Identification of Six Novel Proteins Containing a ZP Module from Nemertean Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens Collection, Gametes Collection, and Fertilization
2.2. RNA Extraction and cDNA Synthesis
2.3. cDNA Representational Difference Analysis
2.4. Searching Genes Coding a ZP-Module in Transcripts
2.5. In Situ Hybridization
2.6. In Vitro mRNA Synthesis and Microinjection
3. Results
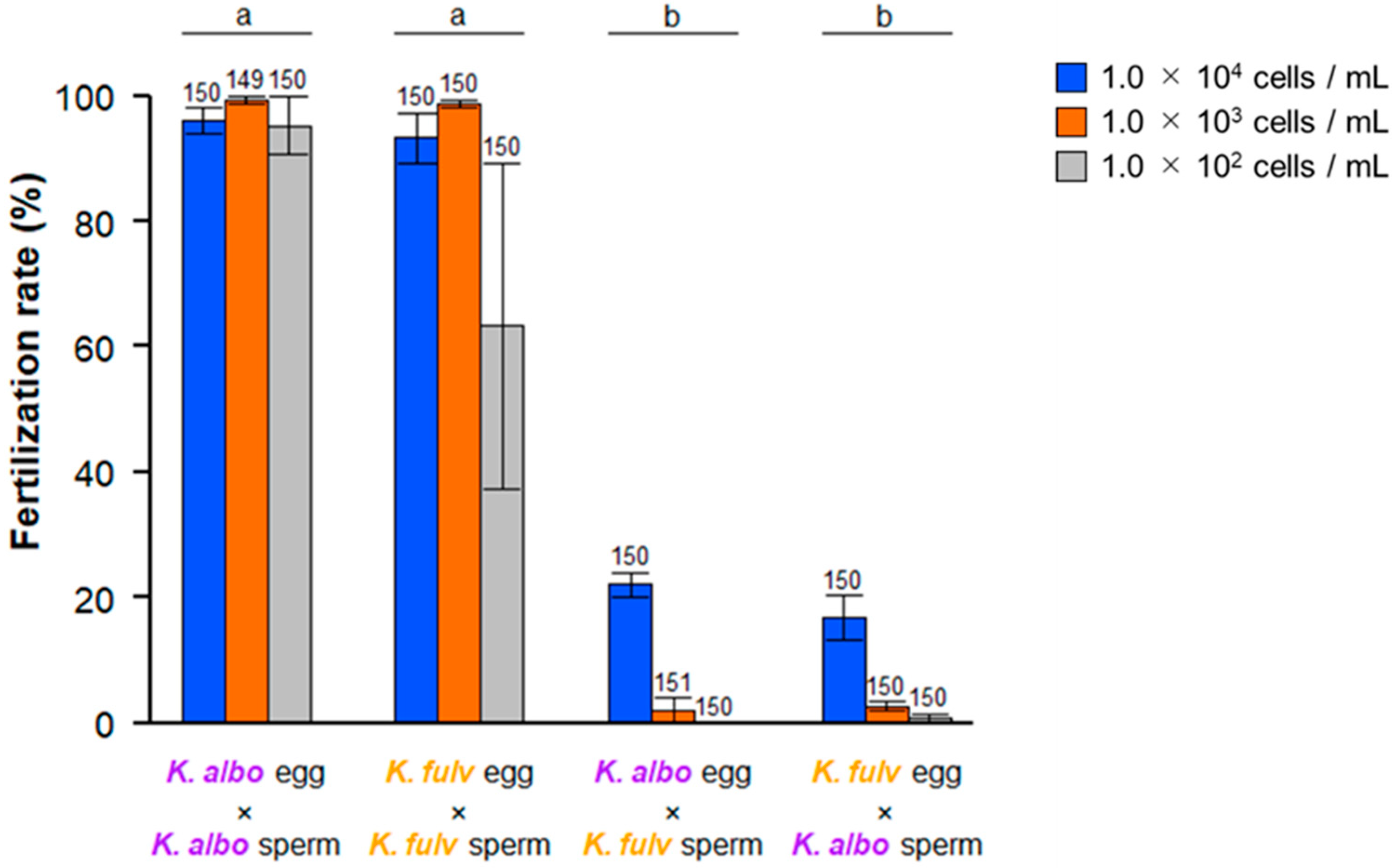
3.1. Cross-Fertilization Ability Between K. alborostrata and K. fulva
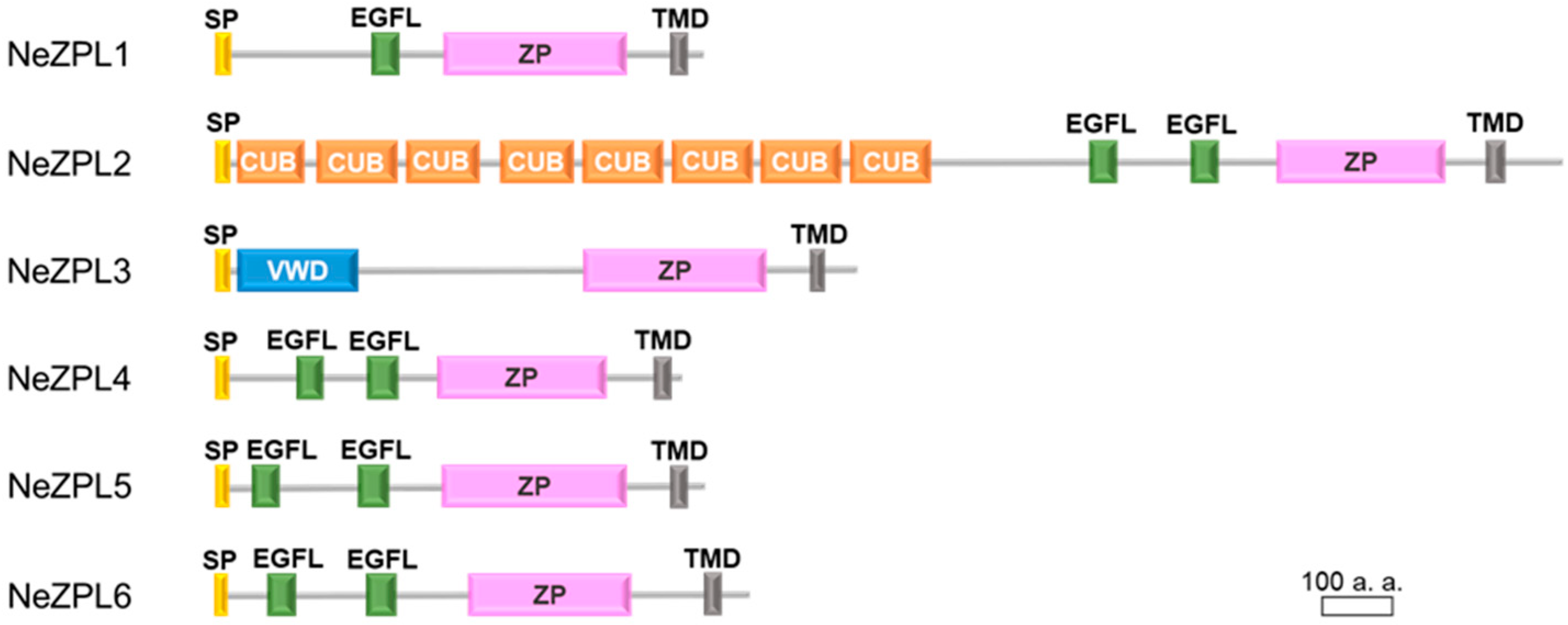
3.2. Six Novel ZP Module-Containing Proteins in K. alborostrata and K. fulva
3.3. Comparison Between Orthologs of NeZPL Genes
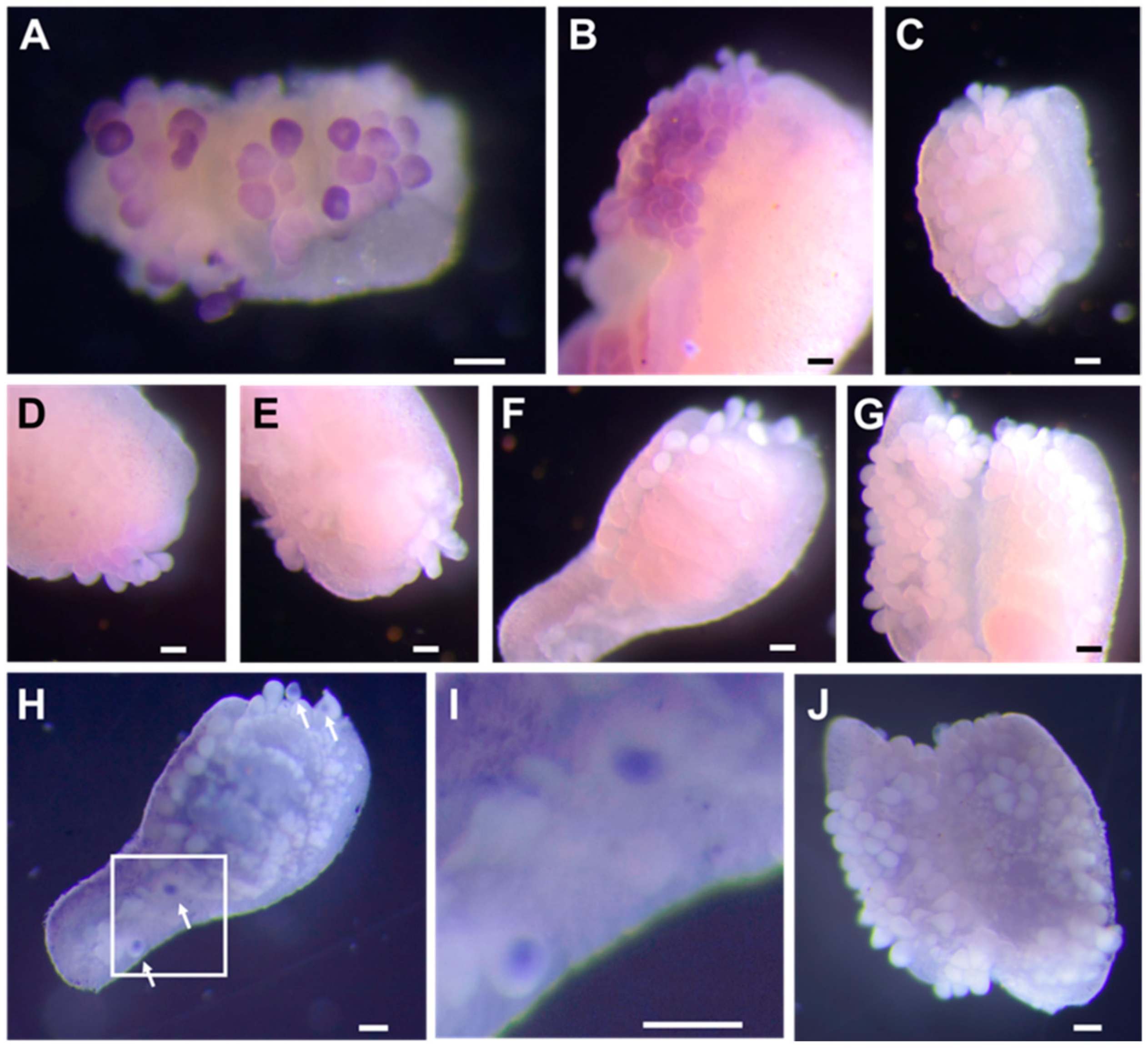
3.4. Expression of NeZPL1–NeZPL6 in Oocyte
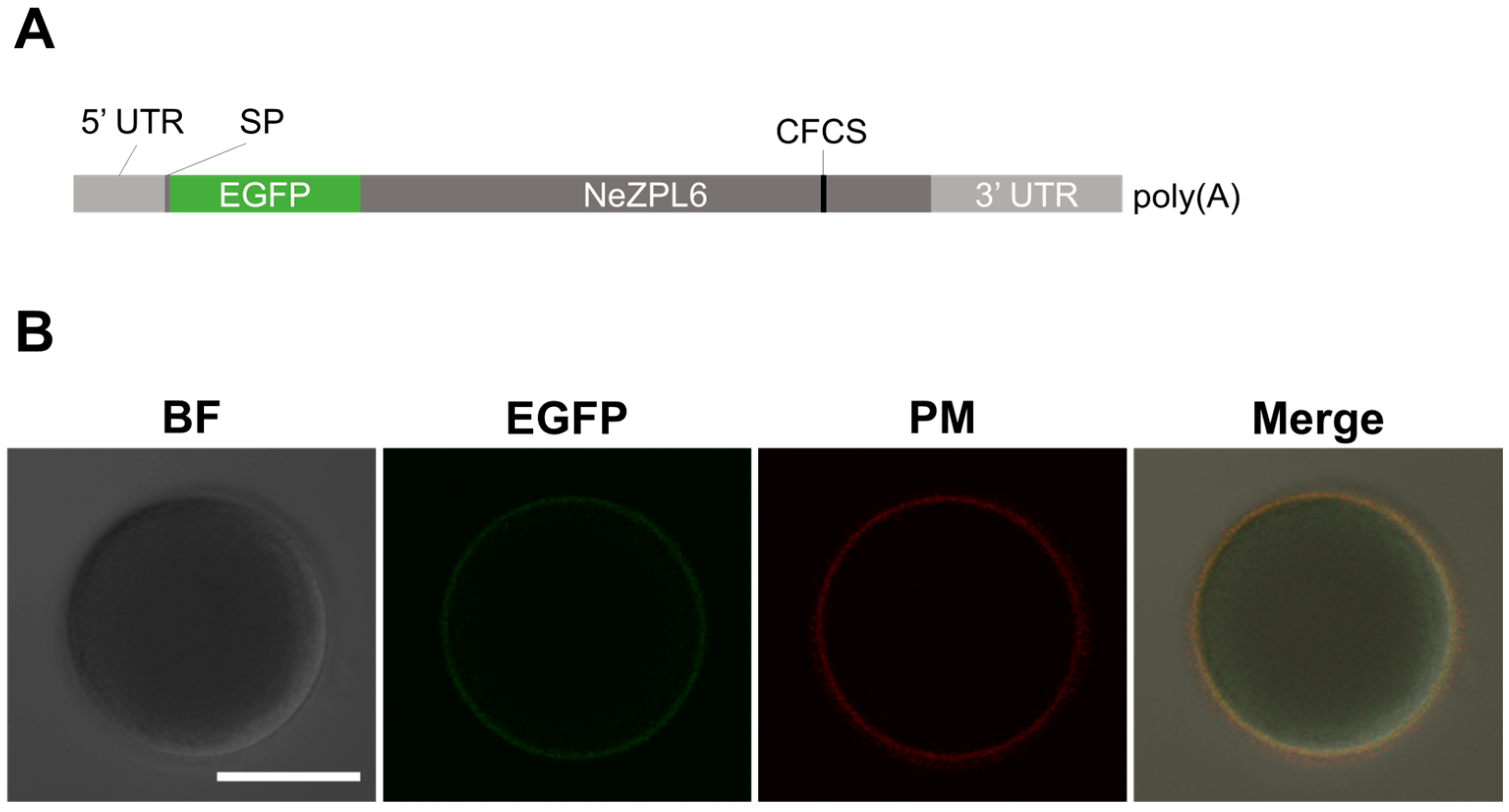
3.5. Localization of NeZPL6 on the Plasma Membrane of Egg
4. Discussion
4.1. Proteins with Both ZP Modules in Other Lophotrochozoans
4.2. Role of NeZPL Proteins on Fertilization
4.3. Effect of the Indelin NeZPL6 on Interaction Between Protein
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gert, K.R.; Pauli, A. Species-specific mechanisms during fertilization. Curr. Top. Dev. Biol. 2020, 140, 121–144. [Google Scholar] [CrossRef] [PubMed]
- Jovine, L.; Qi, H.; Williams, Z.; Litscher, E.S.; Wassarman, P.M. A duplicated motif controls assembly of zona pellucida domain proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 5922–5927. [Google Scholar] [CrossRef] [PubMed]
