Structural and Dynamical Properties of Nucleic Acid Hairpins Implicated in Trinucleotide Repeat Expansion Diseases
Abstract
1. Introduction
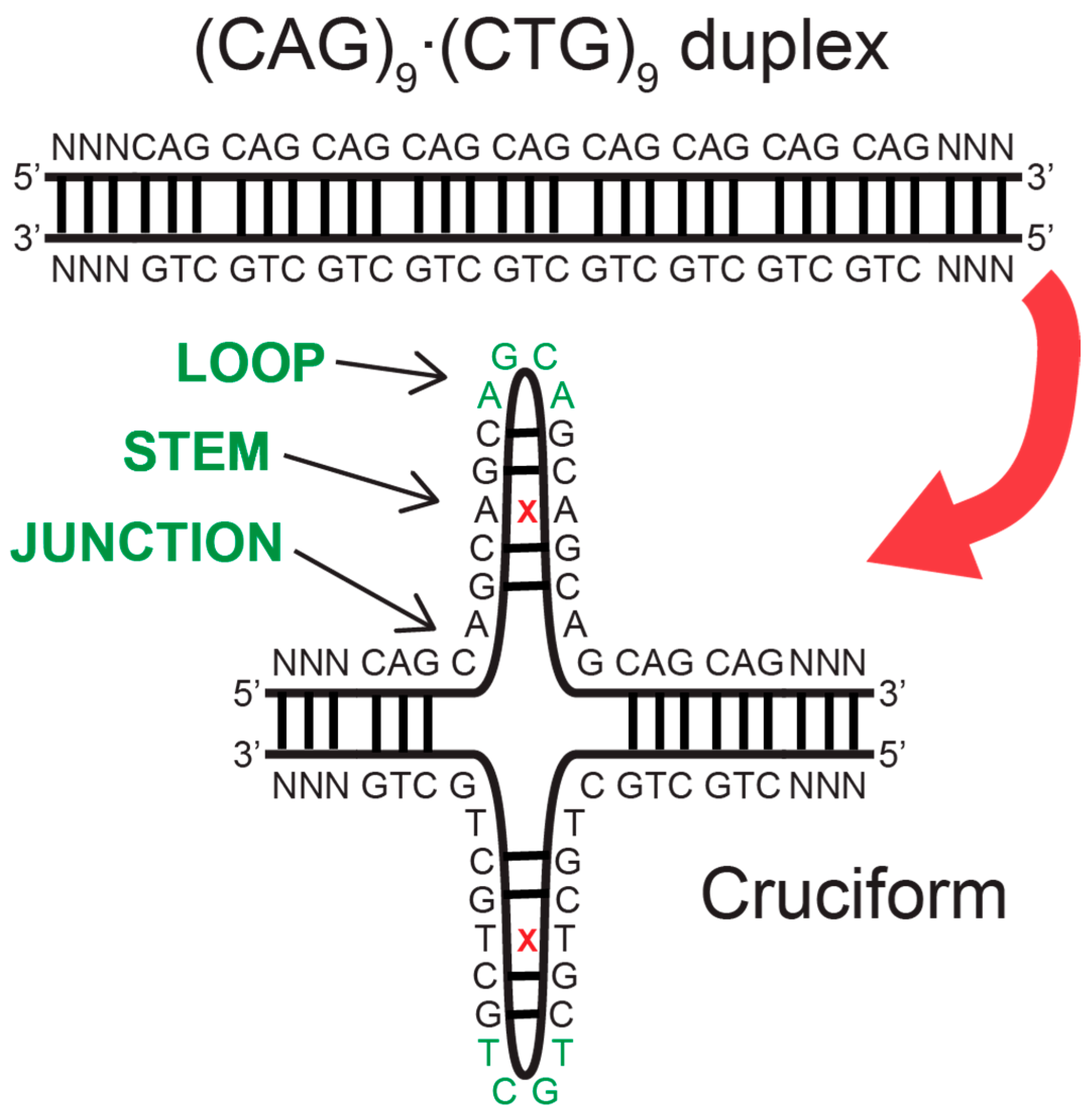
1.1. Trinucleotide Repeat Sequences and Associated Human Diseases
1.2. DNA Mismatch Repair Proteins Are Causative for Disease-Related Expansion of Trinucletoide Repeat Sequences
2. Results
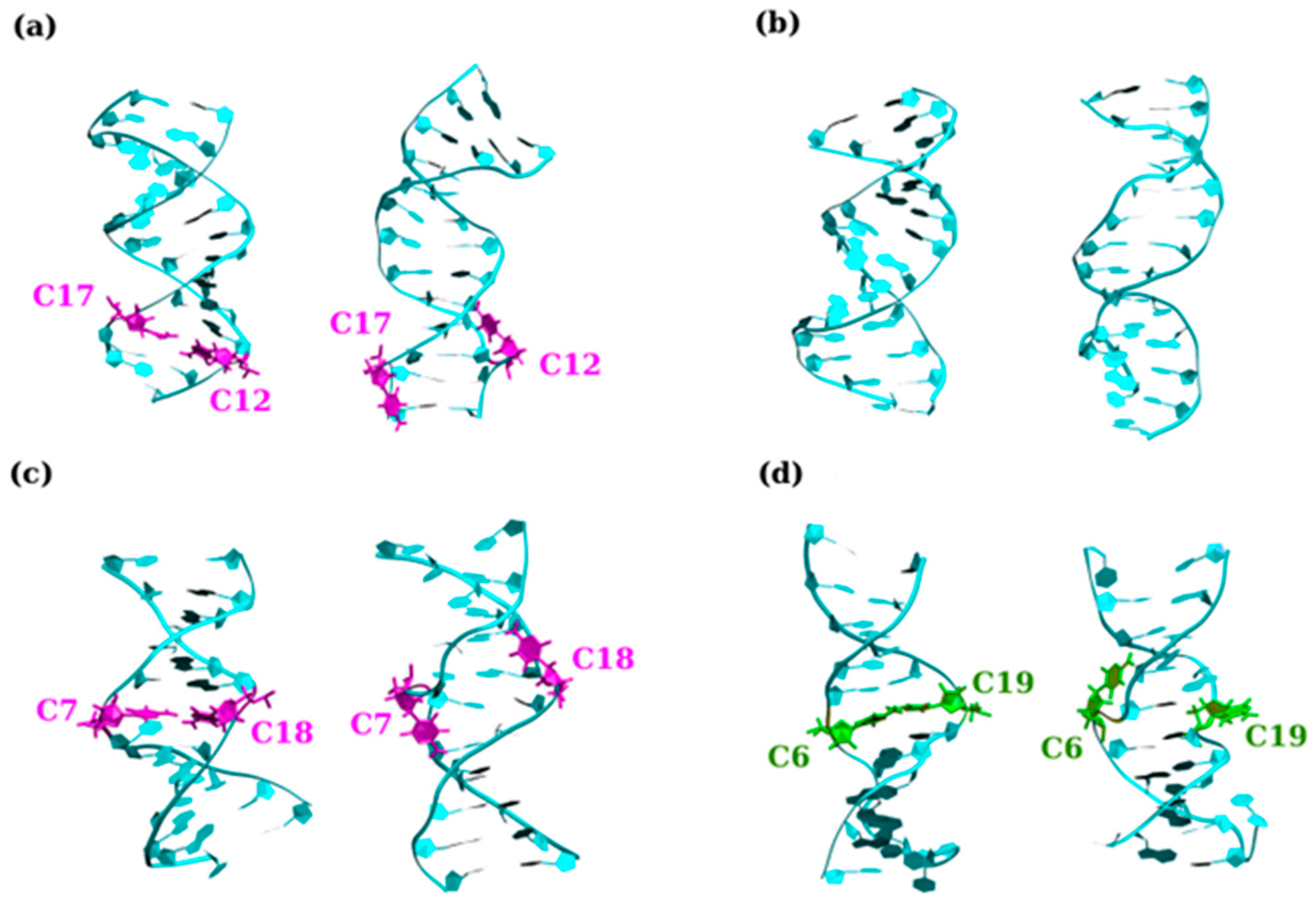
2.1. Hairpins Formed from CAG and GAC TRs
2.1.1. CAG and GAC Homoduplexes
2.1.2. Structure and Dynamics of CAG and GAC Loops
2.2. Hairpins Formed from CTG (CUG for RNA) and GTC (GUG for RNA) Repeats
2.2.1. CTG (CUG for RNA) and GTC (GUC for RNA) Homoduplexes
2.2.2. Structure and Dynamics of CTG and GTC Hairpins
2.3. Hairpins Formed from C and G Exclusive Trinucleotides
2.3.1. CGG, GGC, CCG and GCC Homoduplexes
2.3.2. E-Motif Formed by C- and G-Rich DNA Homoduplexes
2.3.3. Structure and Dynamics of CCG and GGC Hairpins
3. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wells, R.D.; Ashizawa, T. Genetic Instabilities and Neurological Diseases; Elsevier: Boston, MA, USA, 2006. [Google Scholar]
- Ellegren, H. Microsatellites: Simple Sequences with Complex Evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Kashi, Y.; King, D.; Soller, M. Simple Sequence Repeats as a Source of Quantitative Genetic Variation. Trends Genet. 1997, 13, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Caburet, S.; Cocquet, J.; Vaiman, D.; Veitia, R. Coding Repeats and Evolutionary “Agility”. BioEssays 2005, 27, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, B. Uber mytotoische Dystrophie mit Katarakt. Albrecht Von Graefes Arch. Klin. Exp. Optalmol. 1918, 96, 905–914. [Google Scholar]
- Oberle, I.; Rouseau, F.; Heit, D.; Devy, D.; Zengerling, S.; Mandel, J. Molecular-Basis of the Fragile-X Syndrome and Diagnostic Applications. Am. J. Hum. Genet. 1991, 49, 76. [Google Scholar]
- Campuzano, V.; Montermini, L.; Molt, M.; Pianese, L.; Cossee, M.; Cavalcanti, F.; Monros, E.; Rodius, F.; Duclos, F.; Monticelli, A. Friedreich’s Ataxia: Autosomal Recessive Disease Caused by an Intronic GAA Triplet Repeat Expansion. Science 1996, 271, 1423–1427. [Google Scholar] [CrossRef]
- Mirkin, S.M. DNA Structures, Repeat Expansions and Human Hereditary Disorders. Curr. Opin. Struct. Biol. 2006, 16, 351–358. [Google Scholar] [CrossRef]
- Mirkin, S.M. Expandable DNA Repeats and Human Disease. Nature 2007, 447, 932–940. [Google Scholar] [CrossRef]
- Pearson, C.E.; Edamura, K.N.; Cleary, J.D. Repeat Instability: Mechanisms of Dynamic Mutations. Nat. Rev. Genet. 2005, 6, 729–742. [Google Scholar] [CrossRef]
- Pan, F.; Zhang, Y.; Xu, P.; Man, V.H.; Roland, C.; Weninger, K.; Sagui, C. Molecular Conformations and Dynamics of Nucleotide Repeats Associated with Neurodegenerative Diseases: Double Helices and CAG Hairpin Loops. Comput. Struct. Biotechnol. J. 2021, 19, 2819–2832. [Google Scholar] [CrossRef]
- Toth, G.; Gaspari, Z.; Jurka, J. Microsatellites in Different Eukaryotic Genomes: Survey and Analysis. Genome Res. 2000, 10, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Borstnik, B.; Pumpernik, D. Tandem repeats in protein coding regions of primate genes. Genom. Res. 2002, 12, 909–915. [Google Scholar] [CrossRef]
- Bacolla, A.; Larson, J.; Collins, J.; Li, J.; Milosavljevic, A.; Stenson, P.; Cooper, D.; Wells, R. Abundance and Length of Simple Repeats in Vertebrate Genomes Are Determined by Their Structural Properties. Gen. Res. 2008, 18, 1545–1553. [Google Scholar] [CrossRef]
- Clark, R.; Bhaskar, S.; Miyahar, M.; Dalgliesh, G.; Bidichandani, S. Expansion of GAA Trinucleotide Repeats in Mammals. Genomics 2006, 87, 57–67. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Galka-Marciniak, P.; Urbanek, M.O.; Krzyzosiak, W.J. Triplet Repeats in Transcripts: Structural Insights into RNA Toxicity. Biol. Chem. 2012, 393, 1299–1315. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, B.; Spencer, C.; Eberwine, J. CAG Trinucleotide RNA Repeats Interact with RNA- Binding Proteins. Am. J. Hum. Genet. 1996, 59, 561–569. [Google Scholar]
- Timchenko, L.; Timchenko, N.; Caskey, C.; Roberts, R. Novel Proteins with Binding Specificity for DNA CTG Repeats and RNA CUG Repeats: Implications for Myotonic Dystrophy. Hum. Mol. Genet. 1996, 5, 115–121. [Google Scholar] [CrossRef][Green Version]
- Sobczak, K.; Mezer, M.; Michlewski, G.; Krol, J.; Krzyzosiak, W. RNA Structure of Trinucleotide Repeats Associated with Human Neurological Diseases. Nucleic Acids Res. 2003, 31, 5469–5482. [Google Scholar] [CrossRef]
- Timchenko, L. Myotonic Dystrophy: The Role of RNA CUG Triplet Repeats. Am. J. Hum. Genet. 1999, 64, 360–364. [Google Scholar] [CrossRef]
- Mankodi, A.; Takahashi, M.; Jiang, H.; Beck, C.; Bowers, W.; Moxley, R.; Cannon, S.; Thornton, C. Expanded CUG Repeats Trigger Aberrant Splicing of CIC-1 Chloride Channel Pre-MRNA and Hyperexcitability of Skeletal Muscle in Myotonic Dystrophy. Mol. Cell 2002, 10, 35–44. [Google Scholar] [CrossRef]
- Lu, X.; Timchenko, N.; Timchenko, L. Cardiac Delav-Type RNA-Binding Protein (ETR-3) Binds to RNA CUG Repeats Ex-Panded in Myotonic Dystrophy. Hum. Mol. Genet. 1999, 8, 53–60. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Philips, A.; Timchenko, L.; Cooper, T. Disruption of Splicing Regulated by a CUG-Binding Protein in Myotonic Dystrophy. Science 1998, 280, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Tapscott, S.; Thornton, C. Biomedicine—Reconstructing myotonic dystrophy. Science 2001, 293, 816–817. [Google Scholar] [CrossRef] [PubMed]
- Charlet, B.N.; Savkur, R.; Singh, G.; Philips, A.; Grice, E.; Cooper, T. Loss of the Chloride Channel in DM1 Skeletal Muscle Due to Misregulated Alternative Splicing: A Likely Cause of Myotonia. Eur. J. Hum. Genet. 2002, 10, 257–258. [Google Scholar]
- Amack, J.; Mahadevan, M. The Myotonic Dystrophy Expanded CUG Repeat Tract Is Necessary but Not Sufficient to Disrupt C2C12 Myoblast Differentiation. Hum. Mol. Genet. 2021, 10, 1879–1887. [Google Scholar] [CrossRef]
- Amack, J.; Reagan, S.; Mahadevan, M. Mutant DMPK 3’-UTR Transcripts Disrupt C2C12 Myogenic Differentiation by Com-Promising MyoD. J. Cell Biol. 2002, 159, 419–429. [Google Scholar] [CrossRef]
