Bioinformatics Prediction for Network-Based Integrative Multi-Omics Expression Data Analysis in Hirschsprung Disease
Abstract
1. Introduction
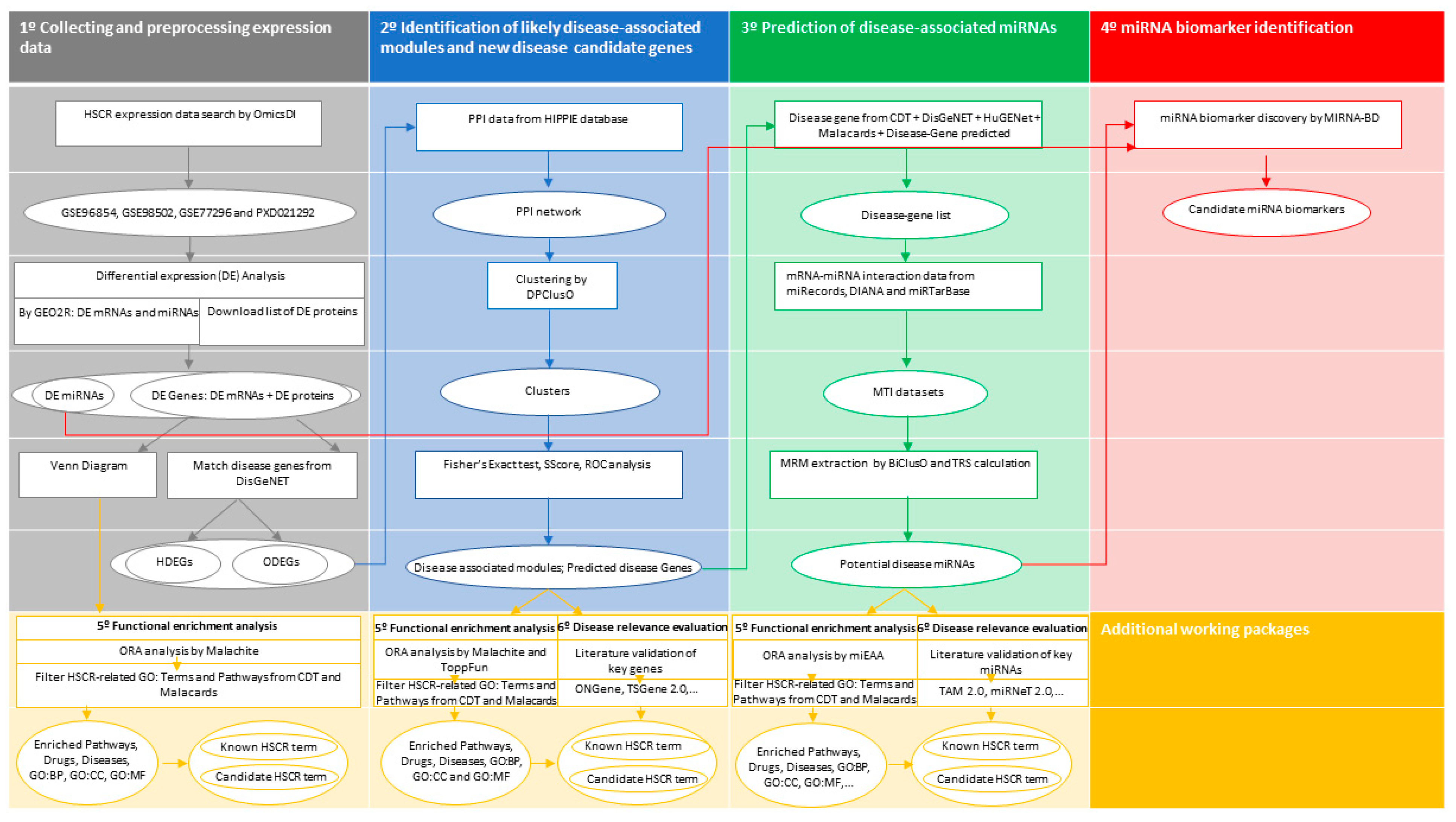
2. Materials and Methods
2.1. Data Collection
2.2. Data Preprocessing and Differential Expression Analysis
2.3. Identification of Novel Candidate Disease Genes and Disease-Associated Modules
2.4. Prediction of Disease-Associated miRNAs
2.5. Identification of Potential miRNA Biomarkers in HSCR
2.6. Functional Enrichment Analysis
2.7. Disease Relevance Evaluation
2.8. Statistical Analysis
3. Results
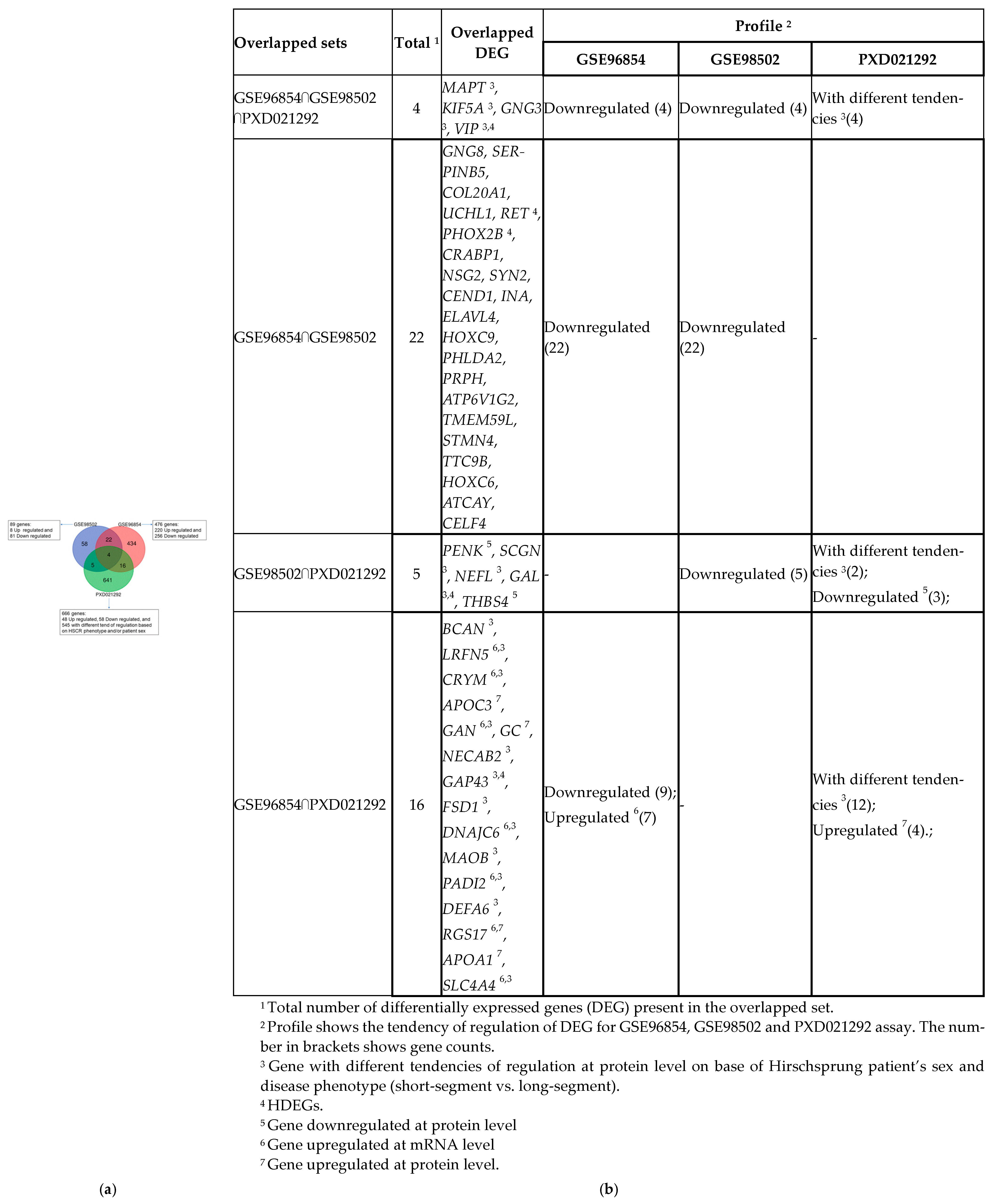
3.1. Identification of DE Genes in HSCR and Gene Enrichment Meta-Analysis
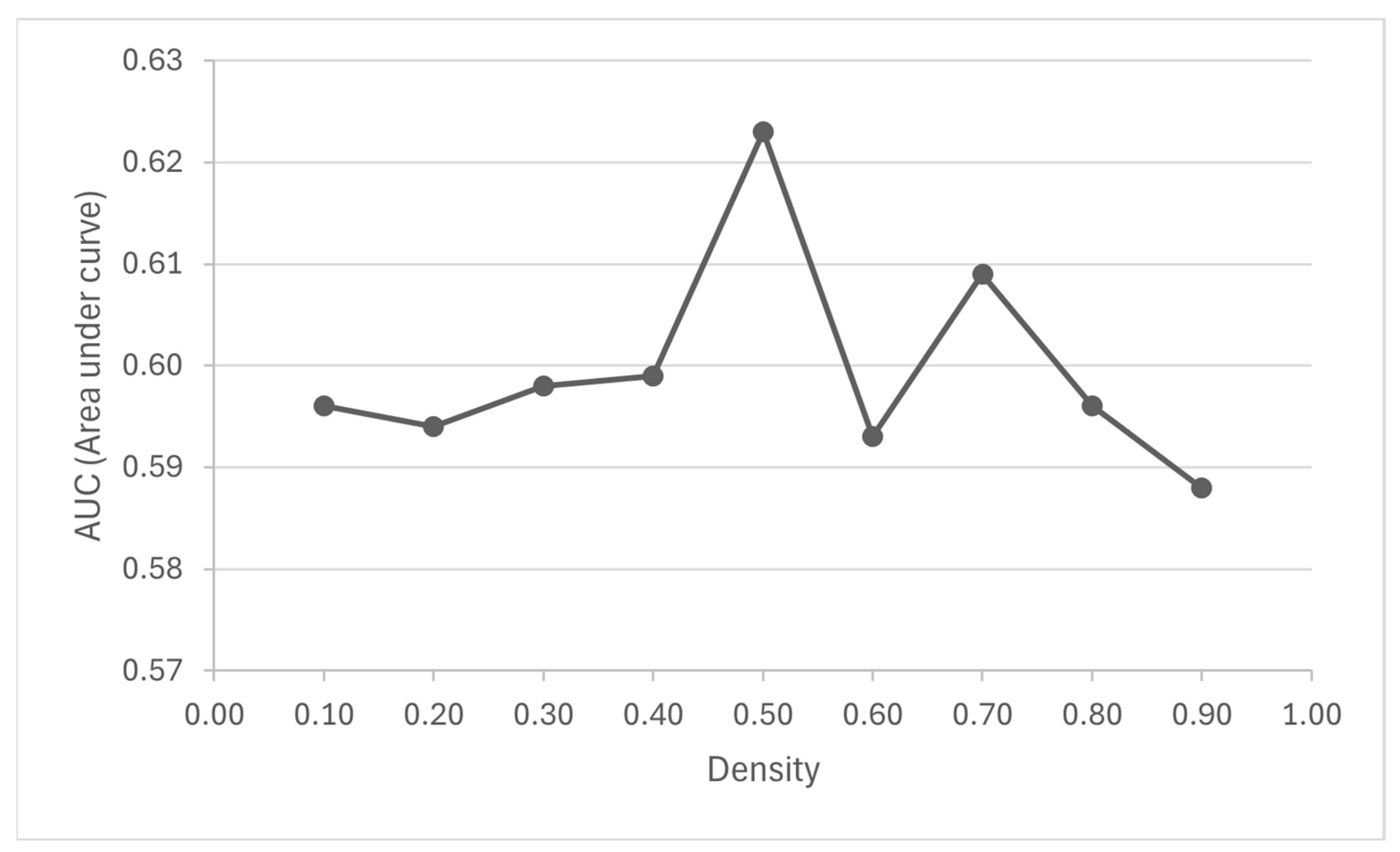
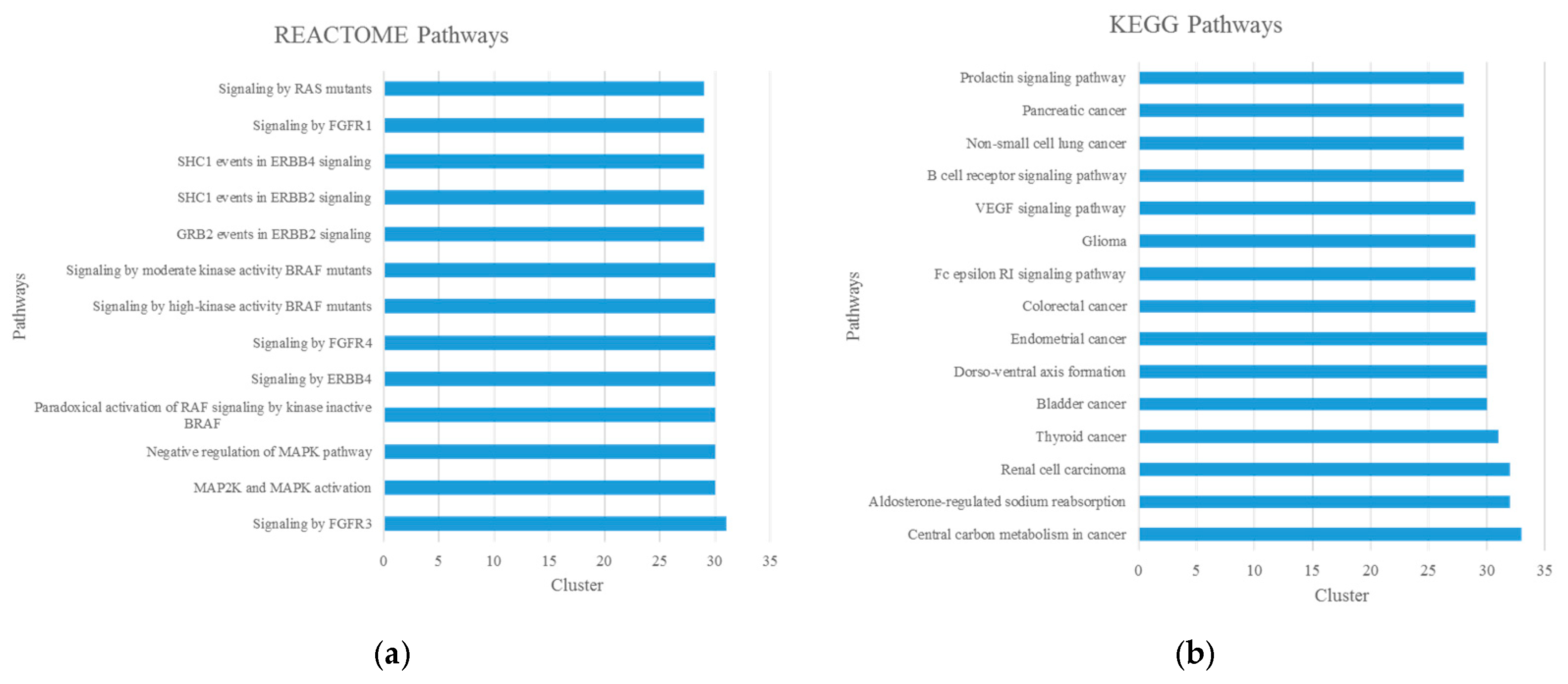
3.2. Identification and Enrichment Analysis of Candidate Disease-Associated Modules and Potential New Disease Genes
3.3. Prediction of HSCR-Associated miRNAs
3.4. Assessment of the Relevance of the Identified Candidate miRNAs in HSCR
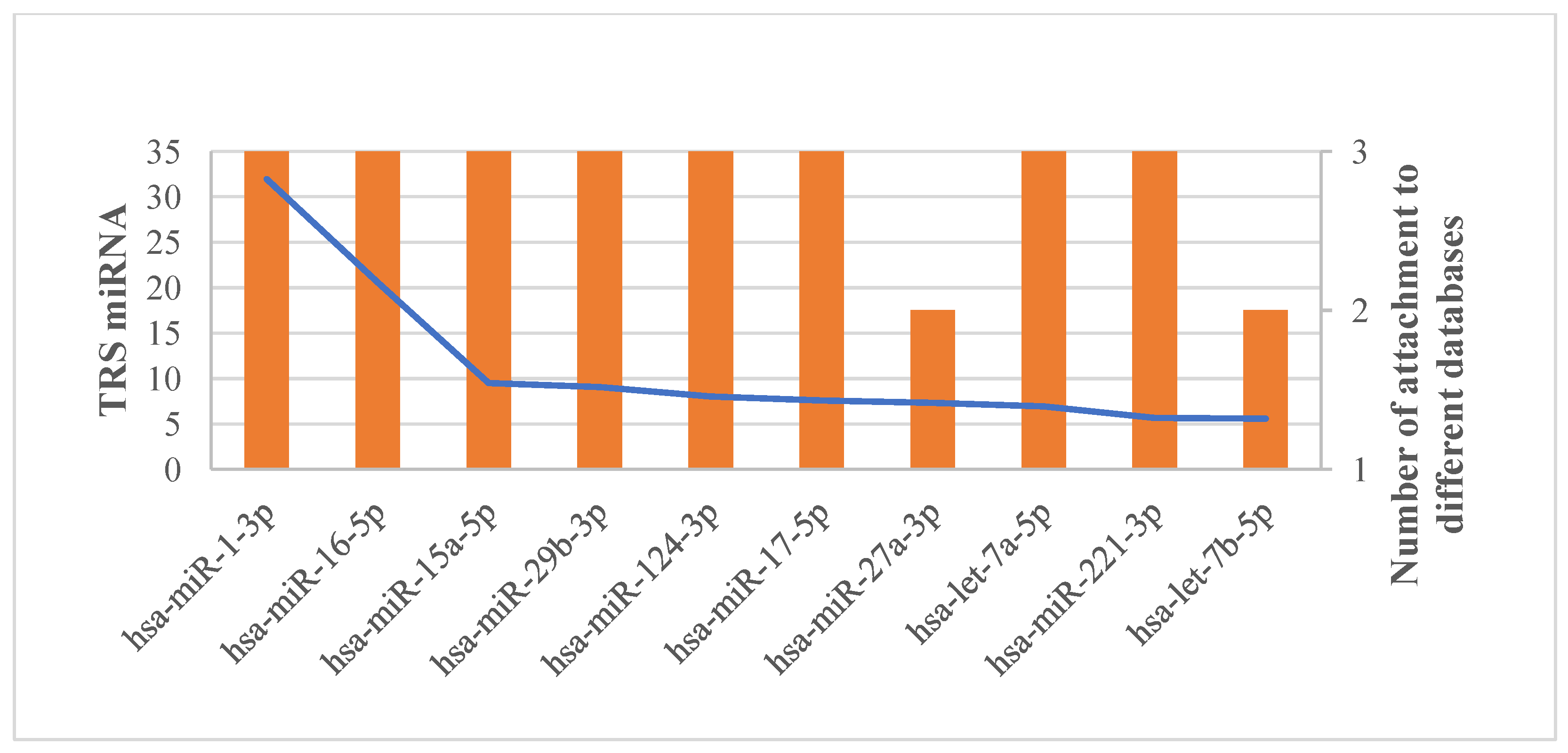
3.5. Potential miRNA Biomarker Identification in HSCR Based on miRNA-Target Regulatory Network Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luzón-Toro, B.; Villalba-Benito, L.; Torroglosa, A.; Fernández, R.M.; Antiñolo, G.; Borrego, S. What is new about the genetic background of Hirschsprung disease? Clin. Genet. 2020, 97, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Tilghman, J.M.; Ling, A.Y.; Turner, T.N.; Sosa, M.X.; Krumm, N.; Chatterjee, S.; Kapoor, A.; Coe, B.P.; Nguyen, K.H.; Gupta, N.; et al. Molecular Genetic Anatomy and Risk Profile of Hirschsprung’s Disease. N. Engl. J. Med. 2019, 380, 1421–1432. [Google Scholar] [CrossRef] [PubMed]
- Singh, V. Multigenic Effects. In Encyclopedia of Animal Cognition and Behavior; Vonk, J., Shackelford, T., Eds.; Springer Nature: Basel, Switzerland, 2017. [Google Scholar]
- Xu, Z.; Yan, Y.; Gu, B.; Cai, W.; Wang, Y. Up-Regulation of microRNA-424 Causes an Imbalance in AKT Phosphorylation and Impairs Enteric Neural Crest Cell Migration in Hirschsprung Disease. Int. J. Mol. Sci. 2023, 24, 6700. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Li, Y.; Li, H.; Zhou, L.; Wang, B.; Zhi, Z.; Tang, W. Molecular function predictions and diagnostic value analysis of plasma exosomal miRNAs in Hirschsprung’s disease. Epigenomics 2020, 12, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, Q.; Chakravarti, A.; Cai, H.; Xu, Z.; Wu, W.; Gu, B.; Li, L.; Cai, W. MicroRNA-4516-mediated regulation of MAPK10 relies on 3′ UTR cis-acting variants and contributes to the altered risk of Hirschsprung disease. J. Med. Genet. 2020, 57, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef] [PubMed]
- Olde Loohuis, N.F.; Kos, A.; Martens, G.J.; Van Bokhoven, H.; Nadif Kasri, N.; Aschrafi, A. MicroRNA networks direct neuronal development and plasticity. Cell. Mol. Life Sci. 2012, 69, 89–102. [Google Scholar] [CrossRef]
- Wang, Y.; Veremeyko, T.; Wong, A.H.; El Fatimy, R.; Wei, Z.; Cai, W.; Krichevsky, A.M. Downregulation of miR-132/212 impairs S-nitrosylation balance and induces tau phosphorylation in Alzheimer’s disease. Neurobiol. Aging 2017, 51, 156–166. [Google Scholar] [CrossRef]