- Jovine, L.; Qi, H.; Williams, Z.; Litscher, E.S.; Wassarman, P.M. The ZP domain is a conserved module for protein polymerization. Nat. Cell Biol. 2002, 4, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Stanisich, J.J.; Zyla, D.S.; Afanasyev, P.; Xu, J.; Kipp, A.; Olinger, E.; Devuyst, O.; Pilhofer, M.; Boehringer, D.; Glockshuber, R. The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism. eLife 2020, 9, e60265. [Google Scholar] [CrossRef]
- Stsiapanava, A.; Xu, C.; Brunati, M.; Zamora-Caballero, S.; Schaeffer, C.; Bokhove, M.; Han, L.; Hebert, H.; Carroni, M.; Yasumasu, S.; et al. Cryo-EM structure of native human uromodulin, a zona pellucida module polymer. EMBO J. 2020, 39, e106807. [Google Scholar] [CrossRef]
- Nishio, S.; Emori, C.; Wiseman, B.; Fahrenkamp, D.; Dioguardi, E.; Zamora-Caballero, S.; Bokhove, M.; Han, L.; Stsiapanava, A.; Algarra, B.; et al. ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat. Cell 2024, 187, 1440–1459.e24. [Google Scholar] [CrossRef] [PubMed]
- Litscher, E.S.; Qi, H.; Wassarman, P.M. Mouse zona pellucida glycoproteins mZP2 and mZP3 undergo carboxy-terminal proteolytic processing in growing oocytes. Biochemistry 1999, 38, 12280–12287. [Google Scholar] [CrossRef]
- Williams, Z.; Wassarman, P.M. Secretion of mouse ZP3, the sperm receptor, requires cleavage of its polypeptide at a consensus furin cleavage-site. Biochemistry 2001, 40, 929–937. [Google Scholar] [CrossRef]
- Bokhove, M.; Jovine, L. Structure of zona pellucida module proteins. Curr. Top. Dev. Biol. 2018, 130, 413–442. [Google Scholar] [CrossRef]
