Lipidomic Characterization of Whey Concentrates Rich in Milk Fat Globule Membranes and Extracellular Vesicles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Procurement of Whey Concentrates
2.3. Mass Spectrometry-Based Lipidomics
2.4. Lipid Quantification from Mass Spectrometric Data
2.5. Statistical Analysis
2.6. Lipid Nomenclature
3. Results and Discussion
3.1. Bulk Composition of Whey Concentrates
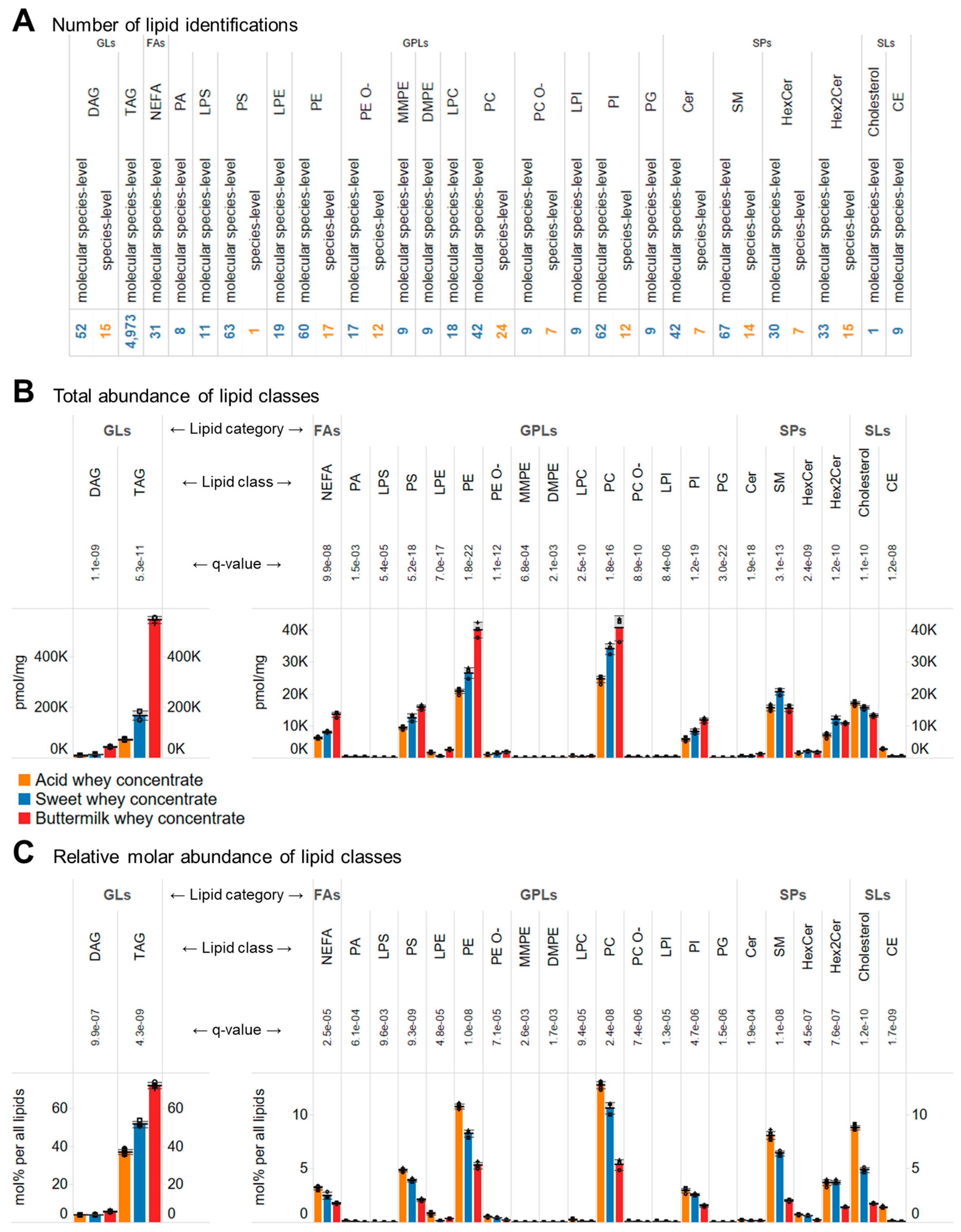
3.2. Comprehensive In-Depth Lipid Profiling of Whey Concentrates
3.3. Molecular Compositions of the Whey Concentrates
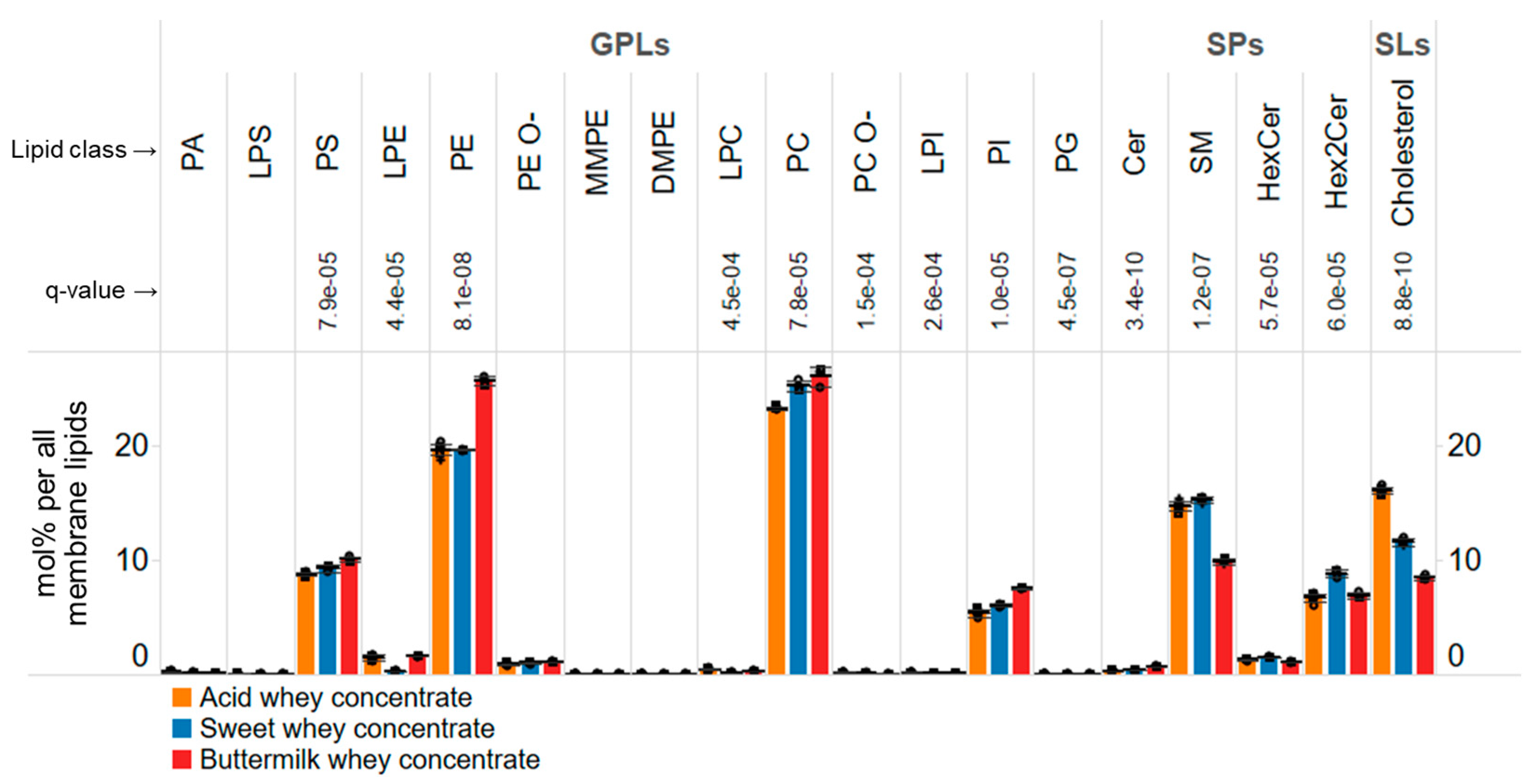
3.4. Membrane Lipid Composition of Whey Concentrates
3.5. Molecular Profiling of TAG Species
3.6. Similar Profiles of Sphingolipids in the Whey Concentrates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, Y.W. Overview of bioactive components in milk and dairy products. In Book Chapter in Bioactive Components in Milk and Dairy Products; Wiley-Blackwell: Hoboken, NJ, USA, 2009; pp. 1–12. [Google Scholar]
- Thorning, T.K.; Raben, A.; Tholstrup, T.; Soedamah-Muthu, S.S.; Givens, I.; Astrup, A. Milk and dairy products: Good or bad for human health? An assessment of the totality of scientific evidence. Food Nutr. Res. 2016, 60, 32527. [Google Scholar] [CrossRef] [PubMed]
- Givens, D.I. MILK Symposium review: The importance of milk and dairy foods in the diets of infants, adolescents, pregnant women, adults, and the elderly. J. Dairy Sci. 2020, 103, 9681–9699. [Google Scholar] [CrossRef] [PubMed]
- Park, K.M.; Fulgoni, V.L., 3rd. The association between dairy product consumption and cognitive function in the National Health and Nutrition Examination Survey. Br. J. Nutr. 2013, 109, 1135–1142. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Radlowski, E.C.; Conrad, M.S.; Li, Y.; Dilger, R.N.; Johnson, R.W. Early supplementation of phospholipids and gangliosides affects brain and cognitive development in neonatal piglets. J. Nutr. 2014, 144, 1903–1909. [Google Scholar] [CrossRef] [PubMed]
