Diversity of Endolysin Domain Architectures in Bacteriophages Infecting Bacilli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Database of Bacillus Phage Genomes
2.2. Database of Bacillus Phage Endolysins
2.3. Classification of Endolysins by Their EAD Types
3. Results
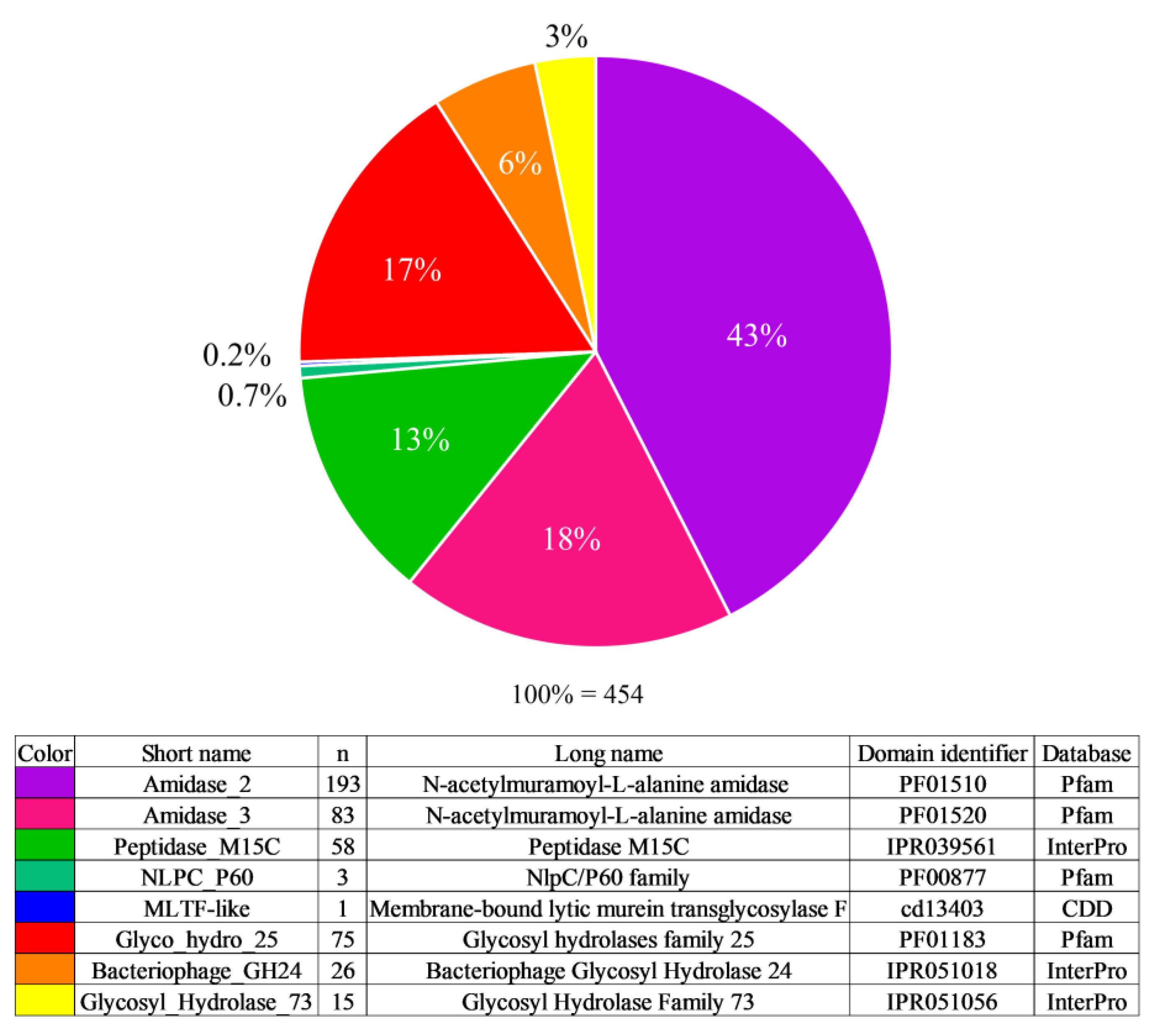
3.1. Enzyme Active Domain Types
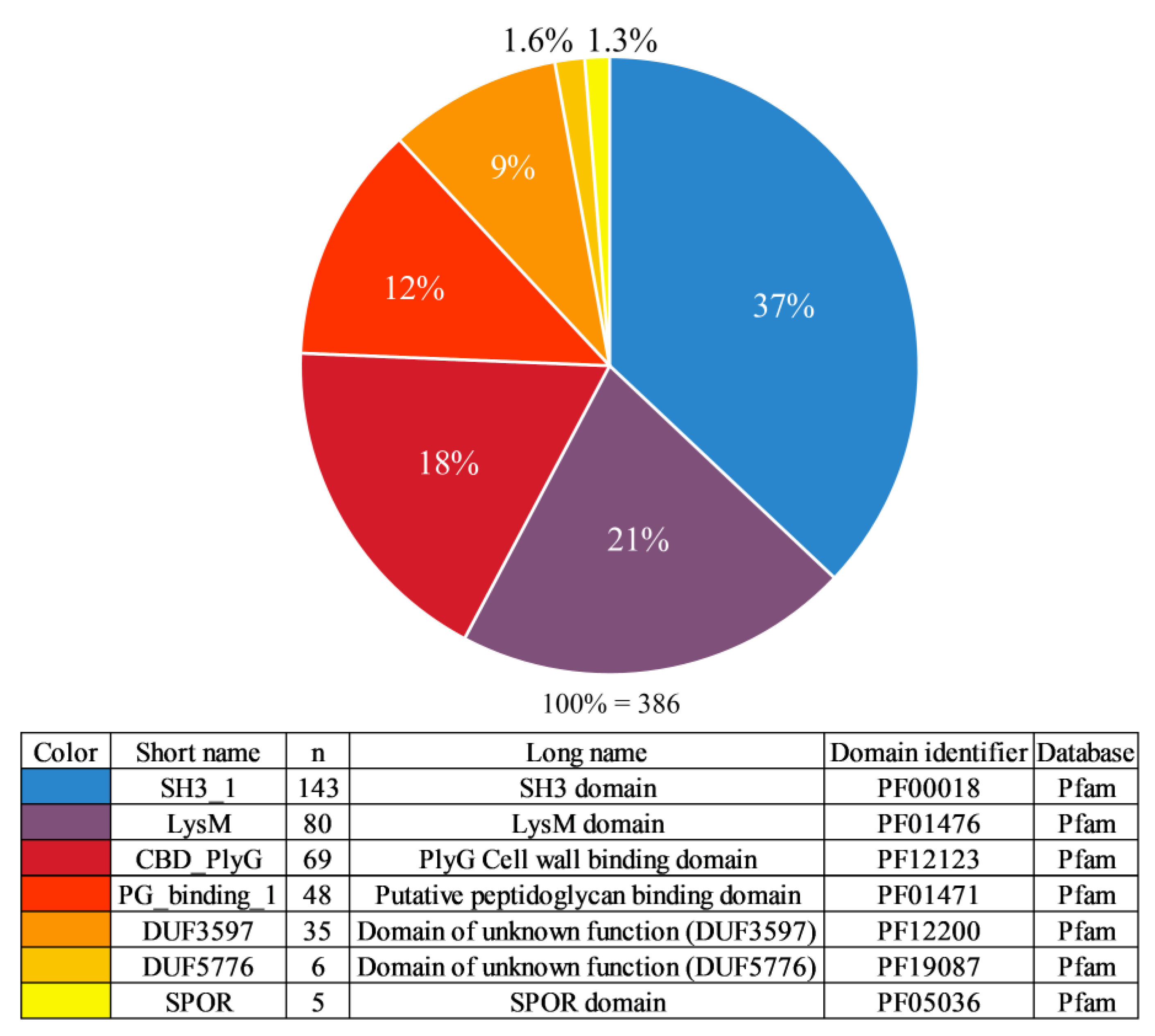
3.2. Cell Wall Binding Domain Types
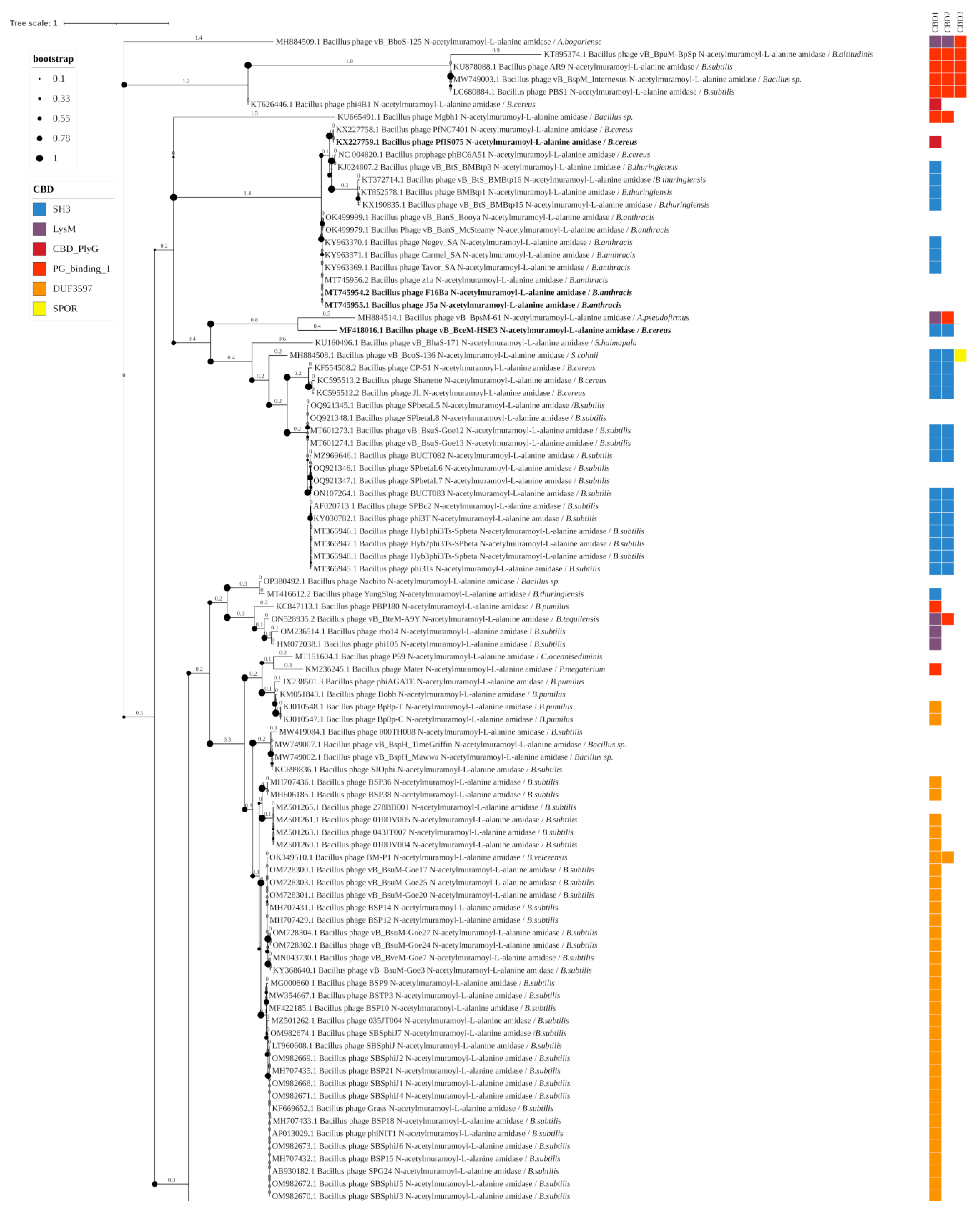
