Cellular Profile of Subfornical Organ Insulin Receptors in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Immunohistochemistry
2.3. Quantification and Statistical Analysis
3. Results
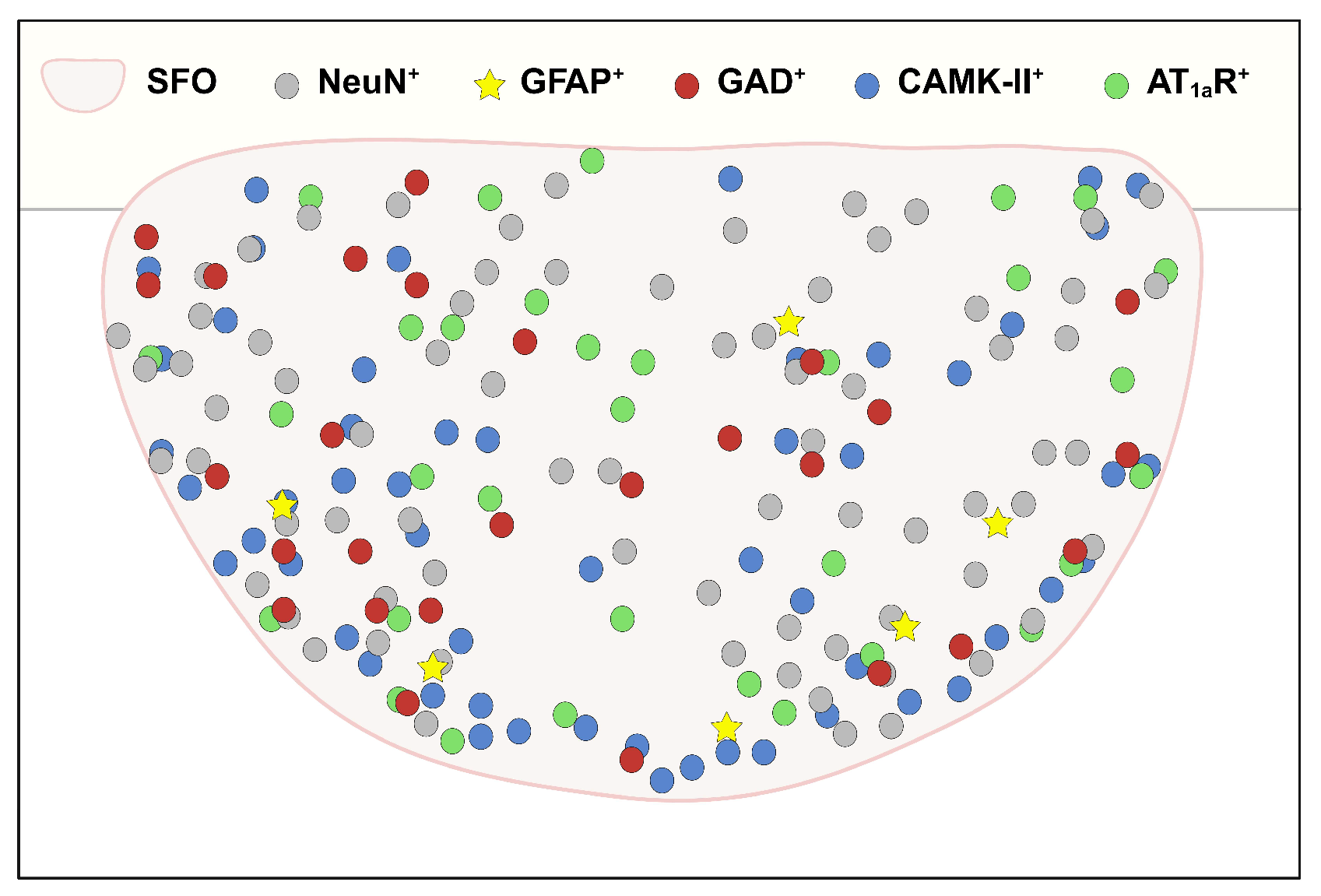
3.1. SFO Insulin Receptors Are Primarily Expressed on Neurons
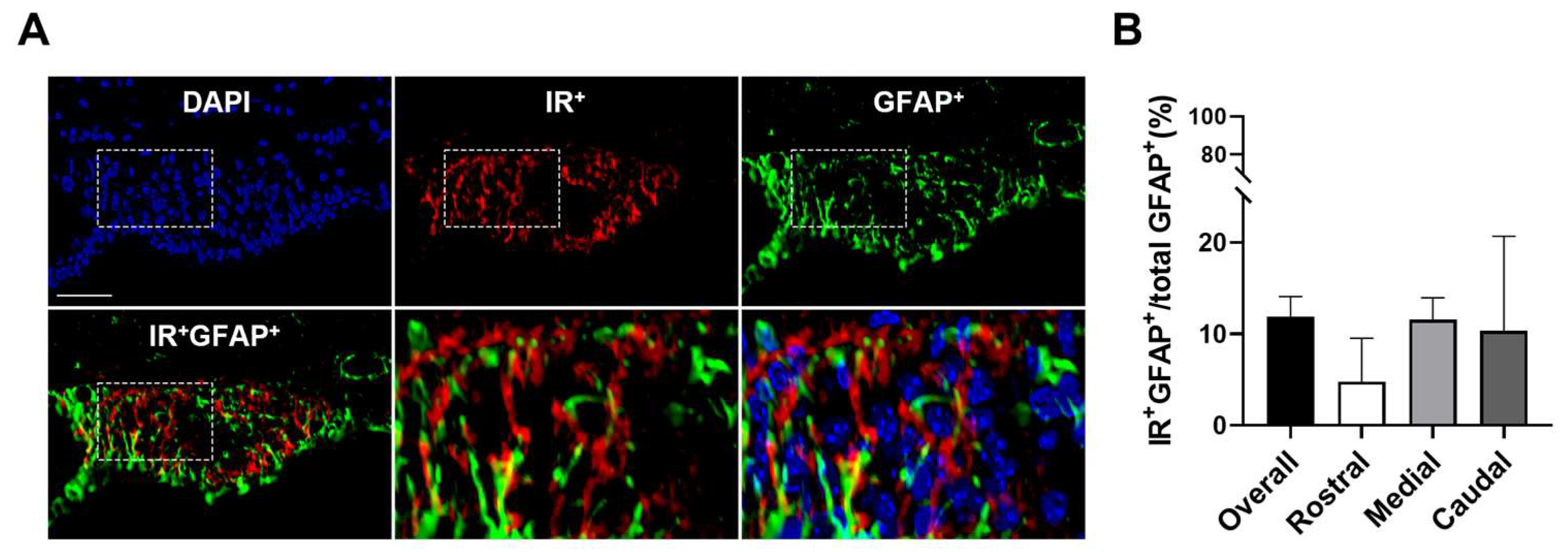
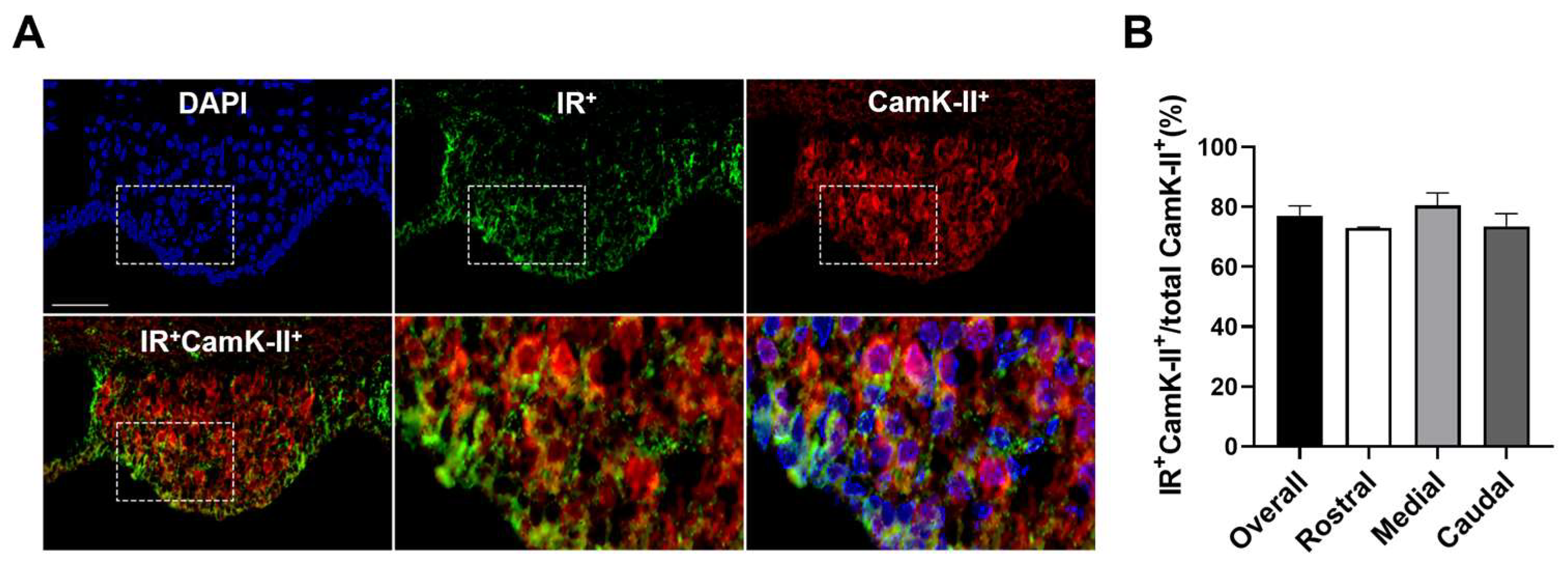
3.2. Insulin Receptors Are Expressed on Inhibitory and Excitatory Neurons in the SFO
3.3. Insulin Receptors Are Expressed on SFO AT1aR Neurons
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bruning, J.C.; Gautam, D.; Burks, D.J.; Gillette, J.; Schubert, M.; Orban, P.C.; Klein, R.; Krone, W.; Muller-Wieland, D.; Kahn, C.R. Role of brain insulin receptor in control of body weight and reproduction. Science 2000, 289, 2122–2125. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Kahn, C.R.; Accili, D. Insulin receptor knockout mice. Annu. Rev. Physiol. 2003, 65, 313–332. [Google Scholar] [CrossRef] [PubMed]
- Cabou, C.; Cani, P.D.; Campistron, G.; Knauf, C.; Mathieu, C.; Sartori, C.; Amar, J.; Scherrer, U.; Burcelin, R. Central insulin regulates heart rate and arterial blood flow: An endothelial nitric oxide synthase-dependent mechanism altered during diabetes. Diabetes 2007, 56, 2872–2877. [Google Scholar] [CrossRef] [PubMed]
- Klockener, T.; Hess, S.; Belgardt, B.F.; Paeger, L.; Verhagen, L.A.; Husch, A.; Sohn, J.W.; Hampel, B.; Dhillon, H.; Zigman, J.M.; et al. High-fat feeding promotes obesity via insulin receptor/PI3K-dependent inhibition of SF-1 VMH neurons. Nat. Neurosci. 2011, 14, 911–918. [Google Scholar] [CrossRef]
- Lim, K.; Burke, S.L.; Head, G.A. Obesity-related hypertension and the role of insulin and leptin in high-fat-fed rabbits. Hypertension 2013, 61, 628–634. [Google Scholar] [CrossRef]
- Hausen, A.C.; Ruud, J.; Jiang, H.; Hess, S.; Varbanov, H.; Kloppenburg, P.; Bruning, J.C. Insulin-Dependent Activation of MCH Neurons Impairs Locomotor Activity and Insulin Sensitivity in Obesity. Cell Rep. 2016, 17, 2512–2521. [Google Scholar] [CrossRef]
- Brown, L.M.; Clegg, D.J.; Benoit, S.C.; Woods, S.C. Intraventricular insulin and leptin reduce food intake and body weight in C57BL/6J mice. Physiol. Behav. 2006, 89, 687–691. [Google Scholar] [CrossRef]
- Diggs-Andrews, K.A.; Zhang, X.; Song, Z.; Daphna-Iken, D.; Routh, V.H.; Fisher, S.J. Brain insulin action regulates hypothalamic glucose sensing and the counterregulatory response to hypoglycemia. Diabetes 2010, 59, 2271–2280. [Google Scholar] [CrossRef]
- Garcia-Caceres, C.; Quarta, C.; Varela, L.; Gao, Y.; Gruber, T.; Legutko, B.; Jastroch, M.; Johansson, P.; Ninkovic, J.; Yi, C.X.; et al. Astrocytic Insulin Signaling Couples Brain Glucose Uptake with Nutrient Availability. Cell 2016, 166, 867–880. [Google Scholar] [CrossRef]