- Timby, N.; Domellof, E.; Hernell, O.; Lonnerdal, B.; Domellof, M. Neurodevelopment, nutrition, and growth until 12 mo of age in infants fed a low-energy, low-protein formula supplemented with bovine milk fat globule membranes: A randomized controlled trial. Am. J. Clin. Nutr. 2014, 99, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.; Pauling, J.K.; Sokol, E.; Hannibal-Bach, H.K.; Ejsing, C.S. Comprehensive lipidome analysis by shotgun lipidomics on a hybrid quadrupole-orbitrap-linear ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 2015, 26, 133–148. [Google Scholar] [CrossRef] [PubMed]
- McManaman, J.L. Formation of milk lipids: A molecular perspective. Clin. Lipidol. 2009, 4, 391–401. [Google Scholar] [CrossRef]
- Osorio, J.S.; Lohakare, J.; Bionaz, M. Biosynthesis of milk fat, protein, and lactose: Roles of transcriptional and posttranscriptional regulation. Physiol. Genom. 2016, 48, 231–256. [Google Scholar] [CrossRef]
- Heid, H.W.; Keenan, T.W. Intracellular origin and secretion of milk fat globules. Eur. J. Cell Biol. 2005, 84, 245–258. [Google Scholar] [CrossRef]
- Blans, K.; Hansen, M.S.; Sorensen, L.V.; Hvam, M.L.; Howard, K.A.; Moller, A.; Wiking, L.; Larsen, L.B.; Rasmussen, J.T. Pellet-free isolation of human and bovine milk extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2017, 6, 1294340. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.C.; Kuksis, A. A comparative study of the lipids of globule membrane and fat core and of the milk serum of cows. Lipids 1967, 2, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Patton, S.; Keenan, T.W. The relationship of milk phospholipids to membranes of the secretory cell. Lipids 1971, 6, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Jensen, R.G.; Ferris, A.M.; Lammi-Keefe, C.J. The composition of milk fat. J. Dairy Sci. 1991, 74, 3228–3243. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Rochfort, S.; Cocks, B. Milk lipidomics: What we know and what we don’t. Prog. Lipid Res. 2018, 71, 70–85. [Google Scholar] [CrossRef] [PubMed]
- Selvalatchmanan, J.; Rukmini, A.V.; Ji, S.; Triebl, A.; Gao, L.; Bendt, A.K.; Wenk, M.R.; Gooley, J.J.; Torta, F. Variability of Lipids in Human Milk. Metabolites 2021, 11, 104. [Google Scholar] [CrossRef] [PubMed]