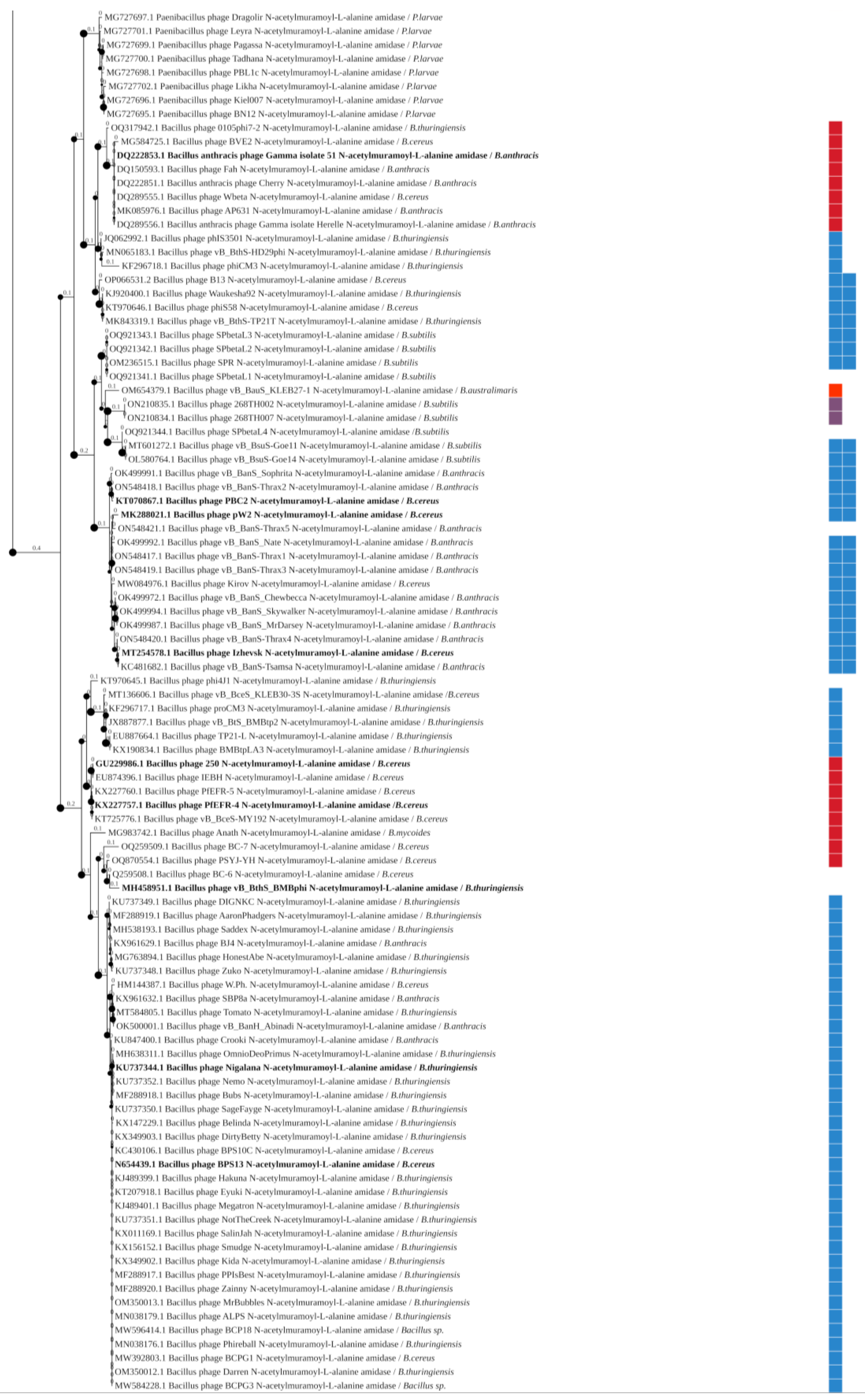
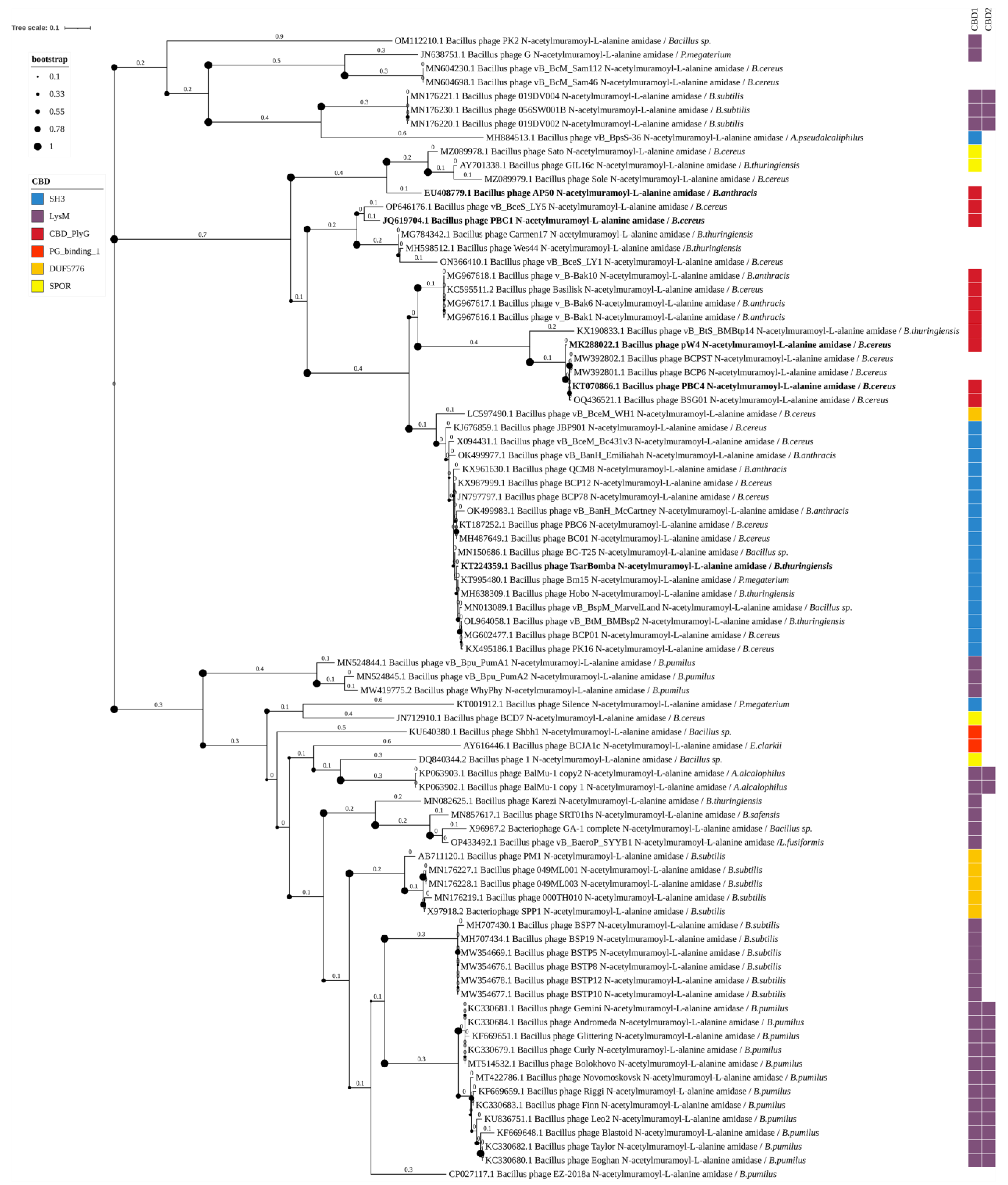
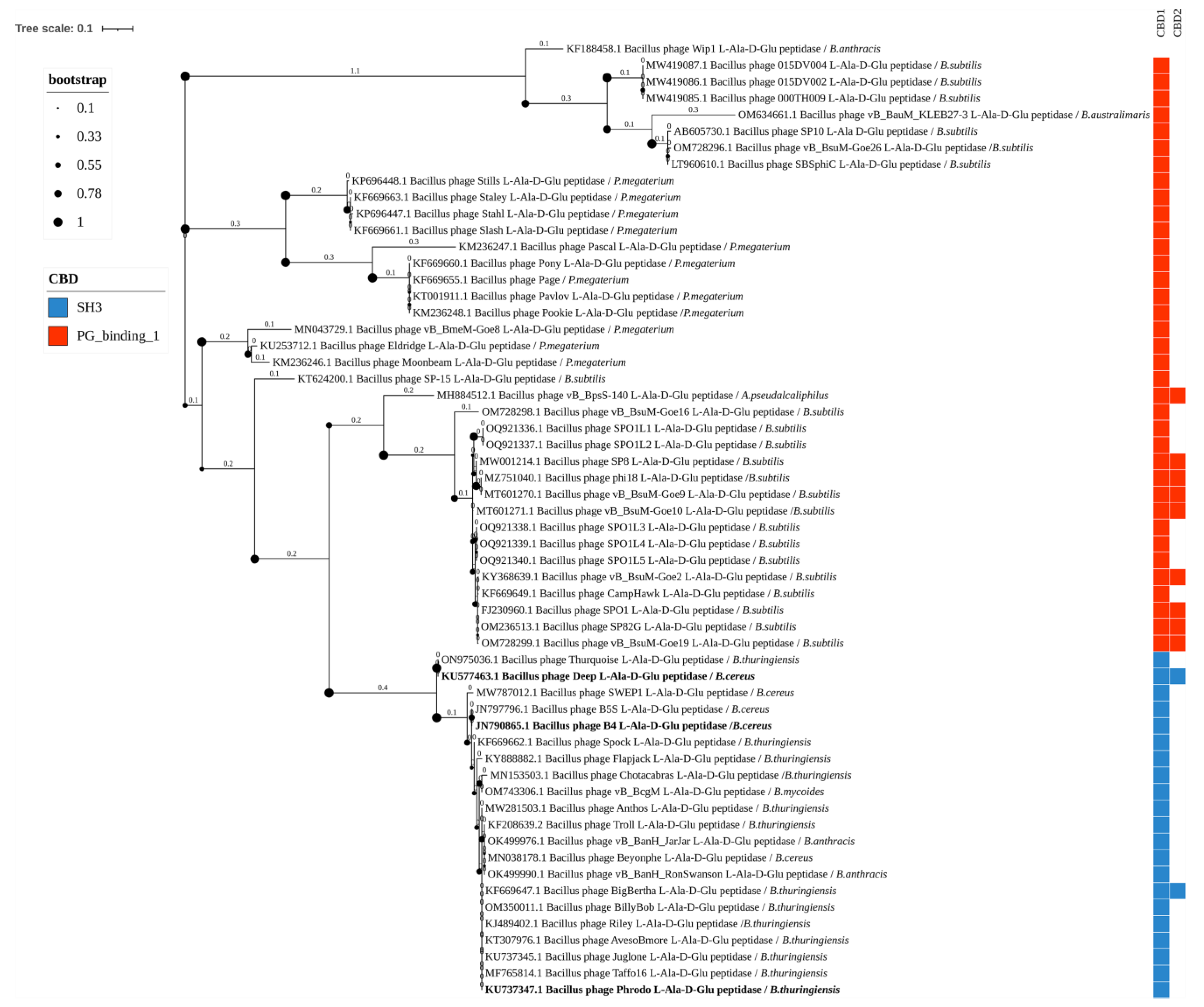
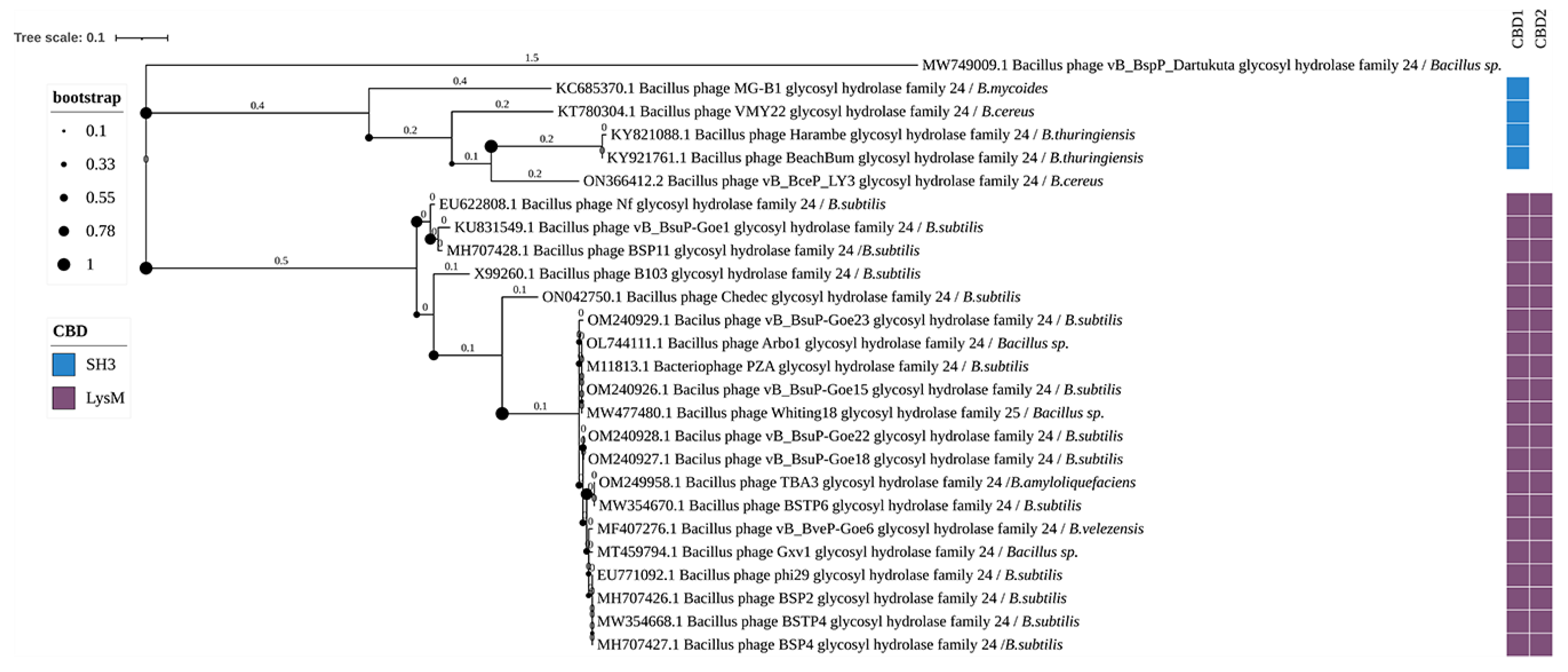
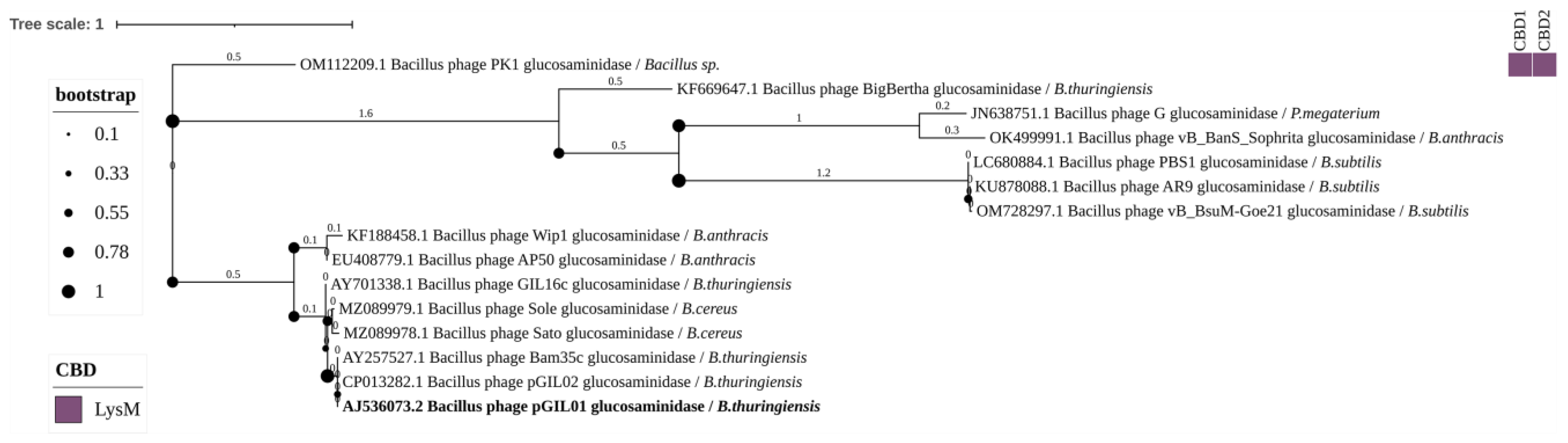
3.3. Phylogenetic Analysis
4. Discussion
4.1. N-Acetylmuramoyl-L-Alanine Amidase
4.1.1. Amidase_2 Domain-Containing Endolysins of Bacillus-Infecting Bacteriophages
4.1.2. Amidase_3 Domain-Containing Endolysins of Bacillus-Infecting Bacteriophages