- Konner, A.C.; Hess, S.; Tovar, S.; Mesaros, A.; Sanchez-Lasheras, C.; Evers, N.; Verhagen, L.A.; Bronneke, H.S.; Kleinridders, A.; Hampel, B.; et al. Role for insulin signaling in catecholaminergic neurons in control of energy homeostasis. Cell Metab. 2011, 13, 720–728. [Google Scholar] [CrossRef]
- Rahmouni, K.; Morgan, D.A.; Morgan, G.M.; Liu, X.; Sigmund, C.D.; Mark, A.L.; Haynes, W.G. Hypothalamic PI3K and MAPK differentially mediate regional sympathetic activation to insulin. J. Clin. Investig. 2004, 114, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Muntzel, M.S.; Morgan, D.A.; Mark, A.L.; Johnson, A.K. Intracerebroventricular insulin produces nonuniform regional increases in sympathetic nerve activity. Am. J. Physiol. 1994, 267, R1350–R1355. [Google Scholar] [CrossRef] [PubMed]
- Pricher, M.P.; Freeman, K.L.; Brooks, V.L. Insulin in the brain increases gain of baroreflex control of heart rate and lumbar sympathetic nerve activity. Hypertension 2008, 51, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Ward, K.R.; Bardgett, J.F.; Wolfgang, L.; Stocker, S.D. Sympathetic response to insulin is mediated by melanocortin 3/4 receptors in the hypothalamic paraventricular nucleus. Hypertension 2011, 57, 435–441. [Google Scholar] [CrossRef]
- Young, C.N.; Deo, S.H.; Chaudhary, K.; Thyfault, J.P.; Fadel, P.J. Insulin enhances the gain of arterial baroreflex control of muscle sympathetic nerve activity in humans. J. Physiol. 2010, 588, 3593–3603. [Google Scholar] [CrossRef]
- Anderson, E.A.; Balon, T.W.; Hoffman, R.P.; Sinkey, C.A.; Mark, A.L. Insulin increases sympathetic activity but not blood pressure in borderline hypertensive humans. Hypertension 1992, 19, 621–627. [Google Scholar] [CrossRef]
- Berne, C.; Fagius, J.; Pollare, T.; Hjemdahl, P. The sympathetic response to euglycaemic hyperinsulinaemia. Evidence from microelectrode nerve recordings in healthy subjects. Diabetologia 1992, 35, 873–879. [Google Scholar] [CrossRef]
- Cassaglia, P.A.; Hermes, S.M.; Aicher, S.A.; Brooks, V.L. Insulin acts in the arcuate nucleus to increase lumbar sympathetic nerve activity and baroreflex function in rats. J. Physiol. 2011, 589, 1643–1662. [Google Scholar] [CrossRef]
- Cassaglia, P.A.; Shi, Z.; Brooks, V.L. Insulin increases sympathetic nerve activity in part by suppression of tonic inhibitory neuropeptide Y inputs into the paraventricular nucleus in female rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 311, R97–R103. [Google Scholar] [CrossRef]
