Abstract
The 18-exon human TKFC gene codes for dual-activity triokinase and FMN cyclase (TKFC) in an ORF, spanning from exon 2 to exon 18. In addition to TKFC-coding transcripts (classified as tkfc type by their intron-17 splice), databases contain evidence for alternative TKFC transcripts, but none of them has been expressed, studied, and reported in the literature. A novel full-ORF transcript was cloned from brain cDNA and sequenced (accession no. DQ344550). It results from an alternative 3′ splice-site in intron 17. The cloned cDNA contains an ORF also spanning from exon 2 to exon 18 of the TKFC gene but with a 56-nt insertion between exons 17 and 18 (classified as tkfc_ins56 type). This insertion introduces an in-frame stop, and the resulting ORF codes for a shorter TKFC variant, which, after expression, is enzymatically inactive. TKFC intron-17 splicing was found to be differentially expressed in human tissues. In a multiple-tissue northern blot using oligonucleotide probes, the liver showed a strong expression of the tkfc-like splice of intron 17, and the heart preferentially expressed the tkfc_ins56-like splice. Through a comparison to global expression data from massive-expression studies of human tissues, it was inferred that the intestine preferentially expresses TKFC transcripts that contain neither of those splices. An analysis of transcript levels quantified by RNA-Seq in the GTEX database revealed an exception to this picture due to the occurrence of a non-coding short transcript with a tkfc-like splice. Altogether, the results support the occurrence of potentially relevant transcript variants of the TKFC gene, differentially expressed in human tissues. (This work is dedicated in memoriam to Professor Antonio Sillero, 1938–2024, for his lifelong mentoring and his pioneering work on triokinase).
1. Introduction
The human TKFC gene (GeneID 26007, Ensembl ID ENSG00000149476) codes for the dual-activity enzyme named triokinase (EC 2.7.1.28) and FMN cyclase (EC 4.6.1.15) [1,2]. The triokinase activity catalyzes the third step of the specific metabolism of fructose (ATP-dependent phosphorylation of D-glyceraldehyde) [3,4,5,6] and the phosphorylation of the exogenous genotoxic dihydroxyacetone [7,8,9,10]. The enzyme also exerts control on the lipogenic potential of fructose and dietary tolerance [11]. The FMN cyclase activity catalyzes the cyclizing lyase splitting of FAD to AMP and cFMN (riboflavin 4′,5′-cyclic phosphate) [12,13]. This unusual flavin nucleotide occurs in nanomolar concentrations in rat livers with an unknown function [14], and it is an abundant component of the posterior flagellum of reproductive cells in the brown alga Scytosiphon lomentaria, where it could be involved in phototaxis [15]. In addition to its enzymatic activities, the TKFC protein interacts with melanoma differentiation antigen 5 (MDA5), an RNA helicase acting as a viral RNA sensor during viral replication, and as an inducer of interferon expression. Through this interaction, TKFC inhibits MDA5-mediated innate antiviral signaling [16,17]; interestingly, this effect can be modulated via the binding of non-coding RNAs (microRNA-122 or circSOBP) to TKFC [18,19]. Additionally, the TKFC promoter is controlled by the carbohydrate response element-binding protein (ChREBP) [20,21,22].
TKFC is a homodimeric protein of two-domain monomers, each containing a K domain (N-terminal) and an L domain (C-terminal). In the homodimer, two triokinase active sites are formed between the K1–L2 and the K2–L1 domains (Scheme 1). In contrast, the FMN cyclase activity requires only the L domain, although full cyclase efficiency also needs the K domain [2]. Interaction of TKFC with MDA5 occurs through the K domain, which, when tested in isolation, is an effective inhibitor of MDA5-mediated innate immunity comparable to the effect of the full TKFC protein [16].
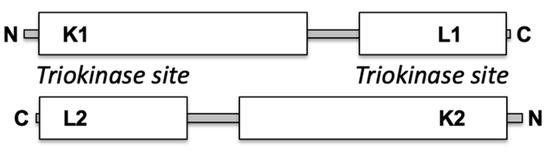
Scheme 1.
Domains in the subunits of homodimeric TKFC with two triokinase active sites.
Human TKFC bi-allelic mutations that inactivate the triokinase activity are present with different phenotypes depending on the protein domain where the altered residue(s) are located. There is a report of an individual with compound heterozygous missense mutations in the K domain (c.574G>C (p.Gly192Arg) and c.682C>T (p.Arg228Trp)), which presents an isolated hair phenotype (hypotrichosis with loose anagen hairs) [23]. On the other hand, several individuals with homozygous variants in the L domain have been reported to display more severe phenotypes. The presence of either c.1628G>T (p.Arg543Ile) or c.1333G>A (p.Gly445Ser), both in homozygosis, have been linked to cataracts and multisystem diseases, including developmental delay, liver dysfunction, microcytic anemia, and fatal cardiomyopathy [24]. Recently, the presence of the homozygous c.1624G>A (p.Gly542Arg) variant has been shown, in one individual, to be associated with a complex primary immunodeficiency in isolation [25].
Very recently, the homozygous c.598G>A p.(Val200Ile) variant of TKFC gene was reported in two neonates with skeletal abnormalities and fatal hypertrophic cardiomyopathy [26]. The relationship of this variant to altered TKFC activities is compounded because no enzyme activity data are available, and the patients also display homozygous variants in the TYR gene.
According to the mRNA NM_015533.4 (which is the MANE Select transcript for the human TKFC gene [27]) aligned to genomic DNA (chromosome 11q12.2), the TKFC gene (ID: 26007) is depicted as containing 18 exons. The ORF coding for TKFC protein (1728 nt; accession no. ABA10576.1) starts in the 3′ end of exon 2 and ends within exon 18, giving rise to a 575 amino acid translation, which, once expressed in homodimeric form, displays triokinase and FMN cyclase activities [1,2,28]. This ORF is present in five transcript variants with different splicing patterns which code for the protein isoform b or X6 (they differ by a single amino acid in position 185 due to a common SNP [28]) (Table 1 and Figure 1). Current evidence indicates that one or more of these transcripts and the protein isoform they encode are the major products of the human TKFC gene. However, as summarized in Table 1 and Figure 1, there is evidence for alternative spliced full-length transcript variants which code for different protein isoforms, although none of them has been expressed and studied so far. Here, we report on the cloning, expression, and characterization of the ORF encoded by transcript variants X12–X14 and their product protein isoform X8.