- Teplyuk, N.M.; Uhlmann, E.J.; Gabriely, G.; Volfovsky, N.; Wang, Y.; Teng, J.; Karmali, P.; Marcusson, E.; Peter, M.; Mohan, A.; et al. Therapeutic potential of targeting microRNA-10b in established intracranial glioblastoma: First steps toward the clinic. EMBO Mol. Med. 2016, 8, 268–287. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Luzón-Toro, B.; Espino-Paisán, L.; Fernández, R.M.; Martín-Sánchez, M.; Antiñolo, G.; Borrego, S. Next-generation-based targeted sequencing as an efficient tool for the study of the genetic background in Hirschsprung patients. BMC Med. Genet. 2015, 16, 89. [Google Scholar] [CrossRef]
- Mederer, T.; Schmitteckert, S.; Volz, J.; Martínez, C.; Röth, R.; Thumberger, T.; Eckstein, V.; Scheuerer, J.; Thöni, C.; Lasitschka, F.; et al. A complementary study approach unravels novel players in the pathoetiology of Hirschsprung disease. PLoS Genet. 2020, 16, e1009106. [Google Scholar] [CrossRef]
- Villalba-Benito, L.; López-López, D.; Torroglosa, A.; Casimiro-Soriguer, C.S.; Luzón-Toro, B.; Fernández, R.M.; Moya-Jiménez, M.J.; Antiñolo, G.; Dopazo, J.; Borrego, S. Genome-wide analysis of DNA methylation in Hirschsprung enteric precursor cells: Unraveling the epigenetic landscape of enteric nervous system development. Clin. Epigenetics 2021, 13, 51. [Google Scholar] [CrossRef]
- Fernández, R.M.; Bleda, M.; Luzón-Toro, B.; García-Alonso, L.; Arnold, S.; Sribudiani, Y.; Besmond, C.; Lantieri, F.; Doan, B.; Ceccherini, I.; et al. Pathways systematically associated to Hirschsprung’s disease. Orphanet J. Rare Dis. 2013, 8, 187. [Google Scholar] [CrossRef][Green Version]
- Fernández, R.M.; Bleda, M.; Núñez-Torres, R.; Medina, I.; Luzón-Toro, B.; García-Alonso, L.; Torroglosa, A.; Marbà, M.; Enguix-Riego, M.V.; Montaner, D.; et al. Four new loci associations discovered by pathway-based and network analyses of the genome-wide variability profile of Hirschsprung’s disease. Orphanet J. Rare Dis. 2012, 7, 103. [Google Scholar] [CrossRef]
- Horgan, R.P.; Kenny, L.C. ‘Omic’ technologies: Genomics, transcriptomics, proteomics and metabolomics. Obstet. Gynaecol. 2011, 13, 189–195. [Google Scholar] [CrossRef]
- Sun, Y.V.; Hu, Y.J. Integrative Analysis of Multi-omics Data for Discovery and Functional Studies of Complex Human Diseases. Adv. Genet. 2016, 93, 147–190. [Google Scholar]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef]
- Agamah, F.E.; Bayjanov, J.R.; Niehues, A.; Njoku, K.F.; Skelton, M.; Mazandu, G.K.; Ederveen, T.H.A.; Mulder, N.; Chimusa, E.R.; ’t Hoen, P.A.C. Computational approaches for network-based integrative multi-omics analysis. Front. Mol. Biosci. 2022, 9, 967205. [Google Scholar] [CrossRef]
- Chakravorty, D.; Banerjee, K.; Saha, S. Integrative Omics for Interactomes. In Synthetic Biology; Singh, S., Ed.; Springer: Singapore, 2018; pp. 39–49. [Google Scholar]
- Zhou, G.; Li, S.; Xia, J. Network-Based Approaches for Multi-omics Integration. Methods Mol. Biol. 2020, 2104, 469–487. [Google Scholar]
- Ranea, J.A.G.; Perkins, J.; Chagoyen, M.; Díaz-Santiago, E.; Pazos, F. Network-Based Methods for Approaching Human Pathologies from a Phenotypic Point of View. Genes 2022, 13, 1081. [Google Scholar] [CrossRef]
- Zuo, Y.; Wei, D.; Zhu, C.; Naveed, O.; Hong, W.; Yang, X. Unveiling the Pathogenesis of Psychiatric Disorders Using Network Models. Genes 2021, 12, 1101. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, M.; da Veiga Leprevost, F.; Squizzato, S.; Park, Y.M.; Haug, K.; Carroll, A.J.; Spalding, D.; Paschall, J.; Wang, M.; et al. Discovering and linking public omics data sets using the Omics Discovery Index. Nat. Biotechnol. 2017, 35, 406–409. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Zorin, A.; Dass, G.; Vu, M.T.; Xu, P.; Glont, M.; Vizcaíno, J.A.; Jarnuczak, A.F.; Petryszak, R.; Ping, P.; et al. Quantifying the impact of public omics data. Nat. Commun. 2019, 10, 3512. [Google Scholar] [CrossRef]