- Avella, M.A.; Baibakov, B.; Dean, J. A single domain of the ZP2 zona pellucida protein mediates gamete recognition in mice and humans. J. Cell Biol. 2014, 205, 801–809. [Google Scholar] [CrossRef]
- Raj, I.; Sadat Al Hosseini, H.; Dioguardi, E.; Nishimura, K.; Han, L.; Villa, A.; de Sanctis, D.; Jovine, L. Structural basis of egg coat-sperm recognition at fertilization. Cell 2017, 169, 1315–1326.e17. [Google Scholar] [CrossRef] [PubMed]
- Galindo, B.E.; Moy, G.W.; Swanson, W.J.; Vacquier, V.D. Full-length sequence of VERL, the egg vitelline envelope receptor for abalone sperm lysin. Gene 2002, 288, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.-J.; Kanda, M.; Koyanagi, R.; Hisata, K.; Akiyama, T.; Sakamoto, H.; Sakamoto, T.; Satoh, N. Nemertean and phoronid genomes reveal lophotrochozoan evolution and the origin of bilaterian heads. Nat. Ecol. Evol. 2018, 2, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Ikenaga, J.; Kajihara, H.; Yoshida, M. Kulikovia alborostrata and Kulikovia fulva comb. nov. (Nemertea: Heteronemertea) are sister species with prezygotic isolating barriers. Zool. Sci. 2021, 38, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Hubank, M.; Schatz, D.G. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994, 22, 5640–5648. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Priyam, A.; Woodcroft, B.J.; Rai, V.; Moghul, I.; Munagala, A.; Ter, F.; Chowdhary, H.; Pieniak, I.; Maynard, L.J.; Gibbins, M.A.; et al. Sequenceserver: A modern graphical user interface for custom BLAST databases. Mol. Biol. Evol. 2019, 36, 2922–2924. [Google Scholar] [CrossRef]