Table 1.
TKFC full-length transcripts (splice variants) and their encoded protein isoforms. The exon structures of the transcript variants are depicted in Figure 1.
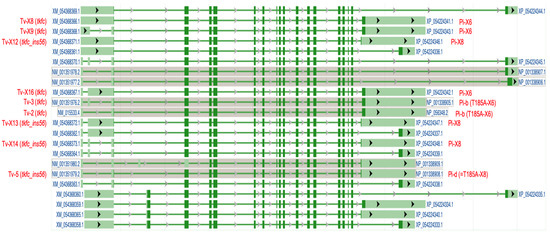
Figure 1.
Exon-intron structure of human full-length TKFC transcript variants as shown in the NCBI Genome Data Viewer (chromosome 11, alternate T2T-CHM13v2.0 genome). Dark green colored regions indicate the coding sequences. The transcript variants labeled in red type are those that code for fully active TKFC protein (isoforms b and X6) or for the inactive isoform cloned and characterized in this manuscript (isoforms d and X8). See also Table 1. Tv, transcript variant; Pi, protein isoform.
2. Materials and Methods
2.1. PCR Cloning, Construction, and Sequencing of Plasmid pGEX-6P-3-hF1
The ORF for human TKFC isoform X8 was amplified with Pfu-Ultra High-Fidelity DNA polymerase (Stratagene) from brain cDNA (Marathon Ready cDNA; BD Biosciences). The design of the primers was based on the sequence of transcript NM_015533 which codes for TKFC isoform b. The forward primer was CATCTGTCGACATGACCTCCAAGAAGCTG, which includes a SalI site (underlined) followed by the first 18 nucleotides of the ORF of isoform b. The reverse primer was TTACAGCGGCCGCCTAGCTCTGCAAGACCTC, which includes a NotI site (underlined) followed by the reverse complement of the 18 last nucleotides of the ORF. An amplicon near the expected size was obtained and inserted in the pGEX-6P-3 plasmid (Amersham) between the SalI and NotI sites. Twelve clones containing an insert were isolated and sequenced. Eleven plasmids contained the 1728 bp ORF coding for TKFC isoform X6 [1], and only one contained the 1784 bp ORF coding for TKFC isoform X8. Plasmid pGEX-6P-3-hF1 was submitted to double-strand sequencing and used for expression. The results of this manuscript were previously undisclosed except in the deposit in GenBank (accession no. DQ344550) on 24 January 2006 by our laboratory.
2.2. Expression of Plasmid pGEX-6P-3-hF1 in BL21 Cells
pGEX-6P-3-hF1 was used to transform BL21 cells, which were seeded in LB agar plates with 0.1 mg/mL ampicillin. One clone was picked and grown overnight at 37 °C in 10 mL of liquid LB medium with ampicillin (amp-LB). Five milliliters were inoculated in 100 mL of amp-LB incubated at 27 °C with shaking. When A600 reached 0.6–0.8, 0.5 mM IPTG was added. After 2 h at 27 °C, the cells were pelleted by 10 min centrifugation at 9000 rpm at 4 °C in a Sorvall SS34 rotor. The cell pellet was resuspended in 10 mL of 20 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1 mM DTT, 50 mM KCl, containing one tablet of CompleteTM protease inhibitor cocktail (Mini, EDTA-free; from Roche). Sonication on ice followed at 170 W for 5 min (0.1 s pulses per second) with three repetitions. The lysate was centrifuged at 15,000 rpm for 10 min at 4 °C in a Sorvall SS34 rotor. The supernatant was used to purify the GST-X8 fusion protein. The precipitate was resuspended in 10 mL of sonication buffer for analytical purposes.
2.3. Purification of TKFC Isoform X8
Ten milliliters of the supernatant containing the GST-X8 fusion protein was applied to a 1.5 mL glutathione-Sepharose column equilibrated in 20 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1 mM DTT, and 50 mM KCl. The column was washed with 15 mL of 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 mM DTT, and 150 mM NaCl to eliminate the unbound protein. After this, 20 µL of PreScissionTM protease (Amersham), diluted in 1.5 mL of wash buffer, was applied to the column, and flow was stopped. After 12 h at 4 °C, the product of in-column specific proteolysis was recovered with 4.5 mL of wash buffer. The fractions collected during and after protease treatment were analyzed by SDS-PAGE.
2.4. Triokinase and FMN Cyclase Assays of Recombinant TKFC Isoform X8
The absence of enzyme activity of isoform X8 was tested using the assay methods described earlier. In brief, triokinase activity was tested with dihydroxyacetone and ATP as substrates using a continuous spectrophotometric assay coupled to glycerol 3-phosphate dehydrogenase as indicator enzyme [2]. The FMN cyclase or FAD-AMP cyclizing lyase activity (FAD → cFMN + AMP) was tested discontinuously by colorimetric assay of the liberation of inorganic phosphate from AMP by alkaline phosphatase, used as a coupling enzyme [13].