- Xiao, S.J.; Zhu, X.C.; Deng, H.; Zhou, W.P.; Yang, W.Y.; Yuan, L.K.; Zhang, J.Y.; Tian, S.; Xu, L.; Zhang, L.; et al. Gene expression profiling coupled with Connectivity Map database mining reveals potential therapeutic drugs for Hirschsprung disease. J. Pediatr. Surg. 2018, 53, 1716–1721. [Google Scholar] [CrossRef]
- Tang, W.; Chen, M.; Guo, X.; Zhou, K.; Wen, Z.; Liu, F.; Liu, X.; Mao, X.; He, X.; Hu, W.; et al. Multiple ‘omics’-analysis reveals the role of prostaglandin E2 in Hirschsprung’s disease. Free Radic. Biol. Med. 2021, 164, 390–398. [Google Scholar] [CrossRef]
- Li, S.; Wang, S.; Guo, Z.; Wu, H.; Jin, X.; Wang, Y.; Li, X.; Liang, S. miRNA Profiling Reveals Dysregulation of RET and RET-Regulating Pathways in Hirschsprung’s Disease. PLoS ONE 2016, 11, 0150222. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, L.; Bai, B.; Li, D.; Xiao, P.; Li, Q.; Zhang, Z.; Wang, H.; Li, L.; Jiang, Q. Quantitative Proteomics Reveals Association of Neuron Projection Development Genes ARF4, KIF5B, and RAB8A With Hirschsprung Disease. Mol. Cell Proteom. 2021, 20, 100007. [Google Scholar] [CrossRef]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; King, B.L.; McMorran, R.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. The Comparative Toxicogenomics Database: Update 2017. Nucleic Acids Res. 2017, 45, D972–D978. [Google Scholar] [CrossRef]
- Piñero, J.; Queralt-Rosinach, N.; Bravo, À.; Deu-Pons, J.; Bauer-Mehren, A.; Baron, M.; Sanz, F.; Furlong, L.I. DisGeNET: A discovery platform for the dynamical exploration of human diseases and their genes. Database 2015, 2015, bav028. [Google Scholar] [CrossRef]
- Yu, W.; Gwinn, M.; Clyne, M.; Yesupriya, A.; Khoury, M.J. A navigator for human genome epidemiology. Nat. Genet. 2008, 40, 124–125. [Google Scholar] [CrossRef]
- Espe, S. Malacards: The Human Disease Database. J. Med. Libr. Assoc. 2018, 106, 140–141. [Google Scholar] [CrossRef]
- Rappaport, N.; Twik, M.; Nativ, N.; Stelzer, G.; Bahir, I.; Stein, T.I.; Safran, M.; Lancet, D. MalaCards: A Comprehensive Automatically-Mined Database of Human Diseases. Curr. Protoc. Bioinform. 2014, 47, 1.24.1–1.24.19. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Shi, J.; Gao, Y.; Cui, C.; Zhang, S.; Li, J.; Zhou, Y.; Cui, Q. HMDD v3.0: A database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019, 47, D1013–D1017. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.H.; Fontaine, J.F.; Vinayagam, A.; Porras, P.; Wanker, E.E.; Andrade-Navarro, M.A. HIPPIE: Integrating Protein Interaction Networks with Experiment Based Quality Scores. PLoS ONE 2012, 7, e31826. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef]
- Xiao, F.; Zuo, Z.; Cai, G.; Kang, S.; Gao, X.; Li, T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009, 37, D105–D110. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Lin, Y.C.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.G.; Jin, C.N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Troup, D.B.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; et al. NCBI GEO: Archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011, 39, D1005–D1010. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Benjamini, Y.; Yekutieli, D. The Control of the False Discovery Rate in Multiple Testing Under Dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Zaru, R.; Orchard, S. UniProt Consortium. UniProt Tools: BLAST, Align, Peptide Search, and ID Mapping. Curr. Protoc. 2023, 3, e697. [Google Scholar] [CrossRef]