- Chong, A.C.; Vogt, M.C.; Hill, A.S.; Bruning, J.C.; Zeltser, L.M. Central insulin signaling modulates hypothalamus-pituitary-adrenal axis responsiveness. Mol. Metab. 2015, 4, 83–92. [Google Scholar] [CrossRef]
- Konner, A.C.; Janoschek, R.; Plum, L.; Jordan, S.D.; Rother, E.; Ma, X.; Xu, C.; Enriori, P.; Hampel, B.; Barsh, G.S.; et al. Insulin action in AgRP-expressing neurons is required for suppression of hepatic glucose production. Cell Metab. 2007, 5, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Garwood, C.J.; Ratcliffe, L.E.; Morgan, S.V.; Simpson, J.E.; Owens, H.; Vazquez-Villasenor, I.; Heath, P.R.; Romero, I.A.; Ince, P.G.; Wharton, S.B. Insulin and IGF1 signalling pathways in human astrocytes in vitro and in vivo; characterisation, subcellular localisation and modulation of the receptors. Mol. Brain 2015, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Xue, C.; Sakaguchi, M.; Konishi, M.; Shirazian, A.; Ferris, H.A.; Li, M.E.; Yu, R.; Kleinridders, A.; Pothos, E.N.; et al. Insulin regulates astrocyte gliotransmission and modulates behavior. J. Clin. Investig. 2018, 128, 2914–2926. [Google Scholar] [CrossRef] [PubMed]
- Kum, W.; Zhu, S.Q.; Ho, S.K.; Young, J.D.; Cockram, C.S. Effect of insulin on glucose and glycogen metabolism and leucine incorporation into protein in cultured mouse astrocytes. Glia 1992, 6, 264–268. [Google Scholar] [CrossRef]
- Okoreeh, A.K.; Bake, S.; Sohrabji, F. Astrocyte-specific insulin-like growth factor-1 gene transfer in aging female rats improves stroke outcomes. Glia 2017, 65, 1043–1058. [Google Scholar] [CrossRef]
- Fernandez, A.M.; Martinez-Rachadell, L.; Navarrete, M.; Pose-Utrilla, J.; Davila, J.C.; Pignatelli, J.; Diaz-Pacheco, S.; Guerra-Cantera, S.; Viedma-Moreno, E.; Palenzuela, R.; et al. Insulin regulates neurovascular coupling through astrocytes. Proc. Natl. Acad. Sci. USA 2022, 119, e2204527119. [Google Scholar] [CrossRef]
- Jeong, J.K.; Horwath, J.A.; Simonyan, H.; Blackmore, K.A.; Butler, S.D.; Young, C.N. Subfornical organ insulin receptors tonically modulate cardiovascular and metabolic function. Physiol. Genom. 2019, 51, 333–341. [Google Scholar] [CrossRef]
- Wilhelm, I.; Nyul-Toth, A.; Suciu, M.; Hermenean, A.; Krizbai, I.A. Heterogeneity of the blood-brain barrier. Tissue Barriers 2016, 4, e1143544. [Google Scholar] [CrossRef]
- Oka, Y.; Ye, M.; Zuker, C.S. Thirst driving and suppressing signals encoded by distinct neural populations in the brain. Nature 2015, 520, 349–352. [Google Scholar] [CrossRef]
- Kawano, H.; Masuko, S. Region-specific projections from the subfornical organ to the paraventricular hypothalamic nucleus in the rat. Neuroscience 2010, 169, 1227–1234. [Google Scholar] [CrossRef]