- Crown, S.B.; Marze, N.; Antoniewicz, M.R. Catabolism of Branched Chain Amino Acids Contributes Significantly to Synthesis of Odd-Chain and Even-Chain Fatty Acids in 3T3-L1 Adipocytes. PLoS ONE 2015, 10, e0145850. [Google Scholar] [CrossRef]
- Green, C.R.; Wallace, M.; Divakaruni, A.S.; Phillips, S.A.; Murphy, A.N.; Ciaraldi, T.P.; Metallo, C.M. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nat. Chem. Biol. 2016, 12, 15–21. [Google Scholar] [CrossRef]
- Mansson, H.L. Fatty acids in bovine milk fat. Food Nutr. Res. 2008, 52, 1821. [Google Scholar] [CrossRef]
- McFadden, J.W.; Rico, J.E. Invited review: Sphingolipid biology in the dairy cow: The emerging role of ceramide. J. Dairy Sci. 2019, 102, 7619–7639. [Google Scholar] [CrossRef]
- Gallego, S.F.; Hojlund, K.; Ejsing, C.S. Easy, Fast, and Reproducible Quantification of Cholesterol and Other Lipids in Human Plasma by Combined High Resolution MSX and FTMS Analysis. J. Am. Soc. Mass Spectrom. 2018, 29, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Husen, P.; Tarasov, K.; Katafiasz, M.; Sokol, E.; Vogt, J.; Baumgart, J.; Nitsch, R.; Ekroos, K.; Ejsing, C.S. Analysis of lipid experiments (ALEX): A software framework for analysis of high-resolution shotgun lipidomics data. PLoS ONE 2013, 8, e79736. [Google Scholar] [CrossRef] [PubMed]
- Pauling, J.K.; Hermansson, M.; Hartler, J.; Christiansen, K.; Gallego, S.F.; Peng, B.; Ahrends, R.; Ejsing, C.S. Proposal for a common nomenclature for fragment ions in mass spectra of lipids. PLoS ONE 2017, 12, e0188394. [Google Scholar] [CrossRef] [PubMed]
- Ellis, S.R.; Paine, M.R.L.; Eijkel, G.B.; Pauling, J.K.; Husen, P.; Jervelund, M.W.; Hermansson, M.; Ejsing, C.S.; Heeren, R.M.A. Automated, parallel mass spectrometry imaging and structural identification of lipids. Nat. Methods 2018, 15, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef] [PubMed]
- Liebisch, G.; Vizcaino, J.A.; Kofeler, H.; Trotzmuller, M.; Griffiths, W.J.; Schmitz, G.; Spener, F.; Wakelam, M.J. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013, 54, 1523–1530. [Google Scholar] [CrossRef] [PubMed]
- Newburg, D.S.; Chaturvedi, P. Neutral glycolipids of human and bovine milk. Lipids 1992, 27, 923–927. [Google Scholar] [CrossRef]