- Seal, R.L.; Braschi, B.; Gray, K.; Jones, T.E.M.; Tweedie, S.; Haim-Vilmovsky, L.; Bruford, E.A. Genenames.org: The HGNC resources in 2023. Nucleic Acids Res. 2023, 51, D1003–D1009. [Google Scholar] [CrossRef]
- Aparicio-Puerta, E.; Hirsch, P.; Schmartz, G.P.; Kern, F.; Fehlmann, T.; Keller, A. miEAA 2023: Updates, new functional microRNA sets and improved enrichment visualizations. Nucleic Acids Res. 2023, 51, W319–W325. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Eguchi, R.; Karim, M.B.; Hu, P.; Sato, T.; Ono, N.; Kanaya, S.; Altaf-Ul-Amin, M. An integrative network-based approach to identify novel disease genes and pathways: A case study in the context of inflammatory bowel disease. BMC Bioinform. 2018, 19, 264. [Google Scholar] [CrossRef]
- Altaf-Ul-Amin, M.; Wada, M.; Kanaya, S. Partitioning a PPI Network into Overlapping Modules Constrained by High-Density and Periphery Tracking. ISRN Biomath. 2012, 2012, 726429. [Google Scholar] [CrossRef]
- Altaf-Ul-Amin, M.; Shinbo, Y.; Mihara, K.; Kurokawa, K.; Kanaya, S. Development and implementation of an algorithm for detection of protein complexes in large interaction networks. BMC Bioinform. 2006, 7, 207. [Google Scholar] [CrossRef]
- Altaf-Ul-Amin, M.; Tsuji, H.; Kurokawa, K.; Asahi, H.; Shinbo, Y.; Kanaya, S. DPClus: A density-periphery based graphclustering software mainly focused on detection of protein complexes ininteraction networks. J. Comput. Aided Chem. 2006, 7, 150–156. [Google Scholar] [CrossRef][Green Version]
- Karim, M.B.; Wakamatsu, N.; Altaf-Ul-Amin, M. DPClusOST: A software tool for general purpose graph clustering. J. Comput. Aided Chem. 2017, 18, 76–93. [Google Scholar] [CrossRef][Green Version]
- Karim, M.B.; Kanaya, S.; Altaf-Ul-Amin, M. DPClusSBO: An integrated software for clustering of simple and bipartite graphs. SoftwareX 2021, 16, 100821S. [Google Scholar] [CrossRef]
- Metz, C.E. Basic principles of ROC analysis. Semin. Nucl. Med. 1978, 8, 283–298. [Google Scholar] [CrossRef]
- Davis, J.; Goadrich, M. The Relationship Between Precision-Recall and ROC Curves. In Proceedings of the 23rd International Conference on Machine Learning (ICML ‘06), Pittsburgh, PA, USA, 25–26 June 2006; Association for Computing Machinery: New York, NY, USA, 2006. [Google Scholar]
- Altaf-Ul-Amin, M.; Karim, M.B.; Hu, P.; Ono, N.; Kanaya, S. Discovery of inflammatory bowel disease-associated miRNAs using a novel bipartite clustering approach. BMC Med. Genom. 2020, 13, 10. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kim, V.; Muth, D.C.; Witwer, K.W. Validated MicroRNA Target Databases: An Evaluation. Drug Dev. Res. 2015, 76, 389–396. [Google Scholar]
- Karim, M.B.; Kanaya, S.; Amin, M.A.U. Comparison of BiClusO with Five Different Biclustering Algorithms Using Biological and Synthetic Data. In Complex Networks and Their Applications VII. Studies in Computational Intelligence; Aiello, L., Cherifi, C., Cherifi, H., Lambiotte, R., Lió, P., Rocha, L., Eds.; Springer: Cham, Switzerland, 2018; Volume 813, pp. 575–585. [Google Scholar]
- Karim, M.B.; Huang, M.; Ono, N.; Kanaya, S.; Amin, M.A. BiClusO: A Novel Biclustering Approach and Its Application to Species-VOC Relational Data. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1955–1965. [Google Scholar] [CrossRef]