- Miselis, R.R. The efferent projections of the subfornical organ of the rat: A circumventricular organ within a neural network subserving water balance. Brain Res. 1981, 230, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Gruber, K.; McRae-Degueurce, A.; Wilkin, L.D.; Mitchell, L.D.; Johnson, A.K. Forebrain and brainstem afferents to the arcuate nucleus in the rat: Potential pathways for the modulation of hypophyseal secretions. Neurosci. Lett. 1987, 75, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Hindmarch, C.C.; Ferguson, A.V. Physiological roles for the subfornical organ: A dynamic transcriptome shaped by autonomic state. J. Physiol. 2016, 594, 1581–1589. [Google Scholar] [CrossRef] [PubMed]
- Young, C.N.; Morgan, D.A.; Butler, S.D.; Rahmouni, K.; Gurley, S.B.; Coffman, T.M.; Mark, A.L.; Davisson, R.L. Angiotensin type 1a receptors in the forebrain subfornical organ facilitate leptin-induced weight loss through brown adipose tissue thermogenesis. Mol. Metab. 2015, 4, 337–343. [Google Scholar] [CrossRef]
- Collister, J.P.; Hendel, M.D. Chronic effects of angiotensin II and at1 receptor antagonists in subfornical organ-lesioned rats. Clin. Exp. Pharmacol. Physiol. 2005, 32, 462–466. [Google Scholar] [CrossRef]
- Hilzendeger, A.M.; Cassell, M.D.; Davis, D.R.; Stauss, H.M.; Mark, A.L.; Grobe, J.L.; Sigmund, C.D. Angiotensin type 1a receptors in the subfornical organ are required for deoxycorticosterone acetate-salt hypertension. Hypertension 2013, 61, 716–722. [Google Scholar] [CrossRef]
- Matsuda, T.; Hiyama, T.Y.; Niimura, F.; Matsusaka, T.; Fukamizu, A.; Kobayashi, K.; Kobayashi, K.; Noda, M. Distinct neural mechanisms for the control of thirst and salt appetite in the subfornical organ. Nat. Neurosci. 2017, 20, 230–241. [Google Scholar] [CrossRef]
- Gonzalez, A.D.; Wang, G.; Waters, E.M.; Gonzales, K.L.; Speth, R.C.; Van Kempen, T.A.; Marques-Lopes, J.; Young, C.N.; Butler, S.D.; Davisson, R.L.; et al. Distribution of angiotensin type 1a receptor-containing cells in the brains of bacterial artificial chromosome transgenic mice. Neuroscience 2012, 226, 489–509. [Google Scholar] [CrossRef]
- Marques-Lopes, J.; Lynch, M.K.; Van Kempen, T.A.; Waters, E.M.; Wang, G.; Iadecola, C.; Pickel, V.M.; Milner, T.A. Female protection from slow-pressor effects of angiotensin II involves prevention of ROS production independent of NMDA receptor trafficking in hypothalamic neurons expressing angiotensin 1A receptors. Synapse 2015, 69, 148–165. [Google Scholar] [CrossRef]
- Leib, D.E.; Zimmerman, C.A.; Poormoghaddam, A.; Huey, E.L.; Ahn, J.S.; Lin, Y.C.; Tan, C.L.; Chen, Y.; Knight, Z.A. The Forebrain Thirst Circuit Drives Drinking through Negative Reinforcement. Neuron 2017, 96, 1272–1281 e1274. [Google Scholar] [CrossRef]