4.2. Peptidases of Bacillus-Infecting Bacteriophages
4.2.1. Peptidases M15
4.2.2. NlpC/P60 Peptidases
4.3. Glycoside Hydrolases
4.3.1. Glycoside Hydrolase Family 24 (GHF24)
4.3.2. Glycoside Hydrolase Family 25 (GHF25)
4.3.3. Glycoside Hydrolase Family 73 (GHF73)
4.4. MLTF Proteins
4.5. CBDs of Endolysins of Bacillus-Infecting Phages
4.5.1. The SH3 Domain
4.5.2. The LysM Domain
4.5.3. The CBD_PlyG Domain
4.5.4. The PG_Binding_1 Domain
4.5.5. The SPOR Domain
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larsson, D.G.J.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Намазoва-Баранoва, Л.С.; Баранoв, А.А. Антибиoтикoрезистентнoсть в сoвременнoм мире. Педиатрическая Фармакoлoгия 2017, 14, 341–354. [Google Scholar]
- Logvin, F.V.; Kondratenko, T.A.; Vodyanitskya, S.Y. Anthrax in the world, CIS and Russian Federation (literature review). Med. Her. South Russ. 2017, 8, 17–22. [Google Scholar] [CrossRef]
- Инфoрмациoннo-аналитический центр управления ветнадзoра ФГБУ «ВНИИЗЖ» Сoстoяние ветеринарных служб субъектoв Рoссийскoй Федерации в 2014 гoду (Анализ инфoрмации вo испoлнение Приказа Рoссельхoзнадзoра oт 20 февраля 2014 г. № 63 «Об инфoрмации o ветеринарных службах субъектoв Рoссийскoй Федерации»). 2015. Available online: https://fsvps.gov.ru/wp-content/uploads/2023/06/vet2014.pdf (accessed on 13 August 2024).
