J.; Theobald-Dietrich, A.; Pizzi, E. Low Complexity Regions Behave as tRNA Sponges to Help Co-Translational Folding of Plasmodial Proteins. FEBS Lett. 2010, 584, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Verra, F.; Hughes, A.L. Biased Amino Acid Composition in Repeat Regions of Plasmodium Antigens. Mol. Biol. Evol. 1999, 16, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L. The Evolution of Amino Acid Repeat Arrays in Plasmodium and Other Organisms. J. Mol. Evol. 2004, 59, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Karlin, S.; Brocchieri, L.; Bergman, A.; Mrázek, J.; Gentles, A.J. Amino Acid Runs in Eukaryotic Proteomes and Disease Associations. Proc. Natl. Acad. Sci. USA 2002, 99, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Rak, R.; Dahan, O.; Pilpel, Y. Repertoires of tRNAs: The Couplers of Genomics and Proteomics. Annu. Rev. Cell Dev. Biol. 2018, 34, 239–264. [Google Scholar] [CrossRef]
- Dittmar, K.A.; Goodenbour, J.M.; Pan, T. Tissue-Specific Differences in Human Transfer RNA Expression. PLoS Genet. 2006, 2, e221. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaramillo Ponce, J.R.; Frugier, M. Plasmodium, the Apicomplexa Outlier When It Comes to Protein Synthesis. Biomolecules 2024, 14, 46. https://doi.org/10.3390/biom14010046
Jaramillo Ponce JR, Frugier M. Plasmodium, the Apicomplexa Outlier When It Comes to Protein Synthesis. Biomolecules. 2024; 14(1):46. https://doi.org/10.3390/biom14010046
Chicago/Turabian StyleJaramillo Ponce, José R., and Magali Frugier. 2024. "Plasmodium, the Apicomplexa Outlier When It Comes to Protein Synthesis" Biomolecules 14, no. 1: 46. https://doi.org/10.3390/biom14010046
APA StyleJaramillo Ponce, J. R., & Frugier, M. (2024). Plasmodium, the Apicomplexa Outlier When It Comes to Protein Synthesis. Biomolecules, 14(1), 46. https://doi.org/10.3390/biom14010046