2.5. Multiple Tissue Northern Blot
A commercial northern membrane containing twelve lanes with 1 µg poly(A) RNA per lane from different human tissues was purchased from BD Biosciences, Clontech (catalog no. 7780-1). For hybridization, two 50-mer oligonucleotides were designed: E17E18 (TGGCCTCGGCTGCAGCTTCGGCACTCTTGACTGCTTTGGTCAGGACTTGT) composed by the reverse complement of the 5′-most 25 nt of TKFC exon 18 (underlined) followed by the reverse complement of the 3′-most 25 nt of TKFC exon 17; INS50 (GATGGGGTGTGGGAGCAGGTTCAAGGGCAGATCACCAGGCCGCCTCCTTC), which is part of the reverse complement of the 56-nt insertion left by the functioning of the alternative 3′-splice site of TKFC intron 17. The probes, E17E18 and INS50, were labeled with digoxigenin-dUTP using the DIG Oligonucleotide Tailing Kit, 2nd Generation (Roche), according to the manufacturer’s instructions. Hybridizations (2 h) were performed essentially as reported [29], except that the (pre)hybridization solutions were supplemented with 50 mg/L polyA and used, like the washing buffer, at 45 °C. Chemiluminescence was recorded by 120 min exposures of X-ray film to the hybridized membrane. First, the membrane was processed with the E17E18 probe. Thereafter, the blot was stripped and reprobed with INS50.
2.6. Molecular Modeling
The most recent molecular model of wild-type homodimeric TKFC, which corresponds to protein isoform X6, has been described by Onoufriadis et al.; it was validated using a combination of homology modeling and molecular dynamics simulation [23]. Using this model of isoform X6, the 50 amino acids lost due the alternative splicing on intron 17 were highlighted by showing them in a different color, or just by hiding them. These digital manipulations were conducted with VMD v.1.9.4a57 software [30].
3. Results
3.1. Cloning and Sequence Analysis of the Coding Sequence of TKFC Protein Isoform X8 Formed by Intervention of an Alternative 3′-Splice Site in TKFC Intron 17
The PCR cloning of human brain TKFC was performed using primers designed to amplify the 1728 nt ORF, from the ATG to the stop codon, based on the MANE Select mRNA NM_015533 [1]. Eleven out of the 12 pGEX-6P-3 clones then sequenced were derived from transcripts constitutively spliced in intron 17 (i.e., they are of tkfc type). The remaining plasmid clone, named pGEX-6P-3-hF1, contained a novel sequence of 1784 nt (GenBank accession no DQ344550) that fits with transcripts alternatively spliced in intron 17, leaving a 56-nt insertion between exons 17 and 18 (i.e., they are of tkfc_ins56 type). The 1784 nt amplicon contained in pGEX-6P-3-hF1 was like the 1728 nt one, except for a silent G/A SNP at position 507 and for a 56 nt insertion (ins56) just after position 1575, i.e., in the junction of TKFC exons 17-18 (Figure 2). The sequence of ins56 aligned exactly with the 3′ end of intron 17 in the genomic TKFC sequence and delimited an alternative 3′-splice site with a weak consensus motif: it had the invariant AG, but its polypyrimidine tract contained several purines, and the nearest fit to the branching point consensus was at a long distance, i.e., 80-nt away from the alternative intron end (Figure 2, larger insert). The functionality of the alternative 3′-splice site denoted by the tkfc_ins56-type splice was confirmed by the neural network predictor NNSPLICE 0.9 [31], which found it as one of four possible 3′-splice sites within intron 17 (Figure 2, larger insert, arrows).
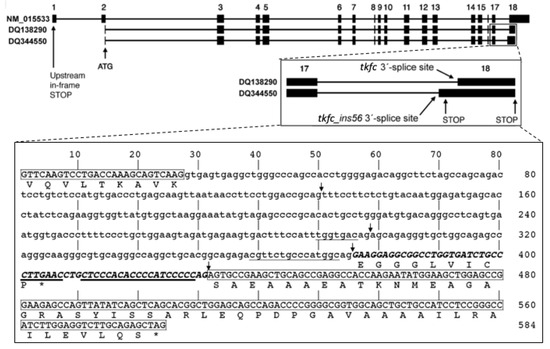
Figure 2.
Exon-intron structure of the human TKFC gene and alternative transcripts produced by the use of different 3′-splice sites in intron 17. NM_015533 is the MANE Select [27] spliced transcript of the TKFC gene used as reference to define the gene structure. DQ138290 is the spliced ORF that encode the 575 amino-acid protein isoform X6 which is active as triokinase and FMN cyclase [1,2,28]. DQ344550 is the novel tkfc_ins56-type spliced sequence reported here, which codes for protein isoform X8. The inserts are enlargements showing the alternative splicing of TKFC intron 17 and the locations of stop codons. The larger insert shows the sequence of intron 17 flanked by exon sequences. Boxed sequences are part of exon 17 (3′-end) and exon 18 (5′-end). The non-boxed part of the sequence corresponds to full intron 17 according to the tkfc-type splicing. The uppercase sequence within intron 17 is the 56 nt insertion (ins56) that remains in the alternative tkfc_ins56-type splicing. The underlined sequences mark the possible branching sites and the polypyrimidine tracts near each 3′-splice site. The arrows mark the acceptor sites identified by the neural network predictor program NNSPLICE 0.9 with scores higher than 0.05 for a 0–1 range: 0.22 (at 130-1), 0.62 (at 298-9), 0.21 (at 375-6; the site used to form tkfc_ins56-type transcripts) and 0.39 (431-2; the site used to form tkfc-type transcripts) [31]. The amino acid translation shown below exon 17 is common to protein isoform X6 and X8. The translation shown below the 56 nt insertion, ending with a proline residue, is the C-terminal sequence unique to protein isoform X8 (accession no. ABC70184), and that below exon 18, ending with a serine residue, is unique to the catalytically active isoform X6 (accession no. ABA10576). Stop codons are indicated by asterisks.
Compared to transcripts that code for fully active TKFC (isoforms b or X6), the insertion of ins56 in the junction of exons 17–18 led to a shorter protein isoform, as the inserted sequence contained an in-frame stop codon. While the ORF of transcripts coding for isoforms b or X6 yielded the 575 amino-acid TKFC [1,2,28], after the 56 nt insertion, the resulting ORF translated to a shorter, 534 amino-acid protein (isoform X8), with the peptide EGGGLVICP substituting for the 50 C-terminal residues of TKFC (Figure 2, larger insert). An interesting feature of this premature termination is that it occurs within the last exon of the transcripts, so that they can escape mRNA degradation via the nonsense-mediated decay pathway, which, in mammals, is related to premature stop signals at least 50–55 nt upstream of the last exon-exon junction [32].