- Lin, Y.; Wu, W.; Sun, Z.; Shen, L.; Shen, B. MiRNA-BD: An evidence-based bioinformatics model and software tool for microRNA biomarker discovery. RNA Biol. 2018, 15, 1093–1105. [Google Scholar]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Pang, Z.; Lu, Y.; Ewald, J.; Xia, J. OmicsNet 2.0: A web-based platform for multi-omics integration and network visual analytics. Nucleic Acids Res. 2022, 50, W527–W533. [Google Scholar] [CrossRef] [PubMed]
- Theodosiou, T.; Efstathiou, G.; Papanikolaou, N.; Kyrpides, N.C.; Bagos, P.G.; Iliopoulos, I.; Pavlopoulos, G.A. NAP: The Network Analysis Profiler, a web tool for easier topological analysis and comparison of medium-scale biological networks. BMC Res. Notes 2017, 10, 278. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef] [PubMed]
- Gershkowitz, G.R.; Abrams, Z.B.; Coombes, C.E.; Coombes, K.R. Malachite: A Gene Enrichment Meta-Analysis (GEM) Tool for ToppGene. bioRxiv 2019. bioRxiv:511527. [Google Scholar] [CrossRef]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]
- Hounkpe, B.W.; Chenou, F.; de Lima, F.; De Paula, E.V. HRT Atlas v1.0 database: Redefining human and mouse housekeeping genes and candidate reference transcripts by mining massive RNA-seq datasets. Nucleic Acids Res. 2021, 49, D947–D955. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Lu, G.; Chen, X.; Zhao, X.M.; Bork, P. OGEE v2: An update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 2017, 45, D940–D944. [Google Scholar] [CrossRef] [PubMed]
- Leskovec, J.; Sosič, R. SNAP: A General Purpose Network Analysis and Graph Mining Library. ACM Trans. Intell. Syst. Technol. 2016, 8, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, J.; Zhao, M. ONGene: A literature-based database for human oncogenes. J. Genet. Genom. 2017, 44, 119–121. [Google Scholar] [CrossRef]
- Zhao, M.; Kim, P.; Mitra, R.; Zhao, J.; Zhao, Z. TSGene 2.0: An updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016, 44, D1023–D1031. [Google Scholar] [CrossRef]
- Rehm, H.L.; Berg, J.S.; Brooks, L.D.; Bustamante, C.D.; Evans, J.P.; Landrum, M.J.; Ledbetter, D.H.; Maglott, D.R.; Martin, C.L.; Nubaum, R.L.; et al. ClinGen--the Clinical Genome Resource. N. Engl. J. Med. 2015, 372, 2235–2242. [Google Scholar] [CrossRef]
- Li, J.; Han, X.; Wan, Y.; Zhang, S.; Zhao, Y.; Fan, R.; Cui, Q.; Zhou, Y. TAM 2.0: Tool for MicroRNA set analysis. Nucleic Acids Res. 2018, 46, W180–W185. [Google Scholar] [CrossRef]
- Hong, M.; Li, X.; Li, Y.; Zhou, Y.; Li, Y.; Chi, S.; Cao, G.; Li, S.; Tang, S. Hirschsprung’s disease: Key microRNAs and target genes. Pediatr. Res. 2022, 92, 737–747. [Google Scholar] [CrossRef]
- Gao, Z.G.; Chen, Q.J.; Shao, M.; Qian, Y.Z.; Zhang, L.F.; Zhang, Y.B.; Xiong, Q.X. Preliminary identification of key miRNAs, signaling pathways, and genes associated with Hirschsprung’s disease by analysis of tissue microRNA expression profiles. World J. Pediatr. 2017, 13, 489–495. [Google Scholar] [CrossRef]
- Xu, W.; Yu, H.; Chen, D.; Pan, W.; Yang, W.; Miao, J.; Jia, W.; Zheng, B.; Liu, Y.; Chen, X.; et al. Identifying the potential transcriptional regulatory network in Hirschsprung disease by integrated analysis of microarray datasets. World J. Pediatr. Surg. 2023, 6, e000547. [Google Scholar] [CrossRef]