- Yu, Z.; Kisner, A.; Bhatt, A.; Polter, A.M.; Marvar, P.J. Central amygdala angiotensin type 1 receptor (Agtr1) expressing neurons contribute to fear extinction. Neuropharmacology 2023, 229, 109460. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.K.; Kim, H.R.; Hwang, S.M.; Park, J.W.; Lee, B.J. Region- and neuronal phenotype-specific expression of NELL2 in the adult rat brain. Mol. Cells 2008, 26, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Hicks, A.I.; Kobrinsky, S.; Zhou, S.; Yang, J.; Prager-Khoutorsky, M. Anatomical Organization of the Rat Subfornical Organ. Front. Cell Neurosci. 2021, 15, 691711. [Google Scholar] [CrossRef] [PubMed]
- Langlet, F.; Mullier, A.; Bouret, S.G.; Prevot, V.; Dehouck, B. Tanycyte-like cells form a blood-cerebrospinal fluid barrier in the circumventricular organs of the mouse brain. J. Comp. Neurol. 2013, 521, 3389–3405. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.M.; Bazzino, P.; Hurh, S.J.; Konanur, V.R.; Roitman, J.D.; Roitman, M.F. Thirst recruits phasic dopamine signaling through subfornical organ neurons. Proc. Natl. Acad. Sci. USA 2020, 117, 30744–30754. [Google Scholar] [CrossRef]
- Kolaj, M.; Renaud, L.P. Metabotropic glutamate receptors in median preoptic neurons modulate neuronal excitability and glutamatergic and GABAergic inputs from the subfornical organ. J. Neurophysiol. 2010, 103, 1104–1113. [Google Scholar] [CrossRef]
- Abbott, S.B.; Machado, N.L.; Geerling, J.C.; Saper, C.B. Reciprocal Control of Drinking Behavior by Median Preoptic Neurons in Mice. J. Neurosci. 2016, 36, 8228–8237. [Google Scholar] [CrossRef]
- Llewellyn, T.; Zheng, H.; Liu, X.; Xu, B.; Patel, K.P. Median preoptic nucleus and subfornical organ drive renal sympathetic nerve activity via a glutamatergic mechanism within the paraventricular nucleus. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, R424–R432. [Google Scholar] [CrossRef]
- Honda, E.; Xu, S.; Ono, K.; Ito, K.; Inenaga, K. Spontaneously active GABAergic interneurons in the subfornical organ of rat slice preparations. Neurosci. Lett. 2001, 306, 45–48. [Google Scholar] [CrossRef]
- Elsaafien, K.; de Kloet, A.D.; Krause, E.G.; Sumners, C. Brain Angiotensin Type-1 and Type-2 Receptors in Physiological and Hypertensive Conditions: Focus on Neuroinflammation. Curr. Hypertens. Rep. 2020, 22, 48. [Google Scholar] [CrossRef]