- Savkur, R.; Philips, A.; Cooper, T. Aberrant Regulation of Insulin Receptor Alternative Splicing Is Associated with Insulin Resistance in Myotonic Dystrophy. Nat. Genet. 2001, 29, 40–47. [Google Scholar] [CrossRef]
- Pearson, C. Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Poten-Tially Nine Toxic Enties. PLoS Genet. 2011, 7, e1002018. [Google Scholar] [CrossRef]
- Castelli, L.; Huang, W.-P.; Chang, K.-Y.; Hautbergue, G. Mechanism of Repeat Associated Non-AUG Translation in Neurological Microsatellite Expansion Disorders. Biochem. Soc. Trans. 2021, 49, 775–792. [Google Scholar] [CrossRef]
- Timchenko, N.; Welm, A.; Lu, X.; Timchenko, L. CUG Repeat Binding Protein (CUGBP1) Interacts with the 5’ Region of C/EBP Beta MRNA and Regulates Translation of C/EBP Beta Isoforms. Nucleic Acids Res. 1999, 27, 4517–4525. [Google Scholar] [CrossRef]
- Wells, R.D.; Warren, S. Genetic Instabilities and Neurological Diseases; Academic Press: San Diego, CA, USA, 1998. [Google Scholar]
- Orr, H.; Zoghbi, H. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007, 30, 575. [Google Scholar] [CrossRef] [PubMed]
- Pearson, C.; Sinden, R. Slipped Strand DNA (S-DNA and SI-DNA), Trinucleotide Repeat Instability and Mismatch Repair: A Short Review. In Proceedings of the Structure, Motion, Interaction and Expression of Biological Macromolecules, Albany, NY, USA, 17–21 June 1998; Sarma, R., Sarma, M., Eds.; US NIH, SUNY: Albany, NY, USA, 1998; Volume 2, pp. 191–207. [Google Scholar]
- Paulson, H. Repeat Expansion Diseases. Handb. Clin. Neurol. 2018, 147, 105–123. [Google Scholar] [PubMed]
- Iyer, R.R.; Pluciennik, A. DNA Mismatch Repair and Its Role in Huntington’s Disease. J. Huntingtons Dis. 2021, 10, 75–94. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Correia, K.; Loupe, J.; Kim, K.H.; Barker, D.; Hong, E.P.; Chao, M.J.; Long, J.D.; Lucente, D.; Vonsattel, J.P.G.; et al. CAG Repeat Not Polyglutamine Length Determines Timing of Huntington’s Disease Onset. Cell 2019, 178, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, Y.P.; Telenius, H.; Hayden, M.R. The Molecular Genetics of Huntington’s Disease. Curr. Opin. Neurol. 1994, 7, 325–332. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, M.E.; Ambrose, C.M.; Duyao, M.P.; Myers, R.H.; Lin, C.; Srinidhi, L.; Barnes, G.; Taylor, S.A.; James, M.; Groot, N.; et al. A Novel Gene Containing a Trinucleotide Repeat That Is Expanded and Unstable on Huntington’s Disease Chromosomes. The Huntington’s Disease Collaborative Research Group. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef]
- Greco, T.M.; Secker, C.; Ramos, E.S.; Federspiel, J.D.; Liu, J.P.; Perez, A.M.; Al-Ramahi, I.; Cantle, J.P.; Carroll, J.B.; Botas, J.; et al. Dynamics of Huntingtin Protein Interactions in the Striatum Identifies Candidate Modifiers of Huntington Disease. Cell Syst. 2022, 13, 304–320. [Google Scholar] [CrossRef]
- Malik, I.; Kelley, C.P.; Wang, E.T.; Todd, P.K. Molecular Mechanisms Underlying Nucleotide Repeat Expansion Disorders. Nat. Rev. Mol. Cell Biol. 2021, 22, 589–607. [Google Scholar] [CrossRef]
- Bunting, E.L.; Hamilton, J.; Tabrizi, S.J. Polyglutamine Diseases. Curr. Opin. Neurobiol. 2022, 72, 39–47. [Google Scholar] [CrossRef]
- Tenchov, R.; Sasso, J.M.; Zhou, Q.A. Polyglutamine (PolyQ) Diseases: Navigating the Landscape of Neurodegeneration. ACS Chem. Neurosci. 2024, 15, 2665–2694. [Google Scholar] [CrossRef]
- Bonsor, M.; Ammar, O.; Schnoegl, S.; Wanker, E.E.; Silva Ramos, E. Polyglutamine Disease Proteins: Commonalities and Differences in Interaction Profiles and Pathological Effects. Proteomics 2024, 24, 2300114. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Mirkin, S. The Balancing Act of DNA Repeat Expansions. Curr. Opin. Genet. Devel 2013, 23, 280–288. [Google Scholar] [CrossRef]
- Khristich, A.N.; Mirkin, S.M. On the Wrong DNA Track: Molecular Mechanisms of Repeat-Mediated Genome Instability. J. Biol. Chem. 2020, 295, 4134–4170. [Google Scholar] [CrossRef] [PubMed]
- Pearson, C.E.; Sinden, R.R. Trinucleotide Repeat DNA Structures: Dynamic Mutations from Dynamic DNA. Curr. Opin. Struct. Biol. 1998, 8, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Wells, R.D.; Dere, R.; Hebert, M.L.; Napierala, M.; Son, L.S. Advances in Mechanisms of Genetic Instability Related to Hereditary Neurological Diseases. Nucleic Acids Res. 2005, 33, 3785–3798. [Google Scholar] [CrossRef]
- Cleary, J.; Walsh; Hofmeister, J.; Shanka, G.; Kuskowski, M.; Selkoe, D.; Ashe, K. Natural Oligomers of the Amyloid-Protein Specifically Disrupt Cognitive Function. Nat. Neurosci. 2005, 8, 79–84. [Google Scholar] [CrossRef]
- López Castel, A.; Cleary, J.D.; Pearson, C.E. Repeat Instability as the Basis for Human Diseases and as a Potential Target for Therapy. Nat. Rev. Mol. Cell Biol. 2010, 11, 165–170. [Google Scholar] [CrossRef]
- Dion, V.; Wilson, J. Instability and Chromatin Structure of Expanded Trinucleotide Repeats. Trends Genet. 2009, 25, 288–297. [Google Scholar] [CrossRef]
- McMurray, C.T. Hijacking of the Mismatch Repair System to Cause CAG Expansion and Cell Death in Neurodegenerative Disease. DNA Repair 2008, 7, 1121–1134. [Google Scholar] [CrossRef]
- McMurray, C.T. DNA Secondary Structure: A Common and Causative Factor for Expansion in Human Disease. Proc. Natl. Acad. Sci. USA 1999, 96, 1823–1825. [Google Scholar] [CrossRef]
- Pearson, C.E.; Sinden, R.R. Alternative Structures in Duplex DNA Formed within the Trinucleotide Repeats of the Myotonic Dystrophy and Fragile X Loci. Biochemistry 1996, 35, 5041–5053. [Google Scholar] [CrossRef]
- Schmidt, M.H.M.; Pearson, C.E. Disease-Associated Repeat Instability and Mismatch Repair. DNA Repair 2016, 38, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, G.B.; Lau, R.; Montgomery, S.E.; Leonard, M.R.; Pearson, C.E. Slipped (CTG)•(CAG) Repeats Can Be Correctly Repaired, Escape Repair or Undergo Error-Prone Repair. Nat. Struct. Mol. Biol. 2005, 12, 654–662. [Google Scholar] [CrossRef]