- Tang, W.; Cai, P.; Huo, W.; Li, H.; Tang, J.; Zhu, D.; Xie, H.; Chen, P.; Hang, B.; Wang, S.; et al. Suppressive action of miRNAs to ARP2/3 complex reduces cell migration and proliferation via RAC isoforms in Hirschsprung disease. J. Cell Mol. Med. 2016, 20, 1266–1275. [Google Scholar] [CrossRef]
- Chen, G.; Peng, L.; Zhu, Z.; Du, C.; Shen, Z.; Zang, R.; Su, Y.; Xia, Y.; Tang, W. LncRNA AFAP1-AS Functions as a Competing Endogenous RNA to Regulate RAP1B Expression by sponging miR-181a in the HSCR. Int. J. Med. Sci. 2017, 14, 1022–1030. [Google Scholar] [CrossRef]
- Wang, G.; Wang, H.; Zhang, L.; Guo, F.; Wu, X.; Liu, Y. MiR-195-5p inhibits proliferation and invasion of nerve cells in Hirschsprung disease by targeting GFRA4. Mol. Cell. Biochem. 2021, 476, 2061–2073. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yuan, X.; Li, Y.; Chen, J.; Wu, S.; Jiang, A.; Miao, X.; Shu, Q. Circulating exosomal microRNA-18a-5p accentuates intestinal inflammation in Hirschsprung-associated enterocolitis by targeting RORA. Am. J. Transl. Res. 2021, 13, 4182–4196. [Google Scholar]
- Su, Y.; Wen, Z.; Shen, Q.; Zhang, H.; Peng, L.; Chen, G.; Zhu, Z.; Du, C.; Xie, H.; Li, H.; et al. Long non-coding RNA LOC100507600 functions as a competitive endogenous RNA to regulate BMI1 expression by sponging miR128-1-3p in Hirschsprung’s disease. Cell Cycle 2018, 17, 459–467. [Google Scholar] [CrossRef]
- Wu, L.; Yuan, W.; Chen, J.; Zhou, Z.; Shu, Y.; Ji, J.; Liu, Z.; Tang, Q.; Zhang, X.; Shu, X. Increased miR-214 expression suppresses cell migration and proliferation in Hirschsprung disease by interacting with PLAGL2. Pediatr. Res. 2019, 86, 460–470. [Google Scholar] [CrossRef]
- Daiyue, Y.; Yang, Y.; Zhaorong, H.; Yi, L.; Chen, W.; Caiyun, L.; Yuqian, S.; Liucheng, Y.; Kai, W. Plasma exosomal miR-199a-3p downregulates cell proliferation and migration in Hirschsprung’s disease by targeting mTOR. Pediatr. Surg. Int. 2022, 39, 54. [Google Scholar] [CrossRef]
- Tang, W.; Li, H.; Tang, J.; Wu, W.; Qin, J.; Lei, H.; Cai, P.; Huo, W.; Li, B.; Rehan, V.; et al. Specific serum microRNA profile in the molecular diagnosis of Hirschsprung’s disease. J. Cell Mol. Med. 2014, 18, 1580–1587. [Google Scholar] [CrossRef]
- Wen, Z.; Shen, Q.; Zhang, H.; Su, Y.; Zhu, Z.; Chen, G.; Peng, L.; Li, H.; Du, C.; Xie, H.; et al. Circular RNA CCDC66 targets DCX to regulate cell proliferation and migration by sponging miR-488-3p in Hirschsprung’s disease. J. Cell. Physiol. 2019, 234, 10576–10587. [Google Scholar] [CrossRef]
- Peng, L.; Chen, G.; Zhu, Z.; Shen, Z.; Du, C.; Zang, R.; Su, Y.; Xie, H.; Li, H.; Xu, X.; et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget 2017, 8, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.G.; Cheng, Y.; Li, D.; Sun, C.; Fang, F.; Guo, W.L. Systematic screen of potential circular RNA biomarkers of Hirschsprung’s disease. Transl. Pediatr. 2022, 11, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.P.; Zhao, F.; Ma, T.D.; Zou, C.J.; Xu, G.; Zhou, C.G. Circ-ITCH overexpression promoted cell proliferation and migration in Hirschsprung disease through miR-146b-5p/RET axis. Pediatr. Res. 2022, 92, 1008–1016. [Google Scholar] [CrossRef]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef]
- Seed-Based d Mapping (SDM, Formerly Signed Differential Mapping) FDR Online Calculator. Available online: https://www.sdmproject.com/utilities/?show=FDR (accessed on 12 June 2023).
- The VENN DIAGRAMS Tool. Available online: https://www.vandepeerlab.org/?q=tools/venn-diagrams (accessed on 12 June 2023).