3.2. Expression and Catalytic Inactivity of the Recombinant Protein Isoform X8
The protein isoform X8 was expressed as a GST fusion protein upon IPTG induction of E. coli BL21 cells transformed with plasmid pGEX-6P-3-hF1. The molecular weight of the induced protein was, as expected, somewhat smaller than the corresponding fusion protein formed with active TKFC (Figure 3a). The GST-tagged protein was adsorbed to a GSH-Sepharose column and was recovered by in-column specific proteolysis that separated it from the GST tag (Figure 3b). In these preparations, the enzyme activities typical of triokinase and FMN cyclase were undetectable, i.e., below 3% of the specific activities measured with active enzyme expressed and prepared in parallel with the same system (Figure 3a).
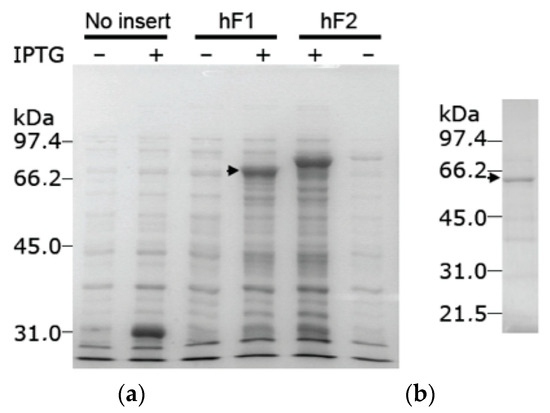
Figure 3.
Expression of the recombinant protein isoform X8, encoded by an alternatively spliced tkfc_ins56-type transcript. (a) IPTG-dependent expression of GST fusion proteins in supernatant lysates of E. coli BL21 cells transformed with plasmids pGEX-6P-3 (without insert), pGEX-6P-3-hF1 (bearing the tkfc_ins56-type ORF) or pGEX-6P-3-hF2 (bearing the tkfc-type ORF [1]). Inserts were cloned in frame with the GST tag. (b) Protein recovered from the GST-tagged expression product after adsorption to GSH-Sepharose and overnight in-column proteolysis with PreScission® protease. The arrowheads mark the GST fusion proteins with isoform X8 (a) or the protein separated from the GST tag (b). Original images of (a,b) can be found in Supplementary Materials.
In the purified isoform X8 (Figure 3b), the enzyme activities typical of triokinase and FMN cyclase were undetectable, below 3% of the rates measured with the active isoform X6. This was shown by assays of the individual isoforms X6 and X8 (Figure 4a) and of X6/X8 mixtures in different proportions (Figure 4b), clearly demonstrating the inactivity of isoform X8.
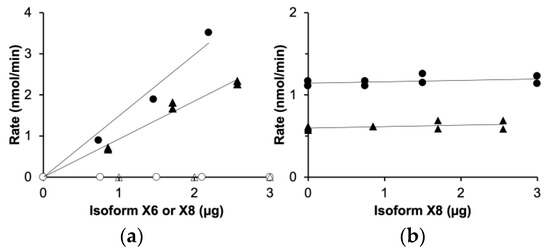
Figure 4.
Inactivity of TKFC isoform X8 compared to isoform X6: assays of dihydroxyacetone kinase (circles) and FMN cyclase (triangles). (a) Assays run with either isoform X6 (filled symbols) or isoform X8 (empty symbols). (b) Assays run with a constant amount of isoform X6 (0.73 µg) and variable concentrations of isoform X8.
The inactivity of isoform X8 is related to the absence of the 50 C-terminal amino acids of the active enzyme (isoform X6). According to the most recent structural model of active TKFC [23], the missing amino acids are part of the L domain, which is required for the FMN cyclase activity [2] and includes a 20 amino-acid loop (residues 535–554) that contains Arg-543 which is essential for triokinase activity [23] (Figure 5a,b), just as it is the conserved residue in the Escherichia coli homolog [33].
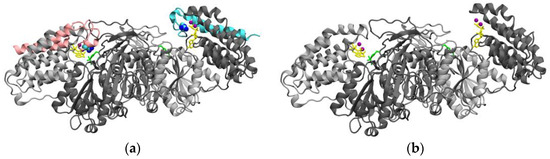
Figure 5.
Comparison of the structure of active TKFC protein (isoform X6) with the catalytically inactive isoform X8 studied in this manuscript. (a) Homodimeric structure of TKFC isoform X6 [1,2,28]. The two subunits are shown in different gray tones, except that the 50 C-terminal amino acids encoded by exon 18 (Figure 2, large insert) are colored either pink or blue in each subunit. The essential Arg543 residues (blue spheres) that interact with ATP (yellow) are highlighted. (b) Parts of isoform X6 that remain in isoform X8. The model is like that of part (a), except that the 50 C-terminal amino acids of each subunit have been removed, showing the disappearing of Arg543 residues. To fully represent the structure of isoform X8, the peptide EGGGLVICP encoded by intron 17 should be added (Figure 2, large insert). Anyhow, since isoform X8 is enzymatically inactive, it is clear that this peptide cannot play the role of Arg543.
3.3. Tissue Expression of TKFC-Gene Transcripts Formed with Alternative 3′-Splice Sites of Intron 17 Studied by Northern Analysis
To estimate the relative levels of human transcripts processed at TKFC intron 17, like tkfc- or tkfc_ins56-type transcripts, a Multiple Tissue Northern (MTN) blot was probed with two 50-mer oligonucleotides one after another: E17E18 and INS50 (Figure 6a,b). According to BLASTN searches run against human genomic sequences, these oligonucleotides were uniquely related to the TKFC gene. The probes were, respectively, designed to hybridize with transcripts containing the exon 17-18 junction like the tkfc-type, or with the 56 nt insertion typical of tkfc_ins56-type transcripts. The E17E18 and INS50 probes disclosed different patterns of expression and, interestingly, they did not correlate with the global expression profile of the TKFC gene according to databases [34,35] (Figure 6c).