- Ikuta, T.; Yoshida, N.; Satoh, N.; Saiga, H. Ciona intestinalis Hox gene cluster: Its dispersed structure and residual colinear expression in development. Proc. Natl. Acad. Sci. USA 2004, 101, 15118–15123. [Google Scholar] [CrossRef]
- Stricker, S.A.; Smythe, T.L. Multiple triggers of oocyte maturation in nemertean worms: The roles of calcium and serotonin. J. Exp. Zool. 2000, 287, 243–261. [Google Scholar] [CrossRef]
- Shimizu, K.; Takeuchi, T.; Negishi, L.; Kurumizaka, H.; Kuriyama, I.; Endo, K.; Suzuki, M. Evolution of epidermal growth factor (EGF)-like and zona pellucida domains containing shell matrix proteins in mollusks. Mol. Biol. Evol. 2022, 39, msac148. [Google Scholar] [CrossRef]
- Marie, B.; Zanella-Cléon, I.; Corneillat, M.; Becchi, M.; Alcaraz, G.; Plasseraud, L.; Luquet, G.; Marin, F. Nautilin-63, a novel acidic glycoprotein from the shell nacre of Nautilus macromphalus. FEBS J. 2011, 278, 2117–2130. [Google Scholar] [CrossRef] [PubMed]
- Marie, B.; Jackson, D.J.; Ramos-Silva, P.; Zanella-Cléon, I.; Guichard, N.; Marin, F. The shell-forming proteome of Lottia gigantea reveals both deep conservations and lineage-specific novelties. FEBS J. 2013, 280, 214–232. [Google Scholar] [CrossRef] [PubMed]