- Ryazanova, A.G.; Skudareva, O.N.; Gerasimenko, D.K.; Logvin, F.V.; Aksenova, L.Y.; Semenova, O.V.; Eremenko, E.I.; Golovinskaya, T.M.; Pechkovsky, G.A.; Kulichenko, A.N. Analysis of the Situation on Anthrax in the World in 2022, the Forecast for the Russian Federation for 2023. Probl. Part. Danger. Infect. 2023, 2, 88–94. [Google Scholar] [CrossRef]
- Carlson, C.J.; Kracalik, I.T.; Ross, N.; Alexander, K.A.; Hugh-Jones, M.E.; Fegan, M.; Elkin, B.T.; Epp, T.; Shury, T.K.; Zhang, W.; et al. The global distribution of Bacillus anthracis and associated anthrax risk to humans, livestock and wildlife. Nat. Microbiol. 2019, 4, 1337–1343. [Google Scholar] [CrossRef] [PubMed]
- Лoбзин, Ю.В.; Вoлжанин, В.М.; Захаренкo, С.М. Сибирская язва. Клиническая Микрoбиoлoгия И Антимикрoбная Химиoтерапия 2002, 4, 104–127. [Google Scholar]
- Veysseyre, F.; Fourcade, C.; Lavigne, J.-P.; Sotto, A. Bacillus cereus infection: 57 case patients and a literature review. Médecine Mal. Infect. 2015, 42, 436–440. [Google Scholar] [CrossRef]
- Glasset, B.; Sperry, M.; Dervyn, R.; Herbin, S.; Brisabois, A.; Ramarao, N. The cytotoxic potential of Bacillus cereus strains of various origins. Food Microbiol. 2021, 98, 103759. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Lereclus, D.; Koehler, T.M. The Bacillus cereus Group: Bacillus Species with Pathogenic Potential. Microbiol. Spectr. 2019, 7, 10–1128. [Google Scholar] [CrossRef]
- Fernández-Ruiz, I.; Coutinho, F.H.; Rodriguez-Valera, F. Thousands of Novel Endolysins Discovered in Uncultured Phage Genomes. Front. Microbiol. 2018, 9, 1033. [Google Scholar] [CrossRef]
- Love, M.J.; Abeysekera, G.S.; Muscroft-Taylor, A.C.; Billington, C.; Dobson, R.C.J. On the catalytic mechanism of bacteriophage endolysins: Opportunities for engineering. Biochim. Biophys. Acta BBA—Proteins Proteom. 2020, 1868, 140302. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F.; Menzi, C.; Moreillon, P.; Resch, G. The multidomain architecture of a bacteriophage endolysin enables intramolecular synergism and regulation of bacterial lysis. J. Biol. Chem. 2021, 296, 100639. [Google Scholar] [CrossRef] [PubMed]
- Criel, B.; Taelman, S.; Van Criekinge, W.; Stock, M.; Briers, Y. PhaLP: A Database for the Study of Phage Lytic Proteins and Their Evolution. Viruses 2021, 13, 1240. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, 5–9. [Google Scholar] [CrossRef]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinforma. 2003, 2, 2.3.1–2.3.22. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Vermassen, A.; Leroy, S.; Talon, R.; Provot, C.; Popowska, M.; Desvaux, M. Cell Wall Hydrolases in Bacteria: Insight on the Diversity of Cell Wall Amidases, Glycosidases and Peptidases Toward Peptidoglycan. Front. Microbiol. 2019, 10, 331. [Google Scholar] [CrossRef] [PubMed]
- Schuch, R.; Nelson, D.; Fischetti, V.A. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 2002, 418, 884–889. [Google Scholar] [CrossRef]
- Fischetti, V. Development of Phage Lysins as Novel Therapeutics: A Historical Perspective. Viruses 2018, 10, 310. [Google Scholar] [CrossRef]
- Yang, H.; Wang, D.-B.; Dong, Q.; Zhang, Z.; Cui, Z.; Deng, J.; Yu, J.; Zhang, X.; Wei, H. Existence of Separate Domains in Lysin PlyG for Recognizing Bacillus anthracis Spores and Vegetative Cells. Antimicrob. Agents Chemother. 2012, 56, 5031–5039. [Google Scholar] [CrossRef]
- Kikkawa, H.S.; Ueda, T.; Suzuki, S.; Yasuda, J. Characterization of the catalytic activity of the γ-phage lysin, PlyG, specific for Bacillus anthracis. FEMS Microbiol. Lett. 2008, 286, 236–240. [Google Scholar] [CrossRef]
- Lee, Y.-D.; Park, J.-H. Genomic sequence of temperate phage 250 isolated from emetic B. cereus and cloning of putative endolysin. Food Sci. Biotechnol. 2010, 19, 1643–1648. [Google Scholar] [CrossRef]
- Geng, P.; Tian, S.; Yuan, Z.; Hu, X. Identification and genomic comparison of temperate bacteriophages derived from emetic Bacillus cereus. PLoS ONE 2017, 12, e0184572. [Google Scholar] [CrossRef]
- Peng, Q.; Yuan, Y. Characterization of a novel phage infecting the pathogenic multidrug-resistant Bacillus cereus and functional analysis of its endolysin. Appl. Microbiol. Biotechnol. 2018, 102, 7901–7912. [Google Scholar] [CrossRef]
- Wan, X.; Geng, P.; Sun, J.; Yuan, Z.; Hu, X. Characterization of two newly isolated bacteriophages PW2 and PW4 and derived endolysins with lysis activity against Bacillus cereus group strains. Virus Res. 2021, 302, 198489. [Google Scholar] [CrossRef] [PubMed]