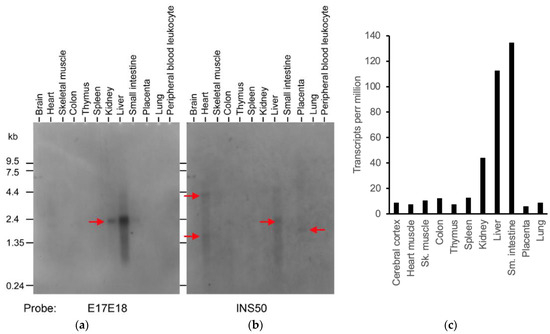
Figure 6.
Differential expression of the alternative splices of TKFC intron 17 in several human tissues. (a) Expression of the tkfc-like splice containing an exon 17-18 junction. (b) Expression of the tkfc_ins56-like splice containing an insertion between exons 17 and 18. The red arrows mark the major RNAs detected. (c) Total expression of the TKFC gene in the human tissues as determined by RNA-Seq (https://www.proteinatlas.org/ENSG00000149476-TKFC/tissue, accessed on 25 August 2024; RNA expression, Consensus dataset of the Human Protein Atlas; accessed on 10 June 2024) [34]. Similar data were found in the Genotype-Tissue Expression (GTEx) database [35] (https://gtexportal.org/home/gene/TKFC; accessed on 25 August 2024). Original images of (a,b) can be found in Supplementary Materials.
Concerning the expression patterns inferred from the Northern blot, hybridization with E17E18 revealed a 2.2 kb transcript strongly expressed in the liver and less so in kidney, spleen, and small intestine (Figure 6a). Reprobing with INS50 gave no strong hybridization signal, but weak ones were observed in the heart (4.3 kb and 1.6 kb), liver (2.2 kb), and placenta (1.8 kb) (Figure 6b). One aspect worthy of comment was the absence of clear E17E18 and INS50 signals in brain tissue, despite the fact that the cDNA clones considered in this and previous work were amplified by PCR from a human brain cDNA library (Section 2.1). This may be explained by an insufficient sensitivity of the northern assay for the relatively low abundance transcripts captured by PCR cloning. Nonetheless, the sensitivity of the northern blot was clearly enough to make conclusions concerning different patterns and relative levels of expression. Particularly noteworthy was the difference between the heart and liver samples. In addition to the different size of the predominant transcripts in each source, the detection of INS50 but not E17E18 signals in the heart makes a strong case for the preferential in vivo use of the ‘alternative’ 3′-splice site of TKFC intron 17, which is contrary to the situation in the liver.
3.4. Tissue Expression of TKFC-Gene Transcripts Quantified by RNA-Seq
Tissue expression data for individual transcript variants, quantified from RNA-Seq experiments using the RSEM software (RNA-Seq by Expectation Maximization [36]), can be found in the GTEx database [35]. Concerning the TKFC gene, besides global expression data from 54 human tissue locations, individual data for 21 Ensembl transcript variants were recorded (https://gtexportal.org/home/gene/TKFC, accessed on 25 August 2024). Out of these, only two transcript variants are clearly related to those deposited in GenBank (Figure 1) and to the two cDNAs from where protein isoforms X6 [1,2,28] and X8 (this work) have been expressed. ENST00000394900.7 is a full-length 4,678 nt transcript, 100% identical to GenBank sequence NM_015533.4 (Tv-2 in Figure 1), which codes for enzymatically active TKFC (protein isoform b or T185A-X6). ENST00000529479.5 is a 1781 nt transcript identical to GenBank sequence DQ344550 (which codes for the inactive TKFC isoform X8), except for three SNPs (with two amino acid substitutions) and for the absence of the initiation codon in the Ensembl transcript. Therefore, ENST00000394900.7 should hybridize with oligonucleotide E17E18 and ENST00000529479.5 with INS50. GTEx data for these two transcripts (Table 2) indicated a 3.3-fold stronger expression of the latter in the heart, in relative agreement with the results of the northern assay (Figure 6a,b). Nonetheless, the same was observed in the GTEx database for the other tissues included in the northern assay, with 1.6–3.9-fold stronger expressions of the transcript encoding the X8-like isoform (Table 2).

Table 2.
Expression data of Ensembl TKFC transcripts theoretically hybridizing with the probes INS50 or E17E18 according to the GTEx database (https://gtexportal.org/home/gene/TKFC; accessed on 25 August 2024).
An intriguing observation in the GTEx database was that the global expression of the TKFC gene in many tissues, including the small intestine, liver, and kidney (tissues showing a strong expression) was >90% attributable to 19 short transcripts unrelated to those coding for isoforms X6 or X8 (see https://gtexportal.org/home/gene/TKFC, accessed on 25 August 2024). Among these short transcripts, the high expression is particularly noticeable of ENST00000525366.5 (Table 2), a non-coding RNA that includes partly exons 14–18 of the TKFC gene model of Figure 2 which is theoretically able to hybridize with the probe E17E18.
4. Discussion
4.1. Possible Role of the Inactive TKFC Isoform X8
The short TKFC isoform X8 studied in this manuscript is inactive as triokinase and FMN cyclase. Therefore, it may not play a metabolic role in the fructose pathway or in the synthesis of cFMN. However, since the N domain of isoform X8 is, at least sequentially, like that of isoform X6, interaction with MDA5 and inhibition of MDA5-mediated innate immunity may still be possible. This should be investigated in future work.