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Kim, Y.; Park, J.H.; Cho, Y.R. Network-Based Approaches for Disease-Gene Association Prediction Using Protein-Protein Interaction Networks. Int. J. Mol. Sci. 2022, 23, 7411. [Google Scholar] [CrossRef]
- Wang, X.; Gulbahce, N.; Yu, H. Network-based methods for human disease gene prediction. Brief. Funct. Genom. 2011, 10, 280–293. [Google Scholar] [CrossRef]
- Gandhi, T.K.; Zhong, J.; Mathivanan, S.; Karthick, L.; Chandrika, K.N.; Mohan, S.S.; Sharma, S.; Pinkert, S.; Nagaraju, S.; Periaswamy, B.; et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat. Genet. 2006, 38, 285–293. [Google Scholar] [CrossRef]
- Lim, J.; Hao, T.; Shaw, C.; Patel, A.J.; Szabó, G.; Rual, J.F.; Fisk, C.J.; Li, N.; Smolyar, A.; Hill, D.E.; et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 2006, 125, 801–814. [Google Scholar] [CrossRef]
- Wood, L.D.; Parsons, D.W.; Jones, S.; Lin, J.; Sjöblom, T.; Leary, R.J.; Shen, D.; Boca, S.M.; Barber, T.; Ptak, J.; et al. The genomic landscapes of human breast and colorectal cancers. Science 2007, 318, 1108–1113. [Google Scholar] [CrossRef]
- Stevens, A.; Cosgrove, K.E.; Padidela, R.; Skae, M.S.; Clayton, P.E.; Banerjee, I.; Dunne, M.J. Can network biology unravel the aetiology of congenital hyperinsulinism? Orphanet J. Rare Dis. 2013, 8, 21. [Google Scholar] [CrossRef]
- Taroni, J.N.; Greene, C.S.; Martyanov, V.; Wood, T.A.; Christmann, R.B.; Farber, H.W.; Lafyatis, R.A.; Denton, C.P.; Hinchcliff, M.E.; Pioli, P.A.; et al. A novel multi-network approach reveals tissue-specific cellular modulators of fibrosis in systemic sclerosis. Genome Med. 2017, 9, 27. [Google Scholar] [CrossRef]
- Buphamalai, P.; Kokotovic, T.; Nagy, V.; Menche, J. Network analysis reveals rare disease signatures across multiple levels of biological organization. Nat. Commun. 2021, 12, 6306. [Google Scholar] [CrossRef]
- Zhang, P.; Itan, Y. Biological Network Approaches and Applications in Rare Disease Studies. Genes 2019, 10, 797. [Google Scholar] [CrossRef]
- Itan, Y.; Zhang, S.Y.; Vogt, G.; Abhyankar, A.; Herman, M.; Nitschke, P.; Fried, D.; Quintana-Murci, L.; Abel, L.; Casanova, J.L. The human gene connectome as a map of short cuts for morbid allele discovery. Proc. Natl. Acad. Sci. USA 2013, 110, 5558–5563. [Google Scholar] [CrossRef]
- Zhu, C.; Kushwaha, A.; Berman, K.; Jegga, A.G. A vertex similarity-based framework to discover and rank orphan disease-related genes. BMC Syst. Biol. 2012, 6, S8. [Google Scholar] [CrossRef]
- Liu, X.; Yang, Z.; Lin, H.; Simmons, M.; Lu, Z. DIGNiFI: Discovering causative genes for orphan diseases using protein-protein interaction networks. BMC Syst. Biol. 2017, 11, 23. [Google Scholar] [CrossRef]
- Nguengang Wakap, S.; Lambert, D.M.; Olry, A.; Rodwell, C.; Gueydan, C.; Lanneau, V.; Murphy, D.; Le Cam, Y.; Rath, A. Estimating cumulative point prevalence of rare diseases: Analysis of the Orphanet database. Eur. J. Hum. Genet. 2020, 28, 165–173. [Google Scholar] [CrossRef]
- Wang, X.; Wang, S.; Jin, X.; Wang, N.; Luo, Y.; Teng, Y. Detection and preliminary screening of the human gene expression profile for Hirschsprung’s disease. Mol. Med. Rep. 2016, 13, 641–650. [Google Scholar] [CrossRef][Green Version]
- Pan, W.-K.; Zhang, Y.-F.; Yu, H.; Gao, Y.; Zheng, B.-J.; Li, P.; Xie, C.; Ge, X. Identifying key genes associated with Hirschsprung’s disease based on bioinformatics analysis of RNA-sequencing data. World J. Pediatr. 2017, 13, 267–273. [Google Scholar] [CrossRef]
- Saeed, A.; Barreto, L.; Neogii, S.G.; Loos, A.; McFarlane, I.; Aslam, A. Identification of novel genes in Hirschsprung disease pathway using whole genome expression study. J. Pediatr. Surg. 2012, 47, 303–307. [Google Scholar] [CrossRef]
- Qin, K.W.; Shi, H.; Zhang, L.; Liu, P.F.; Cai, W.L.; Wu, K.H.; Zhu, X.C.; Lan, F.F. The research on screening differentially expressed genes in Hirschsprung’s disease by using Microarray. J. Pediatr. Surg. 2013, 48, 2281–2288. [Google Scholar] [CrossRef]
- Grissa, D.; Junge, A.; Oprea, T.I.; Jensen, L.J. Diseases 2.0: A weekly updated database of disease-gene associations from text mining and data integration. Database 2022, 2022, baac019. [Google Scholar] [CrossRef]
- Yoneda, A.; Wang, Y.; O’Briain, D.S.; Puri, P. Cell-adhesion molecules and fibroblast growth factor signalling in Hirschsprung’s disease. Pediatr. Surg. Int. 2001, 17, 299–303. [Google Scholar] [CrossRef]
- Zhou, L.; Li, J.; Zhang, X.; Xu, Z.; Yan, Y.; Hu, K. An integrative pan cancer analysis of RET aberrations and their potential clinical implications. Sci. Rep. 2022, 12, 13913. [Google Scholar] [CrossRef]
- Jahangiri, L.; Pucci, P.; Ishola, T.; Pereira, J.; Cavanagh, M.L.; Turner, S.D. Deep analysis of neuroblastoma core regulatory circuitries using online databases and integrated bioinformatics shows their pan-cancer roles as prognostic predictors. Discov. Oncol. 2021, 12, 56. [Google Scholar] [CrossRef]
- Schäfer, M.K.; Altevogt, P. L1CAM malfunction in the nervous system and human carcinomas. Cell. Mol. Life Sci. 2010, 67, 2425–2437. [Google Scholar] [CrossRef]
- Maten, M.V.; Reijnen, C.; Pijnenborg, J.M.A.; Zegers, M.M. L1 Cell Adhesion Molecule in Cancer, a Systematic Review on Domain-Specific Functions. Int. J. Mol. Sci. 2019, 20, 4180. [Google Scholar] [CrossRef]
- Borrego, S.; Eng, C.; Sánchez, B.; Sáez, M.E.; Navarro, E.; Antiñolo, G. Molecular analysis of the ret and GDNF genes in a family with multiple endocrine neoplasia type 2A and Hirschsprung disease. J. Clin. Endocrinol. Metab. 1998, 83, 3361–3364. [Google Scholar] [CrossRef]