- Bork, P.; Beckmann, G. The CUB domain: A widespread module in developmentally regulated proteins. J. Mol. Biol. 1993, 231, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Carrera, E.; Sanz, L.; Töpfer-Petersen, E. Boar spermadhesins AQN-1 and AQN-3: Oligosaccharide and zona pellucida binding characteristics. Biol. Chem. 1996, 377, 521–527. [Google Scholar] [CrossRef]
- Kamei, N.; Glabe, C.G. The species-specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes Dev. 2003, 17, 2502–2507. [Google Scholar] [CrossRef]
- Lieber, T.; Wesley, C.S.; Alcamo, E.; Hassel, B.; Krane, J.F.; Campos-Ortega, J.A.; Young, M.W. Single amino acid substitutions in EGF-like elements of Notch and Delta modify Drosophila development and affect cell adhesion in vitro. Neuron 1992, 9, 847–859. [Google Scholar] [CrossRef]
- Wagner, G.; Wyss, D.F. Cell surface adhesion receptors. Curr. Opin. Struct. Biol. 1994, 4, 841–851. [Google Scholar] [CrossRef]
- Singson, A.; Mercer, K.B.; L’Hernault, S.W.; The, C. The C. Elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell 1998, 93, 71–79. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Krauchunas, A.R.; Marcello, M.R.; Looper, A.; Mei, X.; Putiri, E.; Singaravelu, G.; Ahmed, I.I.; Singson, A. The EGF-motif-containing protein SPE-36 is a secreted sperm protein required for fertilization in C. elegans. Curr. Biol. 2023, 33, 3056–3064.e5. [Google Scholar] [CrossRef]
- Yamamoto, S.; Charng, W.L.; Rana, N.A.; Kakuda, S.; Jaiswal, M.; Bayat, V.; Xiong, B.; Zhang, K.; Sandoval, H.; David, G.; et al. A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science 2012, 338, 1229–1232. [Google Scholar] [CrossRef]
- Zigler, K.S.; McCartney, M.A.; Levitan, D.R.; Lessios, H.A. Sea urchin bindin divergence predicts gamete compatibility. Evolution 2005, 59, 2399–2404. [Google Scholar] [CrossRef] [PubMed]
- Galindo, B.E.; Vacquier, V.D.; Swanson, W.J. Positive selection in the egg receptor for abalone sperm lysin. Proc. Natl. Acad. Sci. USA 2003, 100, 4639–4643. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Kitanobo, S.; Ohki, S.; Shiba, K.; Inaba, K. Positive selection on ADAM10 builds species recognition in the synchronous spawning coral Acropora. Front. Cell Dev. Biol. 2023, 11, 1171495. [Google Scholar] [CrossRef] [PubMed]
- Gert, K.R.; Panser, K.; Surm, J.; Steinmetz, B.S.; Schleiffer, A.; Jovine, L.; Moran, Y.; Kondrashov, F.; Pauli, A. Divergent molecular signatures in fish Bouncer proteins define cross-fertilization boundaries. Nat. Commun. 2023, 14, 3506. [Google Scholar] [CrossRef] [PubMed]
- Savino, S.; Desmet, T.; Franceus, J. Insertions and deletions in protein evolution and engineering. Biotechnol. Adv. 2022, 60, 108010. [Google Scholar] [CrossRef]
- Sawada, H.; Tanaka, E.; Ban, S.; Yamasaki, C.; Fujino, J.; Ooura, K.; Abe, Y.; Matsumoto, K.; Yokosawa, H. Self/nonself recognition in ascidian fertilization: Vitelline coat protein HrVC70 is a candidate allorecognition molecule. Proc. Natl. Acad. Sci. USA 2004, 101, 15615–15620. [Google Scholar] [CrossRef]
- Ban, S.; Harada, Y.; Yokosawa, H.; Sawada, H. Highly polymorphic vitelline-coat protein HaVC80 from the ascidian, Halocynthia aurantium: Structural analysis and involvement in self/nonself recognition during fertilization. Dev. Biol. 2005, 286, 440–451. [Google Scholar] [CrossRef][Green Version]
- Harada, Y.; Sawada, H. Proteins interacting with the ascidian vitelline-coat sperm receptor HrVC70 as revealed by yeast two-hybrid screening. Mol. Reprod. Dev. 2007, 74, 1178–1187. [Google Scholar] [CrossRef]

| Predicted Gene Name | Number of Clones (60 Clones in Total) |
|---|---|
| vitellogenin | 52 |
| abhydrolase | 4 |
| cyclin | 2 |
| ZP-module containing protein | 1 |
| poly-ubiquitin | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikenaga, J.; Yoshida, K.; Yoshida, M. Identification of Six Novel Proteins Containing a ZP Module from Nemertean Species. Biomolecules 2024, 14, 1545. https://doi.org/10.3390/biom14121545
Ikenaga J, Yoshida K, Yoshida M. Identification of Six Novel Proteins Containing a ZP Module from Nemertean Species. Biomolecules. 2024; 14(12):1545. https://doi.org/10.3390/biom14121545
Chicago/Turabian StyleIkenaga, Jumpei, Kaoru Yoshida, and Manabu Yoshida. 2024. "Identification of Six Novel Proteins Containing a ZP Module from Nemertean Species" Biomolecules 14, no. 12: 1545. https://doi.org/10.3390/biom14121545
APA StyleIkenaga, J., Yoshida, K., & Yoshida, M. (2024). Identification of Six Novel Proteins Containing a ZP Module from Nemertean Species. Biomolecules, 14(12), 1545. https://doi.org/10.3390/biom14121545