In genetic studies, the absence of TKFC catalytic activities in biallelic mutants of isoform X6 has been related to several phenotypes including cataracts and multi system disease, developmental delay, liver dysfunction, microcytic anemia, fatal cardiomyopathy [24], hypotrichosis with loose anagen hairs [23], and an isolated, complex primary immunodeficiency [25]. One could speculate about the possible relation between any of those pathologies and the co-expression of the inactive TKFC isoform X8 with the active isoform X6. Since it is known that active TKFC is a homodimer of isoform X6, one wonders what would occur in the event of the formation of mixed dimers X6/X8. Would they show half the activity of X6 homodimers, or would they be inactive? In other words, X8 could show either incomplete dominance over X6 or act as a dominant negative isoform.
4.2. Comparison of the TKFC Transcripts Recorded in Databases with Those Detected in the Northern Blot
The length of the cloned sequence that contains the ORF coding for TKFC protein isoform X6 (DQ138290) is 1728 bp. This protein isoform is the enzymatic protein characterized as triokinase and FMN cyclase [1,2,28]. Figure 6a displays a major hybridization signal of about 2.2 kb, which is long enough to contain the complete coding sequence of isoform X6. Nevertheless, all the full-length RefSeq transcripts that theoretically code for isoform X6 are 4.6–6.0 kb long (Table 1), including the Ensembl transcript ENST00000394900.7. Therefore, although the major 2.2 kb RNA detected by hybridization to E17E18 must contain exons 17 and 18 spliced together by the removal of intron 17, one has to recall that the northern assay demonstrates hybridization to probes but does not prove the full-length character of the detected transcripts. The 2.2 kb RNA may represent artifactually shortened transcripts or tkfc-type transcripts not recorded in the databases. Also worthy of note is the absence of any northern signal around 0.9 kb (Figure 6a), which would correspond to the highly expressed Ensembl transcript ENST00000525366.5 according to the GTEX database (Table 2).
The cloned sequence that contains the ORF translating to TKFC protein isoform X8 (DQ344550) is 1784 bp long. Isoform X8 is the enzymatically inactive protein described in the current manuscript. Figure 6b shows major hybridization signals of about 4.3 kb and 1.6 kb (heart), 2.2 kb (liver), and 1.8 kb (placenta). All of them, except the 1.6 kb signal, can contain the complete coding sequence of isoform X8. However, only the 4.3 kb signal of the heart sample may correspond to one of the full-length RefSeq transcripts, namely, transcripts 5 or X14, which code for the very similar protein isoforms d and X8, respectively, or transcript 6, which codes for protein isoform e (Table 1, Figure 1). The remaining northern blot signals detected with the INS50 probe, either full-length transcripts or not, may represent artifactually shortened transcripts or tkfc_ins56-type transcripts not recorded in the databases. On the other hand, the 1.8 kb signal detected in the placenta is compatible with Ensembl transcript ENST00000529479.5, but this is unlikely to be full-length, as it lacks the initiation codon and any 5′-UTR. Therefore, this Ensembl transcript is probably part of a full-length one.
4.3. Comparison between tkfc-Type and tkfc_ins56-Type Splices of Intron 17 in Different Human Tissues
The distribution of TKFC transcripts in human tissues, detected by Northern blot with specific probes (Figure 6a,b), did not correlate with the global expression profile of the human TKFC gene analyzed by RNA-Seq in the same set of tissues (Figure 6c and GTEx database). The difference between both approaches is better appreciated in terms of the relative levels of liver:intestine TKFC expression. While the liver sample gave a much stronger E17E18 (or INS50) signal than the intestine, the latter tissue displayed the largest frequency of TKFC-derived tags in the RNASeq profile. In addition, comparisons of the heart with other tissues (e.g., brain, skeletal muscle, colon, thymus, spleen, lung) led to a less contrasted but still clear lack of correlation between Figure 6b,c, since the INS50 signals of the heart were stronger than those in the other tissues, while in the RNASeq profile, all of them displayed similar or higher frequencies of TKFC tags than the heart. The major reason for the lack of correlation between the northern assay and global expression data may be that global TKFC expression, assayed by RNA-Seq, reflects a very high proportion sequence tags derived from short transcripts unrelated to those coding for isoforms X6 or X8.
It must be remarked that the Northern blot of Figure 6a,b and the data available in GTEx (Table 2) for the relative levels of tkfc-type and tkfc_ins56-type splices in different tissues do not correlate with each other. This can be attributed to several factors: (i) differences in the sources of RNA: poly(A)-RNA from one donor in the northern assay and poly(A)-selected RNA from many donors in the RNA-Seq assay of the GTEx database; (ii) uncertainty in the estimation of transcript levels due to difficulties in assigning the RNA-Seq reads to a specific transcript; and (iii) the complexity of the transcript set derived from the TKFC gene, namely, 21 Ensemble transcripts in the GTEx database (https://gtexportal.org/home/gene/TKFC, accessed on 25 August 2024) and 22 recorded in GenBank (Figure 1), with few coincidences between both databases.
Altogether, expression data for the TKFC gene seem to imply that, at least in the small intestine, but possibly also in other tissues, the predominant TKFC spliced transcripts did not hybridize with E17E18 (nor with INS50) and therefore had not undergone tkfc- or tkfc_ins56-type splicing of intron-17. An exception to this is the high expression of the short ENST00000525366.5 transcript detected by RNA-Seq (Table 2) but not by northern assay (Figure 6a). Anyhow, the results support the occurrence of potentially relevant transcript variants of the TKFC gene differentially expressed in human tissues. Further experimental research with more quantitative techniques may give a deeper understanding of this matter.