- Wiesner, P.; Leidl, K.; Boettcher, A.; Schmitz, G.; Liebisch, G. Lipid profiling of FPLC-separated lipoprotein fractions by electrospray ionization tandem mass spectrometry. J. Lipid Res. 2009, 50, 574–585. [Google Scholar] [CrossRef]
- Brink, L.R.; Herren, A.W.; McMillen, S.; Fraser, K.; Agnew, M.; Roy, N.; Lonnerdal, B. Omics analysis reveals variations among commercial sources of bovine milk fat globule membrane. J. Dairy Sci. 2020, 103, 3002–3016. [Google Scholar] [CrossRef]
- Gerl, M.J.; Sampaio, J.L.; Urban, S.; Kalvodova, L.; Verbavatz, J.M.; Binnington, B.; Lindemann, D.; Lingwood, C.A.; Shevchenko, A.; Schroeder, C.; et al. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell Biol. 2012, 196, 213–221. [Google Scholar] [CrossRef]


| % of Concentrate 1,2 | % of Total Lipid | ||||||
|---|---|---|---|---|---|---|---|
| Dry Matter | Protein | Total Phospholipid 3 | Total Lipid 4 | Total Fat 5 | Total Phospholipid | Total Fat | |
| Acid whey concentrate | 96 | 76 | 5 | 11 | 6 | 46 | 54 |
| Sweet whey concentrate | 96 | 70 | 8 | 22 | 14 | 35 | 65 |
| Buttermilk whey concentrate | 97 | 34 | 9 | 34 | 25 | 26 | 74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sprenger, R.R.; Ostenfeld, M.S.; Bjørnshave, A.; Rasmussen, J.T.; Ejsing, C.S. Lipidomic Characterization of Whey Concentrates Rich in Milk Fat Globule Membranes and Extracellular Vesicles. Biomolecules 2024, 14, 55. https://doi.org/10.3390/biom14010055
Sprenger RR, Ostenfeld MS, Bjørnshave A, Rasmussen JT, Ejsing CS. Lipidomic Characterization of Whey Concentrates Rich in Milk Fat Globule Membranes and Extracellular Vesicles. Biomolecules. 2024; 14(1):55. https://doi.org/10.3390/biom14010055
Chicago/Turabian StyleSprenger, Richard R., Marie S. Ostenfeld, Ann Bjørnshave, Jan T. Rasmussen, and Christer S. Ejsing. 2024. "Lipidomic Characterization of Whey Concentrates Rich in Milk Fat Globule Membranes and Extracellular Vesicles" Biomolecules 14, no. 1: 55. https://doi.org/10.3390/biom14010055
APA StyleSprenger, R. R., Ostenfeld, M. S., Bjørnshave, A., Rasmussen, J. T., & Ejsing, C. S. (2024). Lipidomic Characterization of Whey Concentrates Rich in Milk Fat Globule Membranes and Extracellular Vesicles. Biomolecules, 14(1), 55. https://doi.org/10.3390/biom14010055