- Nakamori, M.; Panigrahi, G.B.; Lanni, S.; Gall-Duncan, T.; Hayakawa, H.; Tanaka, H.; Luo, J.; Otabe, T.; Li, J.; Sakata, A.; et al. A Slipped-CAG DNA-Binding Small Molecule Induces Trinucleotide-Repeat Contractions in Vivo. Nat. Genet. 2020, 52, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Ranum, L.P.W.; Cooper, T.A. RNA-mediated neuromuscular disorders. Ann. Rev. Neurosci. 2006, 6, 259–277. [Google Scholar] [CrossRef]
- Daughters, R.; Tuttle, D.; Gao, W.; Ikeda, Y.; Moseley, M.; Ebner, T.; Swanson, M.; Ranum, L. RNA Gain-of-Function in Spinocerebellar Ataxia Type 8. PLoS Genet. 2009, 5, e1000600. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Bonini, N. Roles of Trinucleotide-Repeat RNA in Neurological Disease and Degeneration. Trends Neurosci. 2010, 33, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Zarnescu, D.; Zhang, F.; Pearson, C.; Lucchesi, J.; Moses, K.; Warren, S. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron 2003, 39, 739–747. [Google Scholar] [CrossRef]
- Jiang, H.; Mankodi, A.; Swanson, M.; Moxley, R.; Thornton, C. Myotonic Dystrophy Type 1 Is Associated with Nuclear Foci of Mutant RNA, Sequestration of Muscle Blind Proteins and Deregulated Alternative Splicing in Neurons. Hum. Mol. Genet. 2004, 13, 3079–3088. [Google Scholar] [CrossRef]
- Krzyzosiak, W.J.; Sobczak, K.; Wojciechowska, M.; Fiszer, A.; Mykowska, A.; Kozlowski, P. Triplet Repeat RNA Structure and Its Role as Pathogenic Agent and Therapeutic Target. Nucleic Acids Res. 2012, 40, 11–26. [Google Scholar] [CrossRef]
- Campuzano, V.; Montermini, L.; Lutz, Y.; Cova, L.; Hindelan, C.; Jiralerspong, S.; Trottier, Y.; Kish, S.; Faucheux, B.; Trouillas, P.; et al. Frataxin Is Reduced in Friedreich Ataxia Patients and Is Associated with Mitochondrial Membranes. Hum. Mol. Genet. 1997, 6, 1771–1780. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Napierala, M.; Dent, S. Hyperexpansion of GAA Repeats Affects Post-Initiation Steps of FXN Transcription in Friedreich’s Ataxia. Nucl. Acids Res. 2011, 39, 8366–8377. [Google Scholar] [CrossRef] [PubMed]
- Kumari, D.; Biacsi, R.; Usdin, K. Repeat Expansion in Intron 1 of the Frataxin Gene Reduces Transcription Initiation in Friedreich Ataxia. FASEB J. 2011, 25, 895.2. [Google Scholar] [CrossRef]
- Punga, T.; Buehler, M. Long Intronic GAA Repeats Causing Friedreich Ataxia Impede Transcription Elongation. EMBO Mol. Med. 2010, 2, 120–129. [Google Scholar] [CrossRef]
- Iyer, R.R.; Pluciennik, A.; Burdett, V.; Modrich, P.L. DNA Mismatch Repair: Functions and Mechanisms. Chem. Rev. 2006, 106, 302–323. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, T.A.; Erie, D.A. DNA Mismatch Reapir. Annu. Rev. Biochem. 2005, 74, 681–710. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, T.A.; Erie, D.A. Eukaryotic Mismatch Repair in Relation to DNA Replication. Annu. Rev. Genet. 2015, 49, 291–313. [Google Scholar] [CrossRef] [PubMed]
- Modrich, P. Mechanisms in Eukaryotic Mismatch Repair. J. Biol. Chem. 2006, 281, 30305–30309. [Google Scholar] [CrossRef]
- Schofield, M.J.; Hsieh, P. DNA Mismatch Repair: Molecular Mechanisms and Biological Function. Annu. Rev. Microbiol. 2003, 57, 579–608. [Google Scholar] [CrossRef]
- McMurray, C.T. Mechanisms of Trinucleotide Repeat Instability during Human Development. Nat. Rev. Genet. 2010, 11, 786–799. [Google Scholar] [CrossRef]
- Goellner, G.; Tester, D.; Thiboddeau, S.; Almqvist, E.; Goldberg, Y.; Hayden, M.; McMurray, C. Different Mechanisms Underlie DNA Instability in Huntington Disease and Colorectal Cancer. Am. J. Hum. Genet. 1997, 60, 879. [Google Scholar] [PubMed]
- Iyer, R.R.; Pluciennik, A.; Napierala, M.; Wells, R.D. DNA Triplet Repeat Expansion and Mismatch Repair. Annu. Rev. Biochem. 2015, 84, 199–226. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, S.; Donaldson, J.; Flower, M.; Hensman Moss, D.J.; Tabrizi, S.J. Genetic Modifiers of Repeat Expansion Disorders. Emerg. Top. Life Sci. 2023, 7, 325–337. [Google Scholar] [PubMed]
- Jones, L.; Houlden, H.; Tabrizi, S.J. DNA Repair in the Trinucleotide Repeat Disorders. Lancet Neurol. 2017, 16, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Owen, B.A.L.; Yang, Z.; Lai, M.; Gajek, M.; Badger, J.D.; Hayes, J.J.; Edelmann, W.; Kucherlapati, R.; Wilson, T.M.; McMurray, C.T. (CAG)n-Hairpin DNA Binds to Msh2–Msh3 and Changes Properties of Mismatch Recognition. Nat. Struct. Mol. Biol. 2005, 12, 663–670. [Google Scholar] [CrossRef]
- Warren, J.J.; Pohlhaus, T.J.; Changela, A.; Iyer, R.R.; Modrich, P.L.; Beese, L.S. Structure of the Human MutSalpha DNA Lesion Recognition Complex. Mol. Cell 2007, 26, 579–592. [Google Scholar] [CrossRef]
- Gupta, S.; Gellert, M.; Yang, W. Mechanism of Mismatch Recognition Revealed by Human MutSβ Bound to Unpaired DNA Loops. Nat. Struct. Mol. Biol. 2011, 19, 72–79. [Google Scholar] [CrossRef]
- Hsieh, P.; Yamane, K. DNA Mismatch Repair: Molecular Mechanism, Cancer, and Ageing. Mech. Ageing Dev. 2008, 129, 391–407. [Google Scholar] [CrossRef]
- Karran, P.; Offman, J.; Bignami, M. Human Mismatch Repair, Drug-Induced DNA Damage, and Secondary Cancer. Biochimie 2003, 85, 1149–1160. [Google Scholar] [CrossRef]
- Meier, B.; Volkova, N.V.; Hong, Y.; Schofield, P.; Campbell, P.J.; Gerstung, M.; Gartner, A. Mutational Signatures of DNA Mismatch Repair Deficiency in C. Elegans and Human Cancers. Genome Res. 2018, 28, 666–675. [Google Scholar] [CrossRef]
- Fishel, R. The Selection for Mismatch Repair Defects in Hereditary Nonpolyposis Colorectal Cancer: Revising the Mutator Hypothesis. Cancer Res. 2001, 61, 7369–7374. [Google Scholar] [PubMed]
- Li, G.M. DNA Mismatch Repair and Cancer. Front. Biosci. 2003, 8, d997–d1017. [Google Scholar] [CrossRef] [PubMed]
- Modrich, P.; Lahue, R. Mismatch Repair in Replication Fidelity, Genetic Recombination, and Cancer Biology. Annu. Rev. Biochem. 1996, 65, 101–133. [Google Scholar] [CrossRef] [PubMed]
- Muller, A.; Fishel, R. Mismatch Repair and the Hereditary Non-Polyposis Colorectal Cancer Syndrome (HNPCC). Cancer Investig. 2002, 20, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Heinen, C.D. Mismatch Repair Defects and Lynch Syndrome: The Role of the Basic Scientist in the Battle against Cancer. DNA Repair 2016, 38, 127–134. [Google Scholar] [CrossRef]