5. Conclusions
In conclusion: (i) a human brain transcript variant, that contains an alternative 3′-splice of TKFC intron 17, was cloned, sequenced and expressed; (ii) with respect to transcripts coding for active triokinase and FMN cyclase, the new transcript contains a 56-nt insertion between exons 17 and 18 (tkfc_ins56-type transcripts), with an in-frame stop codon; (iii) it encodes a shortened and enzymatically inactive protein identified as TKFC isoform X8, which lacks 50 C-terminal amino acids encoded by TKFC exon 18 and contains instead 9 amino acids encoded by intron 17; (iii) the lack of triokinase activity of isoform X18 was related to the absence of the essential residue Arg543; (iv) the results depict two pathways for TKFC intron 17 splicing: tkfc-type splicing, which leads to the direct junction of exons 17 and 18, and tkfc_ins56-type splicing, which leads to an insertion of 56 nt between exons 17 and 18; (v) in a northern assay, in the liver, the tkfc-type splice is strongly predominant, while in the heart, the tkfc_ins56-type splicing is so; (vi) in the small intestine, TKFC transcripts of tkfc- or tkfc_ins56-type were not detected, in contrast to the strong expression of the TKFC gene determined by RNAseq, what supports that a different pathway of splicing is predominant in the intestine; (vii) northern blot data do not correlate well with the levels of transcripts quantified by RNA-Seq as shown in the GTEx database, what can be explained by technical differences between both approaches; (viii) altogether, this study supports the occurrence of potentially relevant variants of human TKFC transcripts differentially expressed in human tissues.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biom14101288/s1, Original images of Figure 3a,b and Figure 6a,b can be found in Supplementary Materials.
Author Contributions
Conceptualization, M.J.C., J.M.R. and J.C.C.; formal analysis, J.C.C.; investigation, M.J.C., A.C. (Ana Couto), A.C. (Alicia Cabezas), R.M.P., J.M.R. and J.C.C.; writing—original draft preparation, J.C.C.; writing—review and editing, M.J.C. and J.M.R.; visualization, J.M.R.; supervision, J.C.C. and R.M.P.; funding acquisition, J.C.C. and A.C. (Alicia Cabezas). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ministerio de Educación y Ciencia, Spain, and FEDER, grant number BFU2006-00510 (J.C.C.), by the Consejería de Infraestructuras y Desarrollo Tecnológico, Junta de Extremadura, Spain, and FEDER, grant number GRU06031 (J.C.C.), and by Universidad de Extremadura, grant number A7-09 (Alicia Cabezas). A. Co. was supported by a predoctoral fellowship from the Fundação para a Ciência e a Tecnologia, Portugal. The APC was funded by a waiver benefit granted by MDPI.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Cabezas, A.; Costas, M.J.; Pinto, R.M.; Couto, A.; Cameselle, J.C. Identification of human and rat FAD-AMP lyase (cyclic FMN forming) as ATP-dependent dihydroxyacetone kinases. Biochem. Biophys. Res. Commun. 2005, 338, 1682–1689. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.R.; Couto, A.; Cabezas, A.; Pinto, R.M.; Ribeiro, J.M.; Canales, J.; Costas, M.J.; Cameselle, J.C. Bifunctional homodimeric triokinase/FMN cyclase: Contribution of protein domains to the activities of the human enzyme and molecular dynamics simulation of domain movements. J. Biol. Chem. 2014, 289, 10620–10636. [Google Scholar] [CrossRef] [PubMed]
- Hers, H.G.; Kusaka, T. Le metabolisme du fructose-1-phosphate dans le foie. Biochim. Biophys. Acta 1953, 11, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Sillero, M.A.G.; Sillero, A.; Sols, A. Enzymes involved in fructose metabolism in liver and the glyceraldehyde metabolic crossroads. Eur. J. Biochem. 1969, 10, 345–350. [Google Scholar] [CrossRef]
- Herman, M.A.; Birnbaum, M.J. Molecular aspects of fructose metabolism and metabolic disease. Cell Metab. 2021, 33, 2329–2354. [Google Scholar] [CrossRef]
- Hannou, S.A.; Haslam, D.E.; McKeown, N.M.; Herman, M.A. Fructose metabolism and metabolic disease. J. Clin. Investig. 2018, 128, 545–555. [Google Scholar] [CrossRef]
- Hernandez, A.; Hedlich-Dwyer, J.; Hussain, S.; Levi, H.; Sonavane, M.; Suzuki, T.; Kamiya, H.; Gassman, N.R. Acute exposure to dihydroxyacetone promotes genotoxicity and chromosomal instability in lung, cardiac, and liver cell models. Toxicol. Sci. 2024, 201, 85–102. [Google Scholar] [CrossRef]
- Molin, M.; Norbeck, J.; Blomberg, A. Dihydroxyacetone kinases in Saccharomyces cerevisiae are involved in detoxification of dihydroxyacetone. J. Biol. Chem. 2003, 278, 1415–1423. [Google Scholar] [CrossRef]
- Mehta, R.; Sonavane, M.; Migaud, M.E.; Gassman, N.R. Exogenous exposure to dihydroxyacetone mimics high fructose induced oxidative stress and mitochondrial dysfunction. Environ. Mol. Mutagen. 2021, 62, 185–202. [Google Scholar] [CrossRef]
- Beutler, E.; Guinto, E. Dihydroxyacetone metabolism by human erythrocytes: Demonstration of triokinase activity and its characterization. Blood 1973, 41, 559–568. [Google Scholar] [CrossRef]