- Figlewicz, D.P.; Evans, S.B.; Murphy, J.; Hoen, M.; Baskin, D.G. Expression of receptors for insulin and leptin in the ventral tegmental area/substantia nigra (VTA/SN) of the rat. Brain Res. 2003, 964, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Fordahl, S.C.; Jones, S.R. High-Fat-Diet-Induced Deficits in Dopamine Terminal Function Are Reversed by Restoring Insulin Signaling. ACS Chem. Neurosci. 2017, 8, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Yeh, T.C.; Liu, C.P.; Cheng, W.H.; Chen, B.R.; Lu, P.J.; Cheng, P.W.; Ho, W.Y.; Sun, G.C.; Liou, J.C.; Tseng, C.J. Caffeine intake improves fructose-induced hypertension and insulin resistance by enhancing central insulin signaling. Hypertension 2014, 63, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.N.; Lu, P.J.; Lo, W.C.; Lin, C.H.; Hsiao, M.; Tseng, C.J. In situ Akt phosphorylation in the nucleus tractus solitarii is involved in central control of blood pressure and heart rate. Circulation 2004, 110, 2476–2483. [Google Scholar] [CrossRef]
- Hsiao, M.; Lu, P.J.; Huang, H.N.; Lo, W.C.; Ho, W.Y.; Lai, T.C.; Chiang, H.T.; Tseng, C.J. Defective phosphatidylinositol 3-kinase signaling in central control of cardiovascular effects in the nucleus tractus solitarii of spontaneously hypertensive rats. Hypertens. Res. 2008, 31, 1209–1218. [Google Scholar] [CrossRef]
- Watanabe, E.; Hiyama, T.Y.; Shimizu, H.; Kodama, R.; Hayashi, N.; Miyata, S.; Yanagawa, Y.; Obata, K.; Noda, M. Sodium-level-sensitive sodium channel Na(x) is expressed in glial laminate processes in the sensory circumventricular organs. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 290, R568–R576. [Google Scholar] [CrossRef]
- Shimizu, H.; Watanabe, E.; Hiyama, T.Y.; Nagakura, A.; Fujikawa, A.; Okado, H.; Yanagawa, Y.; Obata, K.; Noda, M. Glial Nax channels control lactate signaling to neurons for brain [Na+] sensing. Neuron 2007, 54, 59–72. [Google Scholar] [CrossRef]
- Briant, L.J.; Charkoudian, N.; Hart, E.C. Sympathetic regulation of blood pressure in normotension and hypertension: When sex matters. Exp. Physiol. 2016, 101, 219–229. [Google Scholar] [CrossRef]
- Li, Y.F.; Wang, W.; Mayhan, W.G.; Patel, K.P. Angiotensin-mediated increase in renal sympathetic nerve discharge within the PVN: Role of nitric oxide. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 290, R1035–R1043. [Google Scholar] [CrossRef]
- Buttler, L.; Ribeiro, I.M.; Ferreira-Neto, H.C.; Antunes, V.R. Angiotensin II acting on PVN induces sympathoexcitation and pressor responses via the PI3K-dependent pathway. Auton. Neurosci. 2016, 198, 54–58. [Google Scholar] [CrossRef]
- Capone, C.; Faraco, G.; Peterson, J.R.; Coleman, C.; Anrather, J.; Milner, T.A.; Pickel, V.M.; Davisson, R.L.; Iadecola, C. Central cardiovascular circuits contribute to the neurovascular dysfunction in angiotensin II hypertension. J. Neurosci. 2012, 32, 4878–4886. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.C.; Huang, W.C. Role of angiotensin II in hyperinsulinemia-induced hypertension in rats. J. Hypertens. 1998, 16, 1767–1774. [Google Scholar] [CrossRef] [PubMed]
- Pershadsingh, H.A.; Kurtz, T.W. Insulin-sensitizing effects of telmisartan: Implications for treating insulin-resistant hypertension and cardiovascular disease. Diabetes Care 2004, 27, 1015. [Google Scholar] [CrossRef] [PubMed]
- Fukui, T.; Hirano, T.; Shiraishi, Y.; Nagashima, M.; Adachi, M. Chronic insulin infusion normalizes blood pressure and the gene expressions of angiotensin II type 1 receptor in fructose-fed rats. Hypertens. Res. 2008, 31, 127–133. [Google Scholar] [CrossRef]
- Ogihara, T.; Asano, T.; Ando, K.; Chiba, Y.; Sakoda, H.; Anai, M.; Shojima, N.; Ono, H.; Onishi, Y.; Fujishiro, M.; et al. Angiotensin II-induced insulin resistance is associated with enhanced insulin signaling. Hypertension 2002, 40, 872–879. [Google Scholar] [CrossRef]
- Juan, C.C.; Chien, Y.; Wu, L.Y.; Yang, W.M.; Chang, C.L.; Lai, Y.H.; Ho, P.H.; Kwok, C.F.; Ho, L.T. Angiotensin II enhances insulin sensitivity in vitro and in vivo. Endocrinology 2005, 146, 2246–2254. [Google Scholar] [CrossRef]
- Garcia-Puig, J.; Ruilope, L.M.; Luque, M.; Fernandez, J.; Ortega, R.; Dal-Re, R.; Investigators, A.S.G. Glucose metabolism in patients with essential hypertension. Am. J. Med. 2006, 119, 318–326. [Google Scholar] [CrossRef]
- Hindmarch, C.; Fry, M.; Yao, S.T.; Smith, P.M.; Murphy, D.; Ferguson, A.V. Microarray analysis of the transcriptome of the subfornical organ in the rat: Regulation by fluid and food deprivation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 295, R1914–R1920. [Google Scholar] [CrossRef]
- Paes-Leme, B.; Dos-Santos, R.C.; Mecawi, A.S.; Ferguson, A.V. Interaction between angiotensin II and glucose sensing at the subfornical organ. J. Neuroendocrinol. 2018, 30, e12654. [Google Scholar] [CrossRef]
- Cancelliere, N.M.; Ferguson, A.V. Subfornical organ neurons integrate cardiovascular and metabolic signals. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 312, R253–R262. [Google Scholar] [CrossRef]
- Wei, S.G.; Yu, Y.; Felder, R.B. Blood-borne interleukin-1beta acts on the subfornical organ to upregulate the sympathoexcitatory milieu of the hypothalamic paraventricular nucleus. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2018, 314, R447–R458. [Google Scholar] [CrossRef] [PubMed]
- Simpson, N.J.; Ferguson, A.V. Tumor necrosis factor-alpha potentiates the effects of angiotensin II on subfornical organ neurons. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2018, 315, R425–R433. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wei, S.G.; Weiss, R.M.; Felder, R.B. TNF-alpha receptor 1 knockdown in the subfornical organ ameliorates sympathetic excitation and cardiac hemodynamics in heart failure rats. Am. J. Physiol. Heart Circ. Physiol. 2017, 313, H744–H756. [Google Scholar] [CrossRef] [PubMed]