- Martín-López, J.V.; Fishel, R. The Mechanism of Mismatch Repair and the Functional Analysis of Mismatch Repair Defects in Lynch Syndrome. Fam. Cancer 2013, 12, 159–168. [Google Scholar] [CrossRef]
- Wyatt, M.D.; Pittman, D.L. Methylating Agents and DNA Repair Responses: Methylated Bases and Sources of Strand Break. Chem. Res. Toxicol. 2006, 19, 1580. [Google Scholar] [CrossRef]
- Roos, W.P.; Kaina, B. DNA Damage-Induced Cell Death by Apoptosis. Trends Mol. Med. 2006, 12, 440. [Google Scholar] [CrossRef]
- Kaina, B. DNA Damage-Triggered Apoptosis: Critical Role of DNA Repair, Double-Strand Breaks, Cell Proliferation and Signaling. Biochem. Pharmacol. 2003, 66, 1547–1554. [Google Scholar] [CrossRef]
- Li, Z.; Pearlman, A.; Hsieh, P. DNA Mismatch Repair and the DNA Damage Response. DNA Repair 2016, 38, 94–101. [Google Scholar] [CrossRef]
- Gupta, D.; Heinen, C. The Mismatch Repair-Dependent DNA Damage Response: Mechanism and Implication. DNA Repair 2019, 78, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt, C.; Hensman-Moss, D.; Flower, M.; Wiethoff, S.; Brice, A.; Goizet, C.; Stevanin, G.; Koutsis, G.; Karadima, G.; Panas, M.; et al. DNA Repair Pathways Underlie a Common Genetic Mechanism Modulating Onset in Polyglutamine Diseases. Ann. Neurol. 2016, 79, 983–990. [Google Scholar] [CrossRef] [PubMed]
- Ciosi, M.; Maxwell, A.; Cumming, S.A.; Hensman Moss, D.J.; Alshammari, A.M.; Flower, M.D.; Durr, A.; Leavitt, B.R.; Roos, R.A.C.; Holmans, P.; et al. A Genetic Association Study of Glutamine-Encoding DNA Sequence Structures, Somatic CAG Expansion, and DNA Repair Gene Variants, with Huntington Disease Clinical Outcomes. EBioMedicine 2019, 48, 568–580. [Google Scholar] [CrossRef]
- Lee, J.M.; Wheeler, V.C.; Chao, M.J.; Vonsattel, J.P.G.; Pinto, R.M.; Lucente, D.; Abu-Elneel, K.; Ramos, E.M.; Mysore, J.S.; Gillis, T.; et al. Identification of Genetic Factors That Modify Clinical Onset of Huntington’s Disease. Cell 2015, 162, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, R.; Goold, R.; Coupland, L.; Flower, M.; Tabrizi, S.J. Therapeutic Validation of MMR-Associated Genetic Modifiers in a Human Ex Vivo Model of Huntington Disease. Am. J. Hum. Genet. 2024, 111, 1165–1183. [Google Scholar] [CrossRef] [PubMed]
- Pinto, R.M.; Dragileva, E.; Kirby, A.; Lloret, A.; Lopez, E.; St. Claire, J.; Panigrahi, G.B.; Hou, C.; Holloway, K.; Gillis, T.; et al. Mismatch Repair Genes Mlh1 and Mlh3 Modify CAG Instability in Huntington’s Disease Mice: Genome-Wide and Candidate Approaches. PLoS Genet. 2013, 9, 1003930. [Google Scholar] [CrossRef]
- Dragileva, E.; Hendricks, A.; Teed, A.; Gillis, T.; Lopez, E.T.; Friedberg, E.C.; Kucherlapati, R.; Edelmann, W.; Lunetta, K.L.; MacDonald, M.E.; et al. Intergenerational and Striatal CAG Repeat Instability in Huntington’s Disease Knock-in Mice Involve Different DNA Repair Genes. Neurobiol. Dis. 2009, 33, 37–47. [Google Scholar] [CrossRef]
- Tomé, S.; Manley, K.; Simard, J.P.; Clark, G.W.; Slean, M.M.; Swami, M.; Shelbourne, P.F.; Tillier, E.R.M.; Monckton, D.G.; Messer, A.; et al. MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington’s Disease Mice. PLoS Genet. 2013, 9, e1003280. [Google Scholar] [CrossRef]
- Nakatani, R.; Nakamori, M.; Fujimura, H.; Mochizuki, H.; Takahashi, M.P. Large Expansion of CTG•CAG Repeats Is Exacerbated by MutSβ in Human Cells. Sci. Rep. 2015, 5, 11020. [Google Scholar] [CrossRef]
- Bourn, R.L.; de Biase, I.; Pinto, R.M.; Sandi, C.; Al-Mahdawi, S.; Pook, M.A.; Bidichandani, S.I. Pms2 Suppresses Large Expansions of the (GAA·TTC)n Sequence in Neuronal Tissues. PLoS ONE 2012, 7, e47085. [Google Scholar] [CrossRef]
- Zhao, X.N.; Kumari, D.; Gupta, S.; Wu, D.; Evanitsky, M.; Yang, W.; Usdin, K. Mutsβ Generates Both Expansions and Contractions in a Mouse Model of the Fragile X-Associated Disorders. Hum. Mol. Genet. 2015, 24, 7087–7096. [Google Scholar] [CrossRef] [PubMed]
- Manley, K.; Shirley, T.L.; Flaherty, L.; Messer, A. MSH2 Deficiency Prevents in Vivo Somatic Instability of the CAG Repeat in Huntington Disease Transgenic Mice. Nat. Genet. 1999, 23, 471. [Google Scholar] [CrossRef] [PubMed]
- Kovtun, I.V.; McMurray, C.T. Trinucleotide Expansion in Haploid Germ Cells by Gap Repair. Nat. Genet. 2001, 27, 407–411. [Google Scholar] [CrossRef] [PubMed]
- van den Broek, W.J.; Nelen, M.R.; Wansink, D.G.; Coerwinkel, M.M.; te Riele, H.; Groenen, P.J.; Wieringa, B. Somatic Expansion Behaviour of the (CTG)n Repeat in Myotonic Dystrophy Knock-in Mice Is Differentially Affected by Msh3 and Msh6 Mismatch-Repair Proteins. Hum. Mol. Genet. 2002, 11, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Gonitel, R.; Moffitt, H.; Sathasivam, K.; Woodman, B.; Detloff, P.J.; Faull, R.L.M.; Bates, G.P. DNA Instability in Postmitotic Neurons. Proc. Natl. Acad. Sci. USA 2008, 105, 3467–3472. [Google Scholar] [CrossRef]
- Moore, H.; Greenwell, P.W.; Liu, C.P.; Arnheim, N.; Petes, T.D. Triplet Repeats Form Secondary Structures That Escape DNA Repair in Yeast. Proc. Natl. Acad. Sci. USA 1999, 96, 1504–1509. [Google Scholar] [CrossRef]
- Polak, U.; Mcivor, E.; Dent, S.Y.R.; Wells, R.D.; Napierala, M. Expanded Complexity of Unstable Repeat Diseases. BioFactors 2013, 39, 164–175. [Google Scholar] [CrossRef]
- Usdin, K.; House, N.C.M.; Freudenreich, C.H. Repeat Instability during DNA Repair: Insights from Model Systems. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 142–167. [Google Scholar] [CrossRef]
- Wojciechowska, M.; Napierala, M.; Larson, J.E.; Wells, R.D. Non-B DNA Conformations Formed by Long Repeating Tracts of Myotonic Dystrophy Type 1, Myotonic Dystrophy Type 2, and Friedreich’s Ataxia Genes, Not the Sequences per Se, Promote Mutagenesis in Flanking Regions. J. Biol. Chem. 2006, 281, 24531–24543. [Google Scholar] [CrossRef]
- Zhang, Y.; Roland, C.; Sagui, C. Structural and Dynamical Characterization of DNA and RNA Quadruplexes Obtained from the GGGGCC and GGGCCT Hexanucleotide Repeats Associated with C9FTD/ALS and SCA36 Diseases. ACS Chem. Neurosci. 2018, 9, 1104–1117. [Google Scholar] [CrossRef]
- Zhang, Y.; Roland, C.; Sagui, C. Structure and Dynamics of DNA and RNA Double Helices Obtained from the GGGGCC and CCCCGG Hexanucleotide Repeats That Are the Hallmark of C9FTD/ALS Diseases. ACS Chem. Neurosci. 2017, 8, 578–591. [Google Scholar] [CrossRef]
- Fakharzadeh, A.; Zhang, J.; Roland, C.; Sagui, C. Novel EGZ-Motif Formed by Regularly Extruded Guanine Bases in a Left-Handed Z-DNA Helix as a Major Motif behind CGG Trinucleotide Repeats. Nucleic Acids Res. 2022, 50, 4860–4876. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fakharzadeh, A.; Roland, C.; Sagui, C. RNA as a Major-Groove Ligand: RNA-RNA and RNA-DNA Triplexes Formed by GAA and UUC or TTC Sequences. ACS Omega 2022, 7, 38728–38743. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Man, V.H.; Roland, C.; Sagui, C. Structure and Dynamics of DNA and RNA Double Helices of CAG and GAC Trinucleotide Repeats. Biophys. J. 2017, 113, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Man, V.H.; Roland, C.; Sagui, C. Structure and Dynamics of DNA and RNA Double Helices Obtained from the CCG and GGC Trinucleotide Repeats. J. Phys. Chem. B 2018, 122, 4491–4512. [Google Scholar] [CrossRef]
- Fakharzadeh, A.; Qu, J.; Pan, F.; Sagui, C.; Roland, C. Structure and Dynamics of DNA and RNA Double Helices Formed by d(CTG), d(GTC), r(CUG) and r(GUC) Trinucleotide Repeats and Associated DNA-RNA Hybrids. J. Phys. Chem. B 2023, 127, 7907. [Google Scholar] [CrossRef]
- Zhang, J.; Fakharzadeh, A.; Pan, F.; Roland, C.; Sagui, C. Atypical Structures of GAA/TTC Trinucleotide Repeats Underlying Friedreich’s Ataxia: DNA Triplexes and RNA/DNA Hybrids. Nucleic Acids Res. 2020, 48, 9899–9917. [Google Scholar] [CrossRef]
- Zhang, J.; Fakharzadeh, A.; Pan, F.; Roland, C.; Sagui, C. Construction of DNA/RNA Triplex Helices Based on GAA/TTC Trinucleotide Repeats. Bio Protoc. 2021, 11, e4155. [Google Scholar] [CrossRef]
- Pan, F.; Zhang, Y.; Man, V.H.; Roland, C.; Sagui, C. E-Motif Formed by Extrahelical Cytosine Bases in DNA Homoduplexes of Trinucleotide and Hexanucleotide Repeats. Nucleic Acids Res. 2018, 46, 942–955. [Google Scholar] [CrossRef]
- Xu, P.; Pan, F.; Roland, C.; Sagui, C.; Weninger, K. Dynamics of Strand Slippage in DNA Hairpins Formed by CAG Repeats: Roles of Sequence Parity and Trinucleotide Interrupts. Nucleic Acids Res. 2020, 48, 2232–2245. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, J.; Pan, F.; Mahn, C.; Roland, C.; Sagui, C.; Weninger, K. Frustration Between Preferred States of Complementary Trinucleotide Repeat DNA Hairpins Anticorrelates with Expansion Disease Propensity. J. Mol. Biol. 2023, 435, 168086. [Google Scholar] [CrossRef]
- Weiss, S. Measuring Conformational Dynamics of Biomolecules by Single Molecule Fluorescence Spectroscopy. Nat. Struct. Biol. 2000, 7, 724–729. [Google Scholar] [CrossRef]
- Roy, R.; Hohng, S.; Ha, T. A Practical Guide to Single-Molecule FRET. Nat. Methods 2008, 5, 507–516. [Google Scholar] [CrossRef]
- Götz, M.; Barth, A.; Bohr, S.S.-R.; Börner, R.; Chen, J.; Cordes, T.; Erie, D.A.; Gebhardt, C.; Hadzic, M.C.A.S.; Hamilton, G.L.; et al. A Blind Benchmark of Analysis Tools to Infer Kinetic Rate Constants from Single-Molecule FRET Trajectories. Nat. Commun. 2022, 13, 5402. [Google Scholar] [CrossRef]
- Moradi, M.; Babin, V.; Sagui, C.; Roland, C. Recipes for Free Energy Calculations in Biomolecular Systems. Methods Mol. Biol. 2013, 924, 313–337. [Google Scholar]
- Babin, V.; Roland, C.; Sagui, C. Adaptively Biased Molecular Dynamics for Free Energy Calculations. J. Chem. Phys. 2008, 128, 134101. [Google Scholar] [CrossRef]
- Sugita, Y.; Kitao, A.; Okamoto, Y. Multidimensional Replica-Exchange Method for Free-Energy Calculations. J. Chem. Phys. 2000, 113, 6042–6051. [Google Scholar] [CrossRef]
- Izrailev, S.; Stepaniants, S.; Kosztin, D.; Lu, H.; Molnar, F.; Wriggers, W.; Schulten, K. Steered Molecular Dynamics. In Proceedings of the 2nd International Symposium on Algorithms for Macromolecular Modelling, Berlin, Germany, 21–24 May 1997; pp. 39–65. [Google Scholar]
- Kozlowski, P.; Mezer, M.; Krzyzisuak, W. Trinucleotide Repeats in Human Genome and Exome. Nucl. Acids Res. 2010, 38, 4039. [Google Scholar] [CrossRef]
- Zoghbi, H.Y.; Orr, H.T. Glutamine Repeats and Neurodegeneration. Annu. Rev. Neurosci. 2000, 23, 217–247. [Google Scholar] [CrossRef]
- Sikorski, P.; Atkins, E. New Model for Crystalline Polyglutamine Assemblies and Their Connection with Amyloid Fibrils. Biomacromolecules 2005, 6, 425–432. [Google Scholar] [CrossRef]
- Man, V.H.; Roland, C.; Sagui, C. Structural Determinants of Polyglutamine Protofibrils and Crystallites. ACS Chem. Neurosci. 2015, 6, 632–645. [Google Scholar] [CrossRef] [PubMed]
- Délot, E.; King, L.M.; Briggs, M.D.; Wilcox, W.R.; Cohn, D.H. Trinucleotide Expansion Mutations in the Cartilage Oligomeric Matrix Protein (COMP) Gene. Hum. Mol. Genet. 1999, 8, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Hecht, J.T.; Nelson, L.D.; Crowder, E.; Wang, Y.; Elder, F.F.B.; Harrison, W.R.; Francomano, C.A.; Prange, C.K.; Lennon, G.G.; Deere, M.; et al. Mutations in Exon 17B of Cartilage Oligomeric Matrix Protein (COMP) Cause Pseudoachondroplasia. Nat. Genet. 1995, 10, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Briggs, M.D.; Mortier, G.R.; Cole, W.G.; King, L.M.; Golik, S.S.; Bonaventure, J.; Nuytinck, L.; De Paepe, A.; Leroy, J.G.; Biesecker, L.; et al. Diverse Mutations in the Gene for Cartilage Oligomeric Matrix Protein in the Pseudoachondroplasia-Multiple Epiphyseal Dysplasia Disease Spectrum. Am. J. Hum. Genet. 1998, 62, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Vorlícková, M.; Kejnovská, I.; Tumová, M.; Kypr, J. Conformational Properties of DNA Fragments Containing GAC Trinucleotide Repeats Associated with Skeletal Displasias. Eur. Biophys. J. 2001, 30, 179–185. [Google Scholar] [CrossRef]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Giambasu, G.; et al. AMBER 20; University of California: San Francisco, CA, USA, 2000. [Google Scholar]
- Yildirim, I.; Park, H.; Disney, M.D.; Schatz, G.C.A. Dynamic Structural Model of Expanded RNA CAG Repeats: A Refined X-Ray Structure and Computational Investigations Using Molecular Dynamics and Umbrella Sampling Simulations. J. Am. Chem. Soc. 2012, 40, 4273–4287. [Google Scholar] [CrossRef]
- Amadei, A.; Linssen, A.B.; Berendsen, H.J.C. Essential Dynamics of Proteins. Proteins Struct. Funct. Genet. 1993, 17, 412–425. [Google Scholar] [CrossRef]
- Cadden, G.; Wilkens, S.; Magennis, S. A Single CAA Interrupt in a DNA Three-Way Junction Containing a CAG Repeat Hairpin Results in Parity-Dependent Trapping. Nucl. Acids Res. 2024, 52, 9317–9327. [Google Scholar] [CrossRef]
- Bianco, S.; Hu, T.; Henrich, O.; Magennis, S. Heterogeneous migration routes of DNA triplet repeat slip-outs. Biophys. Rep. 2022, 2, 100070. [Google Scholar] [CrossRef]
- Hu, T.; Morten, M.J.; Magennis, S.W. Conformational and Migrational Dynamics of Slipped-Strand DNA Three-Way Junctions Containing Trinucleotide Repeats. Nat. Commun. 2021, 12, 204. [Google Scholar] [CrossRef]
- Mitchell, M.; Leveille, M.; Solecki, R.; Tran, T.; Cannon, B. Sequence-Dependent Effect of Monovalent Cations on the Structural Dynamics of Trinucleotide-Repeat DNA Hairpins. J. Phys. Chem. B 2018, 122, 11841–11851. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.E.B.; Black, H.F.; Collins, J.A.; Gall-Duncan, T.; Caron, N.S.; Pearson, C.E.; Hayden, M.R. Interrupting Sequence Variants and Age of Onset in Huntington’s Disease: Clinical Implications and Emerging Therapies. Lancet Neurol. 2020, 19, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.E.B.; Collins, J.A.; Kay, C.; McDonald, C.; Dolzhenko, E.; Xia, Q.; Bečanović, K.; Drögemöller, B.I.; Semaka, A.; Nguyen, C.M.; et al. Length of Uninterrupted CAG, Independent of Polyglutamine Size, Results in Increased Somatic Instability, Hastening Onset of Huntington Disease. Am. J. Hum. Genet. 2019, 104, 1116–1126. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Matsuura, T.; Coolbaugh, M.; Zuhlke, C.; Nakamura, K.; Rasmussen, A.; Siciliano, M.; Ashizawa, T.; Lin, X. Instability of Expanded CAG/CAA Repeats in Spinocerebellar Ataxia Typye 17. Eur. J. Hum. Genet. 2008, 16, 215. [Google Scholar] [CrossRef]
- Charles, P.; Camuzat, A.; Benammar, N.; Sellal, F.; Destee, A.; Bonnet, A.; Lesage, S.; Le Ber, I.; Stevanin, G.; Durr, A. Are Interrupted SCA2 CAG Repeat Expansions Responsible for Parkinsonism? Neurology 2007, 69, 1970–1975. [Google Scholar] [CrossRef]
- Elden, A.C.; Kim, H.-J.; Hart, M.P.; Chen-Plotkin, A.S.; Johnson, B.S.; Fang, X.; Armakola, M.; Geser, F.; Greene, R.; Lu, M.M. Ataxin-2 Intermediate-Length Polyglutamine Expansions Are Associated with Increased Risk for ALS. Nature 2010, 466, 1069. [Google Scholar] [CrossRef]
- Choudhry, S.; Mukerji, M.; Srivastava, A.K.; Jain, S.; Brahmachari, S.K. CAG Repeat Instability at SCA2 Locus: Anchoring CAA Interruptions and Linked Single Nucleotide Polymorphisms. Hum. Mol. Genet. 2001, 10, 2437–2446. [Google Scholar] [CrossRef]
- Volker, J.; Breslauer, K. How Sequence Alterations Enhance the Stability and Delay Expansion of DNA Triplet Repeat Domains. QRB Discov. 2023, 4, e8. [Google Scholar] [CrossRef]
- Hartenstine, M.J.; Goodman, M.F.; Petruska, J. Base Stacking and Even/Odd Behavior of Hairpin Loops in DNA Triplet Repeat Slippage and Expansion with DNA Polymerase. J. Biol. Chem. 2000, 275, 18382–18390. [Google Scholar] [CrossRef]
- Brook, J.D.; McCurrach, M.E.; Harley, H.G.; Buckler, A.J.; Church, D.; Aburatani, H.; Hunter, K.; Stanton, V.P.; Thirion, J.-P.; Hudson, T.; et al. Molecular Basis of Myotonic Dystrophy: Expansion of a Trinucleotide (CTG) Repeat at the 3’ End of a Tran-Script Encoding a Protein Kinase Family Member. Cell 1992, 68, 799. [Google Scholar] [CrossRef]
- Mahadevan, M.; Tsilfidi, C.; Sabourin, L.; Shutler, G.; Amemiya, C.; Jansen, G.; Neville, C.; Narang, M.; Barcelo, J.; O.’Hoy, K. Myotonic Dystrophy Mutation: An Unstable CTG Repeat in the Untranslated Region of the Gene. Science 1992, 255, 1253. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.; McCurrach, M.; Taneja, K.; Singer, R.; Housman, D. Expansion of a CUG Trinucleotide Repeat in the 3’ Untranslated Region of Myotonic Dystrophy Protein Kinase Transcripts Results in Nuclear Retention of Transcripts. Proc. Natl. Acad. Sci. USA 1997, 94, 7388. [Google Scholar] [CrossRef] [PubMed]
- Kiliszek, A.; Kierzek, R.; Krzyzosiak, W.J.; Rypniewski, W. Structural Insights into CUG Repeats Containing the ‘Stretched U-U Wobble’: Implications for Myotonic Dystrophy. Nucleic Acids Res. 2009, 37, 4149–4156. [Google Scholar] [CrossRef] [PubMed]
- Coonrod, L.; Lohman, J.R.; Berglund, J.A. Utilizing the GAAA Tetra Loop/Receptor to Facilitate Crystal Packing and the Determination of the Structure of a CUG RNA Helix. Biochemistry 2012, 42, 8330. [Google Scholar]
- Tamjar, J.; Katorcha, E.; Popov, A.; Malinina, L. Structural Dynamics of Double- Helical RNAs Composed of CUG/CUG and CUG/CGG Repeats. J. Biomol. Struct. Dyn. 2012, 30, 505. [Google Scholar] [CrossRef]
- Napierala, M.; Krzyzonsiak, W.J. CUG repeats present in myotonin kinase RNA form metastable “slippery” hairpins. J. Biol. Chem. 1997, 272, 31079. [Google Scholar] [CrossRef]
- Yildirim, I.; Chakraborty, D.; Disney, M.; Wales, D.; Schatz, G. Computational Investigation of RNA CUG Repeats Responsible for Myotonic Dystrophy 1. J. Chem. Theory Comput. 2015, 11, 4943. [Google Scholar] [CrossRef]
- Ni, C.W.; Wei, Y.J.; Shen, Y.I.; Lee, I.R. Long-Range Hairpin Slippage Reconfiguration Dynamics in Trinucleotide Repeat Sequences. J. Phys. Chem. Lett. 2019, 10, 3985–3990. [Google Scholar] [CrossRef]
- Wan, L.; He, A.; Li, J.; Guo, P.; Han, D. High-Resolution NMR Structures of Intra Strand Hairpins Formed by CTG Trinucleotide Repeats. ACS Chem. Neurosci. 2024, 15, 868–876. [Google Scholar] [CrossRef]
- Fu, Y.H.; Kuhl, D.P.; Pizzuti, A.; Pieretti, M.; Sutcliffe, J.S.; Richards, S.; Verkert, A.J.; Holden, J.J., Jr.; RGF, W.; ST, O.; et al. Variation of the CGG Repeat at the Fragile X Site Results in Genetic Instability: Resolution of the Sherman Paradox. Cell 1991, 67, 1047–1058. [Google Scholar] [CrossRef]
- Zhong, N.; Ju, W.; Pietrofesa, J.; Wang, D.; Dobkin, C.; Brown, W.T. Fragile X “Gray Zone” Alleles: AGG Patterns, Expansion Risks, and Associated Haplotypes. Am. J. Med. Genet. 1996, 64, 261–265. [Google Scholar] [CrossRef]