- Liu, L.; Li, T.; Liao, Y.; Wang, Y.; Gao, Y.; Hu, H.; Huang, H.; Wu, F.; Chen, Y.G.; Xu, S.; et al. Triose kinase controls the lipogenic potential of fructose and dietary tolerance. Cell Metab. 2020, 32, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Fraiz, F.J.; Pinto, R.M.; Costas, M.J.; Avalos, M.; Canales, J.; Cabezas, A.; Cameselle, J.C. Enzymic formation of riboflavin 4′,5′-cyclic phosphate from FAD: Evidence for a specific low-Km FMN cyclase in rat liver. Biochem. J. 1998, 330, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Cabezas, A.; Pinto, R.M.; Fraiz, F.; Canales, J.; González-Santiago, S.; Cameselle, J.C. Purification, characterization, and substrate and inhibitor structure-activity studies of rat liver FAD-AMP lyase (cyclizing): Preference for FAD and specificity for splitting ribonucleoside diphosphate-X into ribonucleotide and a five-atom cyclic phosphodiester of X, either a monocyclic compound or a cis-bicyclic phosphodiester-pyranose fusion. Biochemistry 2001, 40, 13710–13722. [Google Scholar] [CrossRef] [PubMed]
- Canales, J.; Cabezas, A.; Pinto, R.M.; Cameselle, J.C. Fluorimetric HPLC detection of endogenous riboflavin 4′,5′-cyclic phosphate in rat liver at nanomolar concentrations. Anal. Biochem. 2005, 341, 214–219. [Google Scholar] [CrossRef]
- Yamano, K.; Saito, H.; Ogasawara, Y.; Fujii, S.; Yamada, H.; Shirahama, H.; Kawai, H. The autofluorescent substance in the posterior flagellum of swarmers of the brown alga Scytosiphon lomentaria. Z. Naturforschung C 1996, 51, 155–159. [Google Scholar] [CrossRef]
- Diao, F.; Li, S.; Tian, Y.; Zhang, M.; Xu, L.G.; Zhang, Y.; Wang, R.P.; Chen, D.; Zhai, Z.; Zhong, B.; et al. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc. Natl. Acad. Sci. USA 2007, 104, 11706–11711. [Google Scholar] [CrossRef]
- Liao, G.; Liu, J.; Yin, L.; He, Y.; Qiao, G.; Song, W.; He, Y.; Deng, Z.; Xiao, J.; Feng, H. DAK inhibits MDA5-mediated signaling in the antiviral innate immunity of black carp. Dev. Comp. Immunol. 2022, 126, 104255. [Google Scholar] [CrossRef]
- Mu, M.; Niu, W.; Chu, F.; Dong, Q.; Hu, S.; Niu, C. CircSOBP suppresses the progression of glioma by disrupting glycolysis and promoting the MDA5-mediated immune response. iScience 2023, 26, 107897. [Google Scholar] [CrossRef]
- Han, J.; Chu, Q.; Huo, R.; Xu, T. Inducible microRNA-122 modulates RIG-I signaling pathway via targeting DAK in miiuy croaker after poly(I:C) stimulation. Dev. Comp. Immunol. 2018, 78, 52–60. [Google Scholar] [CrossRef]
- Lee, H.J.; Cha, J.Y. Recent insights into the role of ChREBP in intestinal fructose absorption and metabolism. BMB Rep. 2018, 51, 429–436. [Google Scholar] [CrossRef]
- Tsukamoto, R.; Watanabe, K.; Kodaka, M.; Iwase, M.; Sakiyama, H.; Inoue, Y.; Suzuki, T.; Yamamoto, Y.; Shimizu, M.; Sato, R.; et al. HNF4α is required for Tkfc promoter activation by ChREBP. Biosci. Biotechnol. Biochem. 2024, 88, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, K. Recent Progress on Fructose Metabolism-Chrebp, Fructolysis, and Polyol Pathway. Nutrients 2023, 15, 1778. [Google Scholar] [CrossRef] [PubMed]
- Onoufriadis, A.; Cabezas, A.; Ng, J.C.F.; Canales, J.; Costas, M.J.; Ribeiro, J.M.; Rodrigues, J.R.; McAleer, M.A.; Castelo-Soccio, L.; Simpson, M.A.; et al. Autosomal recessive hypotrichosis with loose anagen hairs associated with TKFC mutations. Br. J. Dermatol. 2021, 184, 935–943. [Google Scholar] [CrossRef] [PubMed]
- Wortmann, S.B.; Meunier, B.; Mestek-Boukhibar, L.; van den Broek, F.; Maldonado, E.M.; Clement, E.; Weghuber, D.; Spenger, J.; Jaros, Z.; Taha, F.; et al. Bi-allelic variants in TKFC encoding triokinase/FMN cyclase are associated with cataracts and multisystem disease. Am. J. Hum. Genet. 2020, 106, 256–263. [Google Scholar] [CrossRef]
- Tremblay-Laganière, C.; Michaud, C.; Abourjaili-Bilodeau, R.; Cabezas, A.; Canales, J.; Costas, M.J.; Ribeiro, J.M.; Leclerc-Blain, J.; Touzot, F.; Haddad, E.; et al. Homozygous variant in TKFC abolishing triokinase activities is associated with isolated immunodeficiency. J. Med. Genet. 2024, 61, 886–890. [Google Scholar] [CrossRef]
- Ashaat, E.A.; Esmaiel, N.N.; El-Saiedi, S.A.; Ashaat, N.A.; Hussen, D.F.; Ramadan, A.; Al Kersh, M.A.; AbdelHakim, N.S.; Said, I.; Metwally, A.M.; et al. Biallelic TYR and TKFC variants in Egyptian patients with OCA1 and new expanded TKFC features. BMC Genom. 2024, 25, 844. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.; Pujar, S.; Loveland, J.E.; Astashyn, A.; Bennett, R.; Berry, A.; Cox, E.; Davidson, C.; Ermolaeva, O.; Farrell, C.M.; et al. A joint NCBI and EMBL-EBI transcript set for clinical genomics and research. Nature 2022, 604, 310–315. [Google Scholar] [CrossRef]
- Ribeiro, J.M.; Costas, M.J.; Cabezas, A.; Meunier, B.; Onoufriadis, A.; Cameselle, J.C. The TKFC Ala185Thr variant, reported as ‘null’ for fructose metabolism, is fully active as triokinase. FEBS Lett. 2022, 596, 1453–1457. [Google Scholar] [CrossRef]
- Engler-Blum, G.; Meier, M.; Frank, J.; Muller, G.A. Reduction of background problems in nonradioactive Northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal. Biochem. 1993, 210, 235–244. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Maquat, L.E. Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. 2016, 129, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; McDonald, L.; Cui, Q.; Matte, A.; Cygler, M.; Ekiel, I. Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase. Proc. Natl. Acad. Sci. USA 2011, 108, 1302–1307. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).