- Moss, A.M.; Unger, J.W.; Moxley, R.T.; Livingston, J.N. Location of phosphotyrosine-containing proteins by immunocytochemistry in the rat forebrain corresponds to the distribution of the insulin receptor. Proc. Natl. Acad. Sci. USA 1990, 87, 4453–4457. [Google Scholar] [CrossRef] [PubMed]
- Unger, J.; McNeill, T.H.; Moxley, R.T., 3rd; White, M.; Moss, A.; Livingston, J.N. Distribution of insulin receptor-like immunoreactivity in the rat forebrain. Neuroscience 1989, 31, 143–157. [Google Scholar] [CrossRef]
- Werther, G.A.; Hogg, A.; Oldfield, B.J.; McKinley, M.J.; Figdor, R.; Allen, A.M.; Mendelsohn, F.A. Localization and characterization of insulin receptors in rat brain and pituitary gland using in vitro autoradiography and computerized densitometry. Endocrinology 1987, 121, 1562–1570. [Google Scholar] [CrossRef]
- Marks, J.L.; Porte, D., Jr.; Stahl, W.L.; Baskin, D.G. Localization of insulin receptor mRNA in rat brain by in situ hybridization. Endocrinology 1990, 127, 3234–3236. [Google Scholar] [CrossRef]
- van Houten, M.; Posner, B.I.; Kopriwa, B.M.; Brawer, J.R. Insulin-binding sites in the rat brain: In vivo localization to the circumventricular organs by quantitative radioautography. Endocrinology 1979, 105, 666–673. [Google Scholar] [CrossRef]
- Unger, J.W.; Livingston, J.N.; Moss, A.M. Insulin receptors in the central nervous system: Localization, signalling mechanisms and functional aspects. Prog. Neurobiol. 1991, 36, 343–362. [Google Scholar] [CrossRef]



| Insulin Receptor Colocalization (%) | ||||
|---|---|---|---|---|
| Entire SFO | Rostral SFO | Medial SFO | Caudal SFO | |
| NeuN+ | 79.8 ± 3.0 | 89.8 ± 6.2 | 75.3 ± 4.9 | 80.6 ± 7.0 |
| GFAP+ | 11.9 ± 2.2 | 4.8 ± 4.8 | 11.5 ± 2.4 | 10.3 ± 10.3 |
| GAD+ | 37.6 ± 2.9 | 39.4 ± 8.2 | 40.9 ± 5.9 | 17.5 ± 7.5 |
| CamK-II+ | 77.1 ± 3.3 | 73.0 ± 0.3 | 80.4 ± 4.3 | 73.5 ± 4.3 |
| AT1aR+ | 68.5 ± 3.5 | 70.4 ± 7.4 | 66.3 ± 5.8 | 71.3 ± 7.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-R.; Jeong, J.-K.; Young, C.N. Cellular Profile of Subfornical Organ Insulin Receptors in Mice. Biomolecules 2024, 14, 1256. https://doi.org/10.3390/biom14101256
Kim H-R, Jeong J-K, Young CN. Cellular Profile of Subfornical Organ Insulin Receptors in Mice. Biomolecules. 2024; 14(10):1256. https://doi.org/10.3390/biom14101256
Chicago/Turabian StyleKim, Han-Rae, Jin-Kwon Jeong, and Colin N. Young. 2024. "Cellular Profile of Subfornical Organ Insulin Receptors in Mice" Biomolecules 14, no. 10: 1256. https://doi.org/10.3390/biom14101256