- Kong, M.; Na, H.; Ha, N.-C.; Ryu, S. LysPBC2, a Novel Endolysin Harboring a Bacillus cereus Spore Binding Domain. Appl. Environ. Microbiol. 2019, 85, e02462-18. [Google Scholar] [CrossRef] [PubMed]
- Skorynina, A.V.; Piligrimova, E.G.; Kazantseva, O.A.; Kulyabin, V.A.; Baicher, S.D.; Ryabova, N.A.; Shadrin, A.M. Bacillus-infecting bacteriophage Izhevsk harbors thermostable endolysin with broad range specificity. PLoS ONE 2020, 15, e0242657. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Yun, J.; Lim, J.-A.; Kang, D.-H.; Ryu, S. Characterization of an endolysin, LysBPS13, from a Bacillus cereus bacteriophage. FEMS Microbiol. Lett. 2012, 332, 76–83. [Google Scholar] [CrossRef]
- Etobayeva, I.; Linden, S.; Alem, F.; Harb, L.; Rizkalla, L.; Mosier, P.; Johnson, A.; Temple, L.; Hakami, R.; Nelson, D. Discovery and Biochemical Characterization of PlyP56, PlyN74, and PlyTB40—Bacillus Specific Endolysins. Viruses 2018, 10, 276. [Google Scholar] [CrossRef]
- Geng, P.; Hu, Y.; Zhou, G.; Yuan, Z.; Hu, X. Characterization of three autolysins with activity against cereulide-producing Bacillus isolates in food matrices. Int. J. Food Microbiol. 2017, 241, 291–297. [Google Scholar] [CrossRef]
- Nakonieczna, A.; Topolska-Woś, A.; Łobocka, M. New bacteriophage-derived lysins, LysJ and LysF, with the potential to control Bacillus anthracis. Appl. Microbiol. Biotechnol. 2024, 108, 76. [Google Scholar] [CrossRef]
- Yuan, Y.; Peng, Q.; Yang, S.; Zhang, S.; Fu, Y.; Wu, Y.; Gao, M. Isolation of A Novel Bacillus thuringiensis Phage Representing A New Phage Lineage and Characterization of Its Endolysin. Viruses 2018, 10, 611. [Google Scholar] [CrossRef]
- Park, S.; Jun, S.Y.; Kim, C.-H.; Jung, G.M.; Son, J.S.; Jeong, S.T.; Yoon, S.J.; Lee, S.Y.; Kang, S.H. Characterisation of the antibacterial properties of the recombinant phage endolysins AP50-31 and LysB4 as potent bactericidal agents against Bacillus anthracis. Sci. Rep. 2018, 8, 18. [Google Scholar] [CrossRef]
- Kong, M.; Ryu, S. Bacteriophage PBC1 and Its Endolysin as an Antimicrobial Agent against Bacillus cereus. Appl. Environ. Microbiol. 2015, 81, 2274–2283. [Google Scholar] [CrossRef]
- Na, H.; Kong, M.; Ryu, S. Characterization of LysPBC4, a novel Bacillus cereus-specific endolysin of bacteriophage PBC4. FEMS Microbiol. Lett. 2016, 363, fnw092. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Bateman, A. Origins of peptidases. Biochimie 2019, 166, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Son, B.; Yun, J.; Lim, J.-A.; Shin, H.; Heu, S.; Ryu, S. Characterization of LysB4, an endolysin from the Bacillus cereus-infecting bacteriophage B4. BMC Microbiol. 2012, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Leprince, A.; Nuytten, M.; Gillis, A.; Mahillon, J. Characterization of PlyB221 and PlyP32, Two Novel Endolysins Encoded by Phages Preying on the Bacillus cereus Group. Viruses 2020, 12, 1052. [Google Scholar] [CrossRef]
- Anantharaman, V.; Aravind, L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003, 4, R11. [Google Scholar] [CrossRef]
- Shrivastava, S. Introduction to Glycoside Hydrolases: Classification, Identification and Occurrence. In Industrial Applications of Glycoside Hydrolases; Shrivastava, S., Ed.; Springer: Singapore, 2020; pp. 3–84. ISBN 9789811547669. [Google Scholar]
- Premetis, G.E.; Stathi, A.; Papageorgiou, A.C.; Labrou, N.E. Characterization of a glycoside hydrolase endolysin from Acinetobacter baumannii phage Ab TZA1 with high antibacterial potency and novel structural features. FEBS J. 2023, 290, 2146–2164. [Google Scholar] [CrossRef]
- Li, N.; Yuan, X.; Li, C.; Chen, N.; Wang, J.; Chen, B.; Yu, S.; Yu, P.; Zhang, J.; Zeng, H.; et al. A novel Bacillus cereus bacteriophage DLn1 and its endolysin as biocontrol agents against Bacillus cereus in milk. Int. J. Food Microbiol. 2022, 369, 109615. [Google Scholar] [CrossRef]
- Kong, L.; Ding, Y.; Wu, Q.; Wang, J.; Zhang, J.; Li, H.; Yu, S.; Yu, P.; Gao, T.; Zeng, H.; et al. Genome sequencing and characterization of three Bacillus cereus-specific phages, DK1, DK2, and DK3. Arch. Virol. 2019, 164, 1927–1929. [Google Scholar] [CrossRef]
- Yuan, Y.; Peng, Q.; Gao, M. Characteristics of a broad lytic spectrum endolysin from phage BtCS33 of Bacillus thuringiensis. BMC Microbiol. 2012, 12, 297. [Google Scholar] [CrossRef]
- Gong Min-seok. Exploration of bacteriophages, endolysins, and cell wall binding domains of endolysins for control and rapid detection of bacteria. Ph.D. Thesis, College of Agriculture and Life Sciences, Seoul, Republic of Korea, 2015. Available online: https://s-space.snu.ac.kr/bitstream/10371/119497/1/000000053384.pdf (accessed on 13 August 2024).