- Dombrowski, C.; Lévesque, S.; Morel, M.L.; Rouillard, P.; Morgan, K.; Rousseau, F. Pre-Mutation and Intermediate-Size FMR1 Alleles in 10572 Males from the General Population: Loss of an AGG Interruption Is a Late Event in the Generation of Fragile X Syndrome Alleles. Hum. Mol. Genet. 2002, 11, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Hagerman, R.; Leehey, M.; Heinrichs, W.; Tassone, F.; Wilson, R.; Hills, J.; Grigsby, J.; Gage, B.; Hagerman, P. Intention Tremor, Parkinsonism, and Generalized Brain Atrophy in Male Carriers of Fragile X. Neurology 2001, 57, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Sherman, S.L. Premature Ovarian Failure among Fragile X Premutation Carriers: Parent-of-Origin Effect? Am. J. Hum. Genet. 2000, 67, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Glass, I. X linked mental retardation. J. Med. Genet. 1991, 28, 361–371. [Google Scholar] [CrossRef]
- Gu, Y.; Shen, Y.; Gibbs, R.A.; Nelson, D.L. Identification of FMR2, A Novel Gene Associated with the FRAXE CCG Repeat and CpG Island. Nat. Genet. 1996, 13, 109–113. [Google Scholar] [CrossRef]
- Braida, C.; Stefanatos, R.K.; Adam, B.; Mahajan, N.; Smeets, H.J.; Niel, F.; Goizet, C.; Arveiler, B.; Koenig, M.; Lagier-Tourenne, C.; et al. Variant CCG and GGC Repeats within the CTG Ex-Pansion Dramatically Modify Mutational Dynamics and Likely Contribute toward Un- Usual Symptoms in Some Myotonic Dystrophy Type 1 Patients. Hum. Mol. Genet. 2010, 19, 1399–1412. [Google Scholar] [CrossRef]
- Kuma, A.; Fang, P.; Park, H.; Guo, M.; Nettles, K.W.; Disney, M. A Crystal Structure of a Model of the Repeating r(CGG) Transcript Found in Fragile X Syndrome. ChemBioChem 2011, 12, 2140–2142. [Google Scholar] [CrossRef]
- Kiliszek, A.; Kierzek, R.; Krzyzosiak, W.J.; Rypniewski, W. Crystallographic Characterization of CCG Repeats. Nucl. Acids Res. 2012, 40, 8155–8162. [Google Scholar] [CrossRef]
- Man, V.H.; Pan, F.; Sagui, C.; Roland, C. Comparative Melting and Healing of B-DNA and Z-DNA by an Infrared Laser Pulse. J. Chem. Phys. 2016, 144, 145101. [Google Scholar] [CrossRef]
- Gao, X.; Huang, X.; Smith, G.; Zheng, M.; Liu, H. New Antiparallel Duplex Motif of DNA CCG Repeats That Is Stabilized by Extrahelical Basis Symmetrically Located in the Minor-Groove. J. Am. Chem. Soc. 1995, 117, 8883–8884. [Google Scholar] [CrossRef]
- Assi, H.; Garavis, M.; Gonzalez, C.; Damha, M. I-Motif DNA: Structural Features and Significance to Cell Biology. Nucl. Acids Res. 2018, 46, 8038. [Google Scholar] [PubMed]
- Chen, Y.W.; Jhan, C.R.; Neidle, S.; Hou, M.H. Structural Basis for the Identification of an I-Motif Tetraplex Core with a Parallel-Duplex Junction as a Structural Motif in CCG Triplet Repeats. Angew. Chem. Int. Ed. Engl. 2014, 53, 10682. [Google Scholar] [CrossRef] [PubMed]
- Zeraati, M.; Langley, D.; Schofield, P.; Moye, A.; Rouet, R.; Hughes, W.; Bryan, T.; Dinger, M.; Christ, D. I-Motif DNA Structures Are Formed in the Nuclei of Human Cells. Nat. Chem. 2018, 10, 631. [Google Scholar] [CrossRef] [PubMed]
- Fojtik, P.; Kejnovska, I.; Vorlickova, M. The Guanine-Rich Fragile X-Chromosome Repeats Are Reluctant to Form Tetramers. Nucl. Acids Res. 2004, 32, 298. [Google Scholar] [CrossRef]
- Renciuk, D.; Kypr, J.; Vorlickova, M. CGG Repeats Associated with Fragile X Chromosome Form Left-Handed Z-DNA Structure. Biopolymers 2011, 95, 174. [Google Scholar] [CrossRef]
- Shen, Y.-I.; Cheng, K.-C.; Wei, Y.-J.; Lee, I.-R. Structural Dynamics Role of AGG Interruptions in Inhibition CGG Repeat Expansions Associated with Fragile X Syndrome. ACS Chem. Neurosci. 2024, 15, 230–235. [Google Scholar] [CrossRef]
- Eichler, E.E.; Holden, J.J.; Popovich, B.W.; Reiss, A.L.; Snow, K.; Thibodeau, S.N.; Richards, C.S.; Ward, P.A.; Nelson, D.L. Length of Uninterrupted CGG Repeats Determines Instability in the FMR1 Gene. Nat. Genet. 1994, 8, 88–94. [Google Scholar] [CrossRef]
- Falik-Zaccai, T.C.; Shachek, E.; Yalon, M.; Lis, Z.; Borochowitz, Z.; Macpherson, J.N.; Nelson, D.L.; Eichler, E.E. Predisposition to the Fragile X-Syndrome in Jews of Tunisian Descent Is Due to the Absence of AGG Interruptions on a Rare Mediterranean Haplotype. Am. J. Hum. Genet. 1997, 60, 103–112. [Google Scholar]
- Yrigollen, C.M.; Durbin-Johnson, B.; Gane, L.; Nelson, D.L.; Hagerman, R.; Hagerman, P.J.; Tassone, F. AGG Interruptions with the Maternal FMR1 Gene Reduces the Irish Offspring with Fragile X Syndrome. Genet. Med. Off. J. Am. Coll. Med. Genet. 2012, 14, 729–736. [Google Scholar]
- Latham, G.J.; Coppinger, J.; Hadd, A.G.; Nolin, S.L. The Role of AGG Interruptions in Fragile X Repeat Expansions: A Twenty Year Perspective. Front. Genet. 2014, 5, 244. [Google Scholar] [CrossRef] [PubMed]
- Jarem, D.A.; Huckaby, L.V.; Delaney, S. AGG Interruptions in (CGG)n DNA Repeat Tracts Modulate the Structure and Thermodynamics of Non-B Conformations in Vitro. Biochemistry 2010, 49, 6826–6837. [Google Scholar] [CrossRef] [PubMed]
- Rajan-Babu, I.-S.; Dolzenko, E.; Eberle, M.; Friendman, J. Sequence Composition Changes in Short Tandem Repeats: Heterogeneity Detection, Mechanisms and Clinical Implications. Nat. Rev. Genet. 2024, 25, 476–499. [Google Scholar] [CrossRef] [PubMed]
- Matos-Rodrigues, G.; Hisey, J.; Nussenzweig, A.; Mirkin, S. Detection of Alternative DNA Structures and Its Implications for Human Disease. Mol. Cell 2023, 83, 3622–3641. [Google Scholar] [CrossRef] [PubMed]
- Darlow, J.M.; Leach, D.R.F. The Effects of Trinucleotide Repeats Found in Human Inherited Disorders on Palindrome Inviability in Escherichia Coli Suggest Hairpin Folding Preferences in Vivo. Genetics 1995, 141, 825–832. [Google Scholar] [CrossRef]
- Zhao, X.N.; Usdin, K. FAN1 Protects against Repeat Expansions in a Fragile X Mouse Model. DNA Repair 2018, 69, 1–5. [Google Scholar] [CrossRef]
- Goold, R.; Flower, M.; Moss, D.M.; Medway, C.; Wood-Kaczmar, A.; Andre, R.; Farshim, P.; Bates, G.P.; Holmans, P.; Jones, L.; et al. FAN1 Modifies Huntington’s Progression by Stabilizing the Expanded HTT CAG Repeat. Hum. Mol. Genet. 2019, 28, 650–661. [Google Scholar] [CrossRef]
- Goold, R.; Hamilton, J.; Menneteau, T.; Flower, M.; Bunting, E.L.; Aldous, S.G.; Porro, A.; Vicente, J.R.; Allen, N.D.; Wilkinson, H.; et al. FAN1 Controls Mismatch Repair Complex Assembly via MLH1 Retention to Stabilize CAG Repeat Expansion in Huntington’s Disease. Cell Rep. 2021, 36, 109649. [Google Scholar] [CrossRef]
- Loupe, J.M.; Pinto, R.M.; Kim, K.H.; Gillis, T.; Mysore, J.S.; Andrew, M.A.; Kolvalenko, M.; Murtha, R.; Seong, I.; Gusella, J.F.; et al. Promotion and Somatic CAG Repeat Expansion by FAN1 Knock-out in Huntington’s Disease Knock-in Mice Is Blocked by MIh1 Knock-Out. Hum. Mol. Genet. 2020, 29, 3044–3053. [Google Scholar] [CrossRef]
- Desmukh, A.L.; Porro, A.; Mohiuddun, M.; Lanni, S.; Panigrahi, G.B.; Caron, M.C.; Masson, J.Y.; Sartori, A.A.; Pearson, C.E. FAN1, a DNA Repair Nuclease as a Modifier of Repeat Expansion Disorders. Hungtons Dis. 2021, 10, 95–122. [Google Scholar]
- Phadte, A.S.; Bhatia, M.; Ebert, H.; Abdulla, H.; Elrazaq, E.A.; Komolov, K.E.; Pluciennik, A. FAN1 Removes Triplet Repeat Extrusions via a PCNA- and RFC-Dependent Mechanism. Proc. Natl. Acad. Sci. USA 2023, 120, e2302103120. [Google Scholar] [CrossRef] [PubMed]
- Kadyrova, L.Y.; Mieczkowski, P.A.; Kadyrov, F.A. Genome-Wide Contributions of the MutSalpha and MutSbeta-Dependent DNA Mismatch Repair Pathways to the Maintenance of Genetic Stability in Saccharomyces Cerevisiae. J. Biol. Chem. 2023, 299, 104705. [Google Scholar] [CrossRef] [PubMed]
- Tabrizi, S.J.; Flower, M.D.; Ross, C.A.; Wild, E.J. Huntington Disease: New Insights into Molecular Pathogenesis and Therapeutic Opportunities. Nat Rev. Neurol. 2020, 16, 529–546. [Google Scholar] [CrossRef] [PubMed]
- Tabrizi, S.J.; Estevez-Fraga, C.; van Roon-Mom, W.M.C.; Flower, M.D.; Scahill, R.I.; Wild, E.J.; Muñoz-Sanjuan, I.; Sampaio, C.; Rosser, A.E.; Leavitt, B.R. Potential Disease-Modifying Therapies for Huntington’s Disease: Lessons Learned and Future Opportunities. Lancet Neurol. 2022, 21, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, H.; Yang, W. Tandem MutSβ Binding to Long Extruded DNA Trinucleotide Repeats Underpins Pathogenic Expansions. bioRxiv, 2023; preprint. [Google Scholar]
- Lee, J.-H.; Thomsen, M.; Daub, H.; Steinbacher, S.C.; Sztyler, A.; Thieulin-Pardo, G.; Neudegger, T.; Plotnikov, N.V.; Iyer, R.R.; Wilkinson, H.A.; et al. Conformational Dynamics and DNA Recognition by Human MutSβ. bioRxiv, 2023; preprint. [Google Scholar]
- Lang, W.H.; Coats, J.E.; Majka, J.; Hura, G.L.; Lin, Y.; Rasnik, I.; McMurray, C.T. Conformational Trapping of Mismatch Recognition Complex MSH2/MSH3 on Repair-Resistant DNA Loops. Proc. Natl. Acad. Sci. USA 2011, 108, E837–E844. [Google Scholar] [CrossRef]
- Kadyrova, L.Y.; Gujar, V.; Burdett, V.; Modrich, P.L.; Kadyrov, F.A. Human MutLγ, the MLH1–MLH3 Heterodimer, Is an Endonuclease That Promotes DNA Expansion. Proc. Natl. Acad. Sci. USA 2020, 117, 3535–3542. [Google Scholar] [CrossRef]
- Huang, T.Y.; Chang, C.K.; Kao, Y.F.; Chin, C.H.; Ni, C.W.; Hsu, H.Y.; Hu, N.J.; Hsieh, L.C.; Chou, S.H.; Lee, I.R.; et al. Parity-Dependent Hairpin Configurations of Repetitive DNA Sequence Promote Slippage Associated with DNA Expansion. Proc. Natl. Acad. Sci. USA 2017, 114, 9535–9540. [Google Scholar] [CrossRef]
- O’Reilly, D.; Belgrad, J.; Ferguson, C.; Summers, A.; Sapp, E.; McHugh, C.; Mathews, E.; Boudi, A.; Buchwald, J.; Ly, S.; et al. Di-Valent SiRNA-Mediated Silencing of MSH3 Blocks Somatic Repeat Expansion in Mouse Models of Huntington’s Disease. Mol. Ther. 2023, 31, 1661–1674. [Google Scholar] [CrossRef] [PubMed]
- Aldous, S.G.; Smith, E.J.; Landles, C.; Osborne, G.F.; Cañibano-Pico, M.; Nita, I.M.; Phillips, J.; Zhang, Y.; Jin, B.; Hirst, M.B.; et al. A CAG Repeat Threshold for Therapeutics Targeting Somatic Instability in Huntington’s Disease. Brain 2024, 147, 1784–1798. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.E.; Shin, J.W.; Zeng, S.; Hong, E.P.; Jang, J.H.; Loupe, J.M.; Wheeler, V.C.; Stutzman, H.E.; Kleinstiver, B.; Lee, J.M. Base Editing Strategies to Convert CAG to CAA Diminish the Disease-Causing Mutation in Huntington’s Disease. Elife 2024, 12, RP89782. [Google Scholar] [CrossRef] [PubMed]
- Gall-Duncan, T.; Luo, J.; Jurkovic, C.M.; Fisher, L.A.; Fujita, K.; Desmukh, A.L.; Harding, R.J.; Tran, S.; Mehkary, M.; Li, V.; et al. Antagonistic Roles of Canonical and Alternative-RPA in Disease-Associated Tandem CAG Repeat Instability. Cell 2023, 186, 4898–4919. [Google Scholar] [CrossRef] [PubMed]
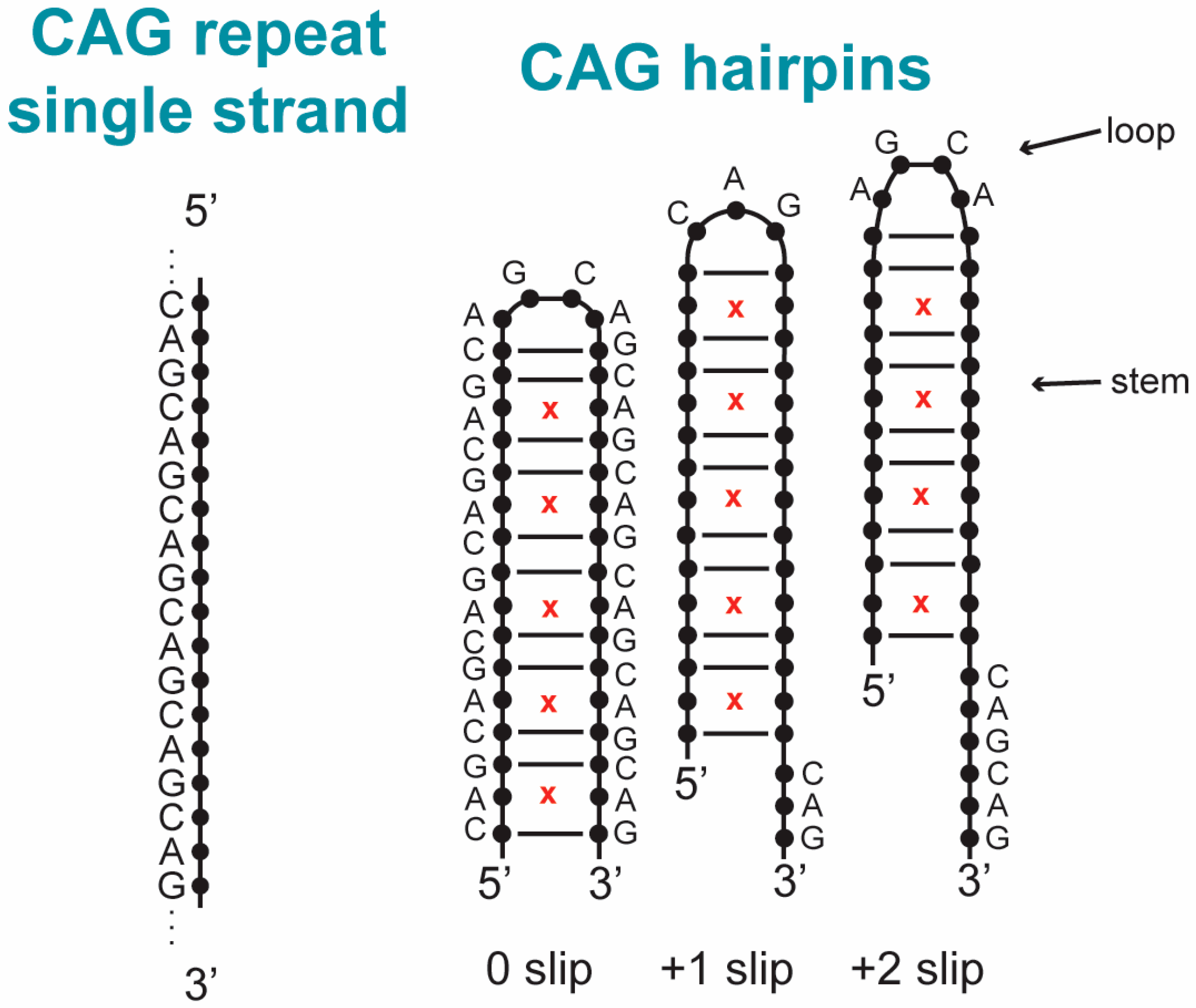
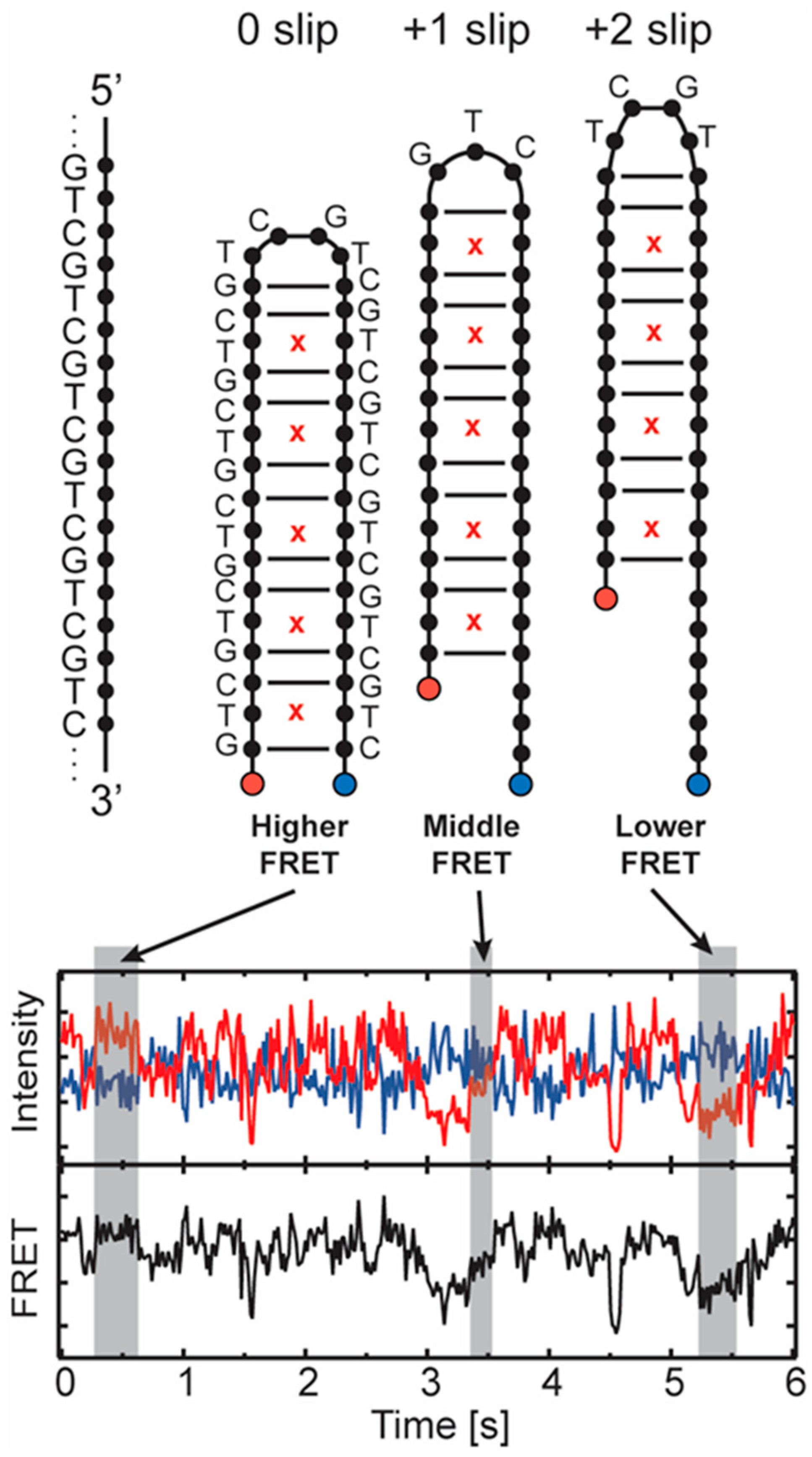
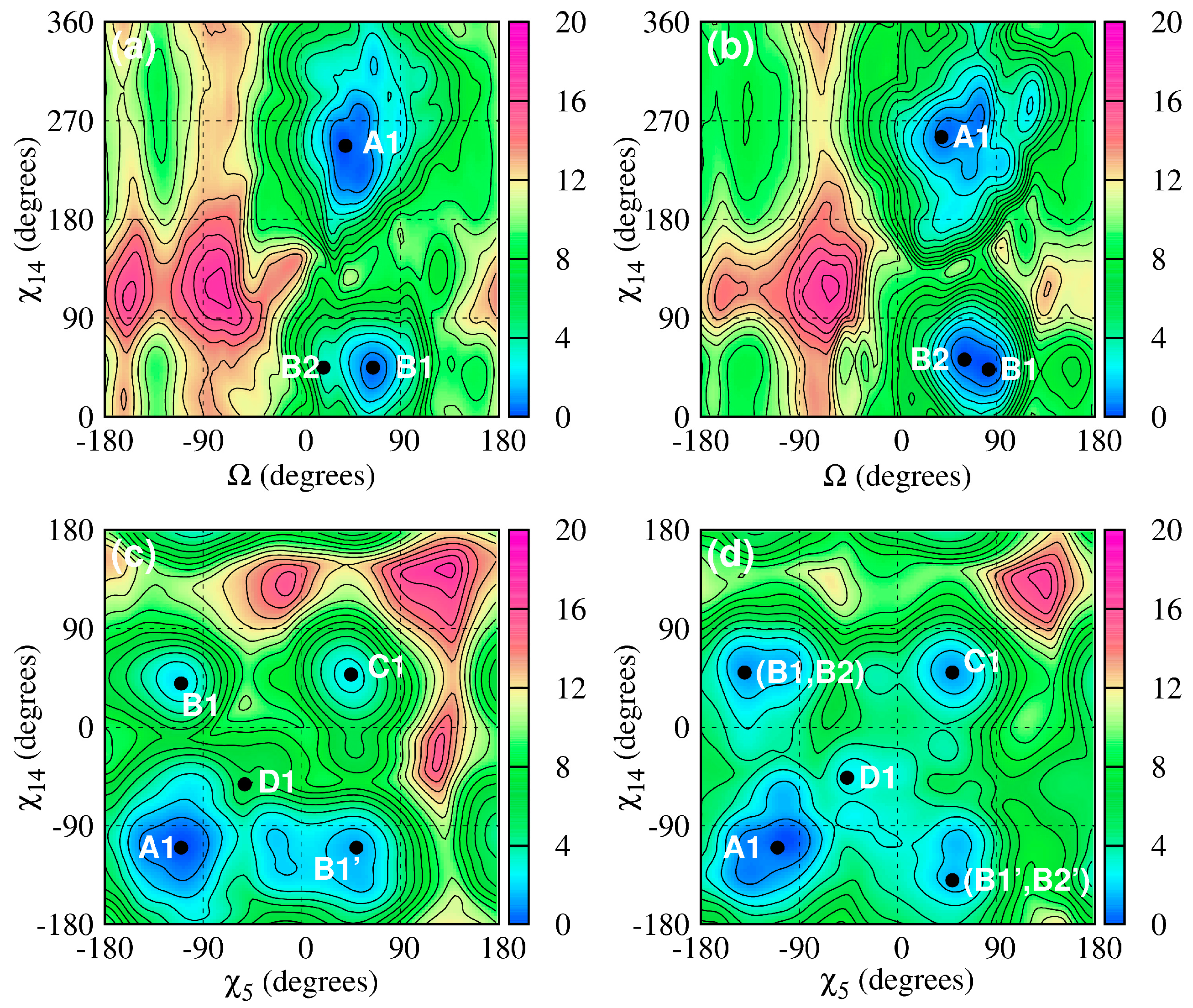
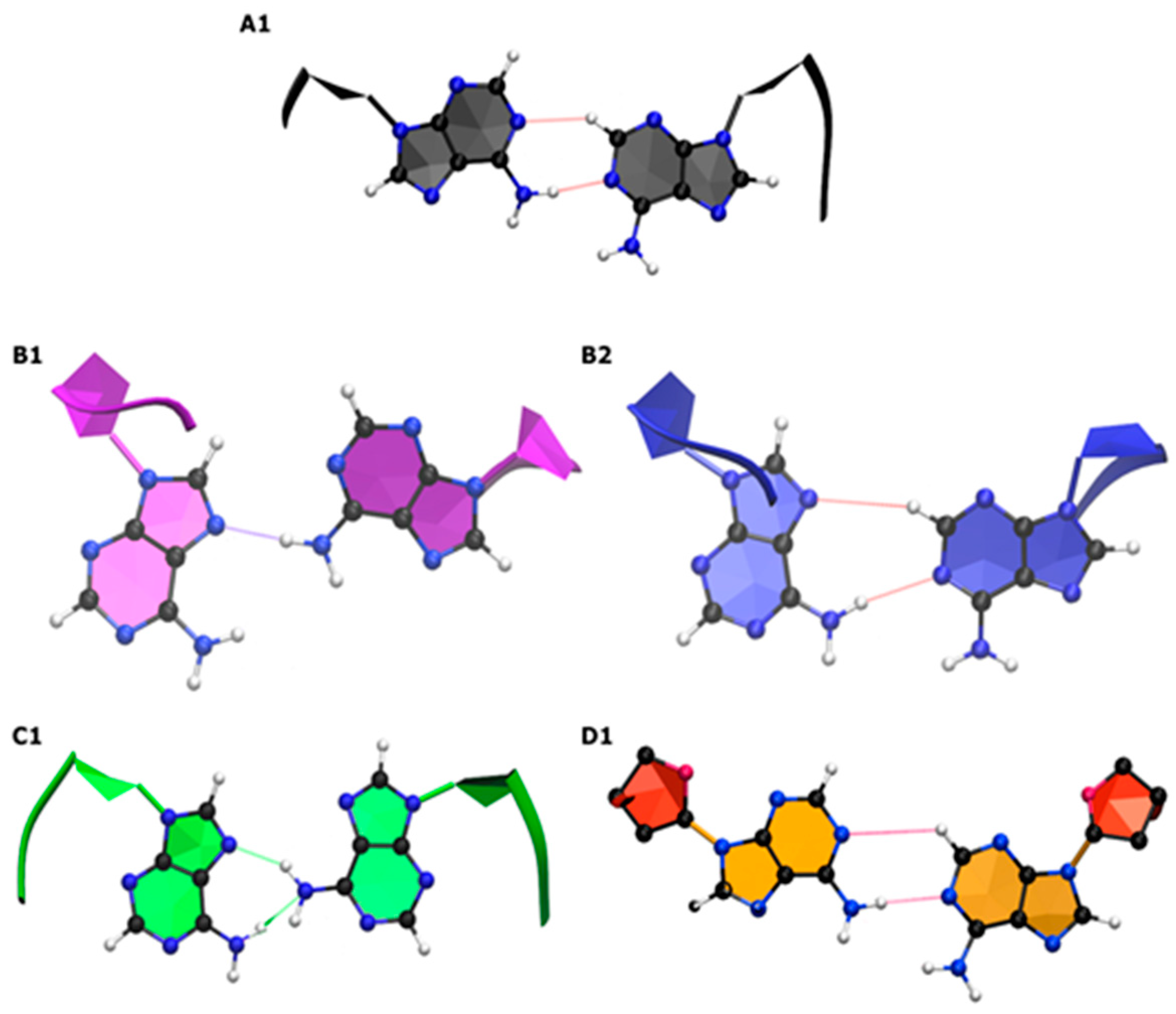

| Location on Gene | Repeat | Disease |
|---|---|---|
| 5′-UTR | CGG | FRAXA, FXTAS |
| GCC | FRAXE | |
| CAG | SCA12 | |
| EXON | CAG | HD, HDL2, SBMA |
| DRPLA, SCA1, SCA2, SCA3 | ||
| SCA6, SCA7, SCA17 | ||
| GAC | MSD | |
| GCG | SPD, HFG, ISSX, CCD | |
| HPES, OPMD, CCHS, BPES | ||
| INTRON | CAA | FRDA |
| CCTG | DM2 | |
| ATTCT | SCA10 | |
| TGGAA | SCA31 | |
| GGCCTG | SCA36 | |
| GGGGCC | ALS | |
| 3′-UTR | CTG | DM1, HDL2, SCA8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, F.; Xu, P.; Roland, C.; Sagui, C.; Weninger, K. Structural and Dynamical Properties of Nucleic Acid Hairpins Implicated in Trinucleotide Repeat Expansion Diseases. Biomolecules 2024, 14, 1278. https://doi.org/10.3390/biom14101278
Pan F, Xu P, Roland C, Sagui C, Weninger K. Structural and Dynamical Properties of Nucleic Acid Hairpins Implicated in Trinucleotide Repeat Expansion Diseases. Biomolecules. 2024; 14(10):1278. https://doi.org/10.3390/biom14101278
Chicago/Turabian StylePan, Feng, Pengning Xu, Christopher Roland, Celeste Sagui, and Keith Weninger. 2024. "Structural and Dynamical Properties of Nucleic Acid Hairpins Implicated in Trinucleotide Repeat Expansion Diseases" Biomolecules 14, no. 10: 1278. https://doi.org/10.3390/biom14101278
APA StylePan, F., Xu, P., Roland, C., Sagui, C., & Weninger, K. (2024). Structural and Dynamical Properties of Nucleic Acid Hairpins Implicated in Trinucleotide Repeat Expansion Diseases. Biomolecules, 14(10), 1278. https://doi.org/10.3390/biom14101278