- Jacobs, S.E.; Tiusaba, L.; Bokova, E.; Al-Shamaileh, T.; Russell, T.L.; Rutan, E.C.; Haroyan, H.; Wang, Y.; Feng, C.; Badillo, A.; et al. A Deeper Curse: A Hirschsprung Patient’s Evaluation Unmasks a Rare Association with Congenital Central Hypoventilation Syndrome and Neuroblastoma. Eur. J. Pediatr. Surg. Rep. 2022, 10, e156–e159. [Google Scholar] [CrossRef]
- Zhao, J.; Zhu, Y.; Xie, X.; Yao, Y.; Zhang, J.; Zhang, R.; Huang, L.; Cheng, J.; Xia, H.; He, J.; et al. Pleiotropic effect of common PHOX2B variants in Hirschsprung disease and neuroblastoma. Aging 2019, 11, 1252–1261. [Google Scholar] [CrossRef]
- Zen, Y.; Vara, R.; Portmann, B.; Hadzic, N. Childhood hepatocellular carcinoma: A clinicopathological study of 12 cases with special reference to EpCAM. Histopathology 2014, 64, 671–682. [Google Scholar] [CrossRef]
- Aguiar, T.; Teixeira, A.; Scliar, M.O.; Sobral de Barros, J.; Lemes, R.B.; Souza, S.; Tolezano, G.; Santos, F.; Tojal, I.; Cypriano, M.; et al. Unraveling the Genetic Architecture of Hepatoblastoma Risk: Birth Defects and Increased Burden of Germline Damaging Variants in Gastrointestinal/Renal Cancer Predisposition and DNA Repair Genes. Front. Genet. 2022, 13, 858396. [Google Scholar] [CrossRef]
- Valera, E.T.; Ferraz, S.T.; Brassesco, M.S.; Zhen, X.; Shen, Y.; dos Santos, A.C.; Neder, L.; Oliveira, R.S.; Scrideli, C.A.; Tone, L.G. Mowat-Wilson syndrome: The first report of an association with central nervous system tumors. Childs Nerv. Syst. 2013, 29, 2151–2155. [Google Scholar] [CrossRef]
- Shanthini, T.; Balaji, S.; Kim, U.; Muthukkaruppan, V.; Vanniarajan, A. Genetic characterization of a patient with an unusual presentation of Waardenburg syndrome Type 4 and retinoblastoma. Pediatr. Blood Cancer 2021, 68, e28553. [Google Scholar] [CrossRef]
- Zaborowski, A.G.; Kruse, C.H.; Kavonic, S.; Pergorano, R.J. Retinoblastoma and Hirschsprung disease with a 13q14 to 22 deletion. J. Pediatr. Ophthalmol. Strabismus 2008, 45, 366–367. [Google Scholar] [CrossRef]
- Doanes, A.M.; Hegland, D.D.; Sethi, R.; Kovesdi, I.; Bruder, J.T.; Finkel, T. VEGF stimulates MAPK through a pathway that is unique for receptor tyrosine kinases. Biochem. Biophys. Res. Commun. 1999, 255, 545–548. [Google Scholar] [CrossRef]
- Takahashi, T.; Ueno, H.; Shibuya, M. VEGF activates protein kinase C-dependent, but Ras-independent Raf-MEK-MAP kinase pathway for DNA synthesis in primary endothelial cells. Oncogene 1999, 18, 2221–2230. [Google Scholar] [CrossRef]
- Celia, G.; Osol, G. Mechanism of VEGF-induced uterine venous hyperpermeability. J. Vasc. Res. 2005, 42, 47–54. [Google Scholar] [CrossRef]
- Suda, K.; Yamada, S.; Miyahara, K.; Fujiwara, N.; Kosaka, S.; Abe, K.; Seo, S.; Nakamura, S.; Lane, G.J.; Yamataka, A. High intestinal vascular permeability in a murine model for Hirschsprung’s disease: Implications for postoperative Hirschsprung-associated enterocolitis. Pediatr. Surg. Int. 2022, 39, 15. [Google Scholar] [CrossRef]
- Schrenk, S.; Schuster, A.; Klotz, M.; Schleser, F.; Lake, J.; Heuckeroth, R.O.; Kim, Y.J.; Laschke, M.W.; Menger, M.D.; Schäfer, K.H. Vascular and neural stem cells in the gut: Do they need each other? Histochem. Cell Biol. 2015, 143, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Theintz, G.; Duhamel, D.; Cox, J.; Nusslé, D.; Le Coultre, C.I.; Pizzolato, F.; Sizonenko, P.C. Cytochemical determination of immunoreactive prolactin in normal and abnormal intestinal mucosa. Pediatr. Res. 1986, 20, 703. [Google Scholar] [CrossRef][Green Version]
- Malsure, S.; Wang, Q.; Charles, R.P.; Sergi, C.; Perrier, R.; Christensen, B.M.; Maillard, M.; Rossier, B.C.; Hummler, E. Colon-specific deletion of epithelial sodium channel causes sodium loss and aldosterone resistance. J. Am. Soc. Nephrol. 2014, 25, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Kunzelmann, K.; Mall, M. Electrolyte transport in the mammalian colon: Mechanisms and implications for disease. Physiol. Rev. 2002, 82, 245–289. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebrouck, S.; Van Laere, D.; Fryns, J.P.; Theyskens, C. Pseudo-Bartter syndrome due to Hirschsprung disease in a neonate with an extra ring chromosome 8. Am. J. Med. Genet. A 2007, 143A, 2469–2472. [Google Scholar] [CrossRef] [PubMed]
- Bugaj, V.; Mironova, E.; Kohan, D.E.; Stockand, J.D. Collecting duct-specific endothelin B receptor knockout increases ENaC activity. Am. J. Physiol. Cell Physiol. 2012, 302, C188–C194. [Google Scholar] [CrossRef]
- Sutthatarn, P.; Lapidus-Krol, E.; Smith, C.; Halaweish, I.; Rialon, K.; Ralls, M.W.; Rentea, R.M.; Madonna, M.B.; Haddock, C.; Rocca, A.M.; et al. Hirschsprung-associated inflammatory bowel disease: A multicenter study from the APSA Hirschsprung disease interest group. J. Pediatr. Surg. 2023, 58, 856–861. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Lim, T.; Puri, P. Inflammatory bowel disease in patients with Hirschsprung’s disease: A systematic review and meta-analysis. Pediatr. Surg. Int. 2018, 34, 149–154. [Google Scholar] [CrossRef]
- Matsuyama, H.; Suzuki, H.I. Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis. Int. J. Mol. Sci. 2019, 21, 132. [Google Scholar] [CrossRef] [PubMed]
- Sergi, C.M.; Caluseriu, O.; McColl, H.; Eisenstat, D.D. Hirschsprung’s disease: Clinical dysmorphology, genes, micro-RNAs, and future perspectives. Pediatr. Res. 2017, 81, 177–191. [Google Scholar] [CrossRef]
- Sharan, A.; Zhu, H.; Xie, H.; Li, H.; Tang, J.; Tang, W.; Zhang, H.; Xia, Y. Down-regulation of miR-206 is associated with Hirschsprung disease and suppresses cell migration and proliferation in cell models. Sci. Rep. 2015, 5, 9302. [Google Scholar] [CrossRef]
- Wu, Q.; Zhao, J.; Zheng, Y.; Xie, X