- Schuch, R.; Pelzek, A.J.; Nelson, D.C.; Fischetti, V.A. The PlyB Endolysin of Bacteriophage vB_BanS_Bcp1 Exhibits Broad-Spectrum Bactericidal Activity against Bacillus cereus Sensu Lato Isolates. Appl. Environ. Microbiol. 2019, 85, e00003-19. [Google Scholar] [CrossRef]
- Yuan, Y.; Gao, M. Proteomic Analysis of a Novel Bacillus Jumbo Phage Revealing Glycoside Hydrolase as Structural Component. Front. Microbiol. 2016, 7, 745. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, D.; Salzmann, M. Mannosyl-glycoprotein endo-beta-N-acetyl glucosaminidase. In Enzyme Handbook 4; Schomburg, D., Salzmann, M., Eds.; Springer: Berlin/Heidelberg, Germany, 1991; pp. 499–505. ISBN 978-3-642-48986-0. [Google Scholar]
- Verheust, C.; Fornelos, N.; Mahillon, J. The Bacillus thuringiensis phage GIL01 encodes two enzymes with peptidoglycan hydrolase activity. FEMS Microbiol. Lett. 2004, 237, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, J.M.; Moynihan, P.J.; Clarke, C.A.; Vandenende, C.; Clarke, A.J. Control of Lytic Transglycosylase Activity Within Bacterial Cell Walls. In Glycomicrobiology; CaisterAcademic Press: Norfolk, UK, 2012; pp. 55–68. [Google Scholar]
- Broendum, S.S.; Buckle, A.M.; McGowan, S. Catalytic diversity and cell wall binding repeats in the phage-encoded endolysins. Mol. Microbiol. 2018, 110, 879–896. [Google Scholar] [CrossRef] [PubMed]
- Jarábková, V.; Tišáková, L.; Godány, A. Phage Endolysin: A Way to Understand A Binding Function of C-Terminal Domains A Mini Review. Nova Biotechnol. Chim. 2015, 14, 117–134. [Google Scholar] [CrossRef]
- Mayer, B.J. SH3 domains: Complexity in moderation. J. Cell Sci. 2001, 114, 1253–1263. [Google Scholar] [CrossRef]
- Denyes, J. Bacteriophage proteins as affinity molecules for the detection of Salmonella, Cronobacter and Bacillus. Ph.D. Thesis, ETH Zurich, Guelph, Canada, 2015. [Google Scholar] [CrossRef]
- Buist, G.; Steen, A.; Kok, J.; Kuipers, O.P. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 2008, 68, 838–847. [Google Scholar] [CrossRef]
- Huang, L.; Xu, L.; Han, G.; Crickmore, N.; Song, F.; Xu, J. Characterization of CwlC, an autolysin, and its role in mother cell lysis of Bacillus thuringiensis subsp. israelensis. Lett. Appl. Microbiol. 2022, 74, 92–102. [Google Scholar] [CrossRef]
- Mehta, K.K.; Paskaleva, E.E.; Azizi-Ghannad, S.; Ley, D.J.; Page, M.A.; Dordick, J.S.; Kane, R.S. Characterization of AmiBA2446, a Novel Bacteriolytic Enzyme Active against Bacillus Species. Appl. Environ. Microbiol. 2013, 79, 5899–5906. [Google Scholar] [CrossRef]
- Tišáková, L.; Vidová, B.; Farkašovská, J.; Godány, A. Bacteriophage endolysin Lyt μ1/6: Characterization of the C-terminal binding domain. FEMS Microbiol. Lett. 2014, 350, 199–208. [Google Scholar] [CrossRef]
- Yahashiri, A.; Jorgenson, M.A.; Weiss, D.S. The SPOR Domain, a Widely Conserved Peptidoglycan Binding Domain That Targets Proteins to the Site of Cell Division. J. Bacteriol. 2017, 199, 10. [Google Scholar] [CrossRef]
- Swift, S.M.; Reid, K.P.; Donovan, D.M.; Ramsay, T.G. Thermophile Lytic Enzyme Fusion Proteins that Target Clostridium perfringens. Antibiotics 2019, 8, 214. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.-Y.; You, R.-I.; Lai, M.-J.; Lin, N.-T.; Chen, L.-K.; Chang, K.-C. Highly potent antimicrobial modified peptides derived from the Acinetobacter baumannii phage endolysin LysAB2. Sci. Rep. 2017, 7, 11477. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.J.; Garefalaki, V.; Spoerl, R.; Narbad, A.; Meijers, R. Structure-Based Modification of a Clostridium difficile-targeting Endolysin Affects Activity and Host Range. J. Bacteriol. 2011, 193, 5477–5486. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koposova, O.N.; Kazantseva, O.A.; Shadrin, A.M. Diversity of Endolysin Domain Architectures in Bacteriophages Infecting Bacilli. Biomolecules 2024, 14, 1586. https://doi.org/10.3390/biom14121586
Koposova ON, Kazantseva OA, Shadrin AM. Diversity of Endolysin Domain Architectures in Bacteriophages Infecting Bacilli. Biomolecules. 2024; 14(12):1586. https://doi.org/10.3390/biom14121586
Chicago/Turabian StyleKoposova, Olga N., Olesya A. Kazantseva, and Andrey M. Shadrin. 2024. "Diversity of Endolysin Domain Architectures in Bacteriophages Infecting Bacilli" Biomolecules 14, no. 12: 1586. https://doi.org/10.3390/biom14121586
APA StyleKoposova, O. N., Kazantseva, O. A., & Shadrin, A. M. (2024). Diversity of Endolysin Domain Architectures in Bacteriophages Infecting Bacilli. Biomolecules, 14(12), 1586. https://doi.org/10.3390/biom14121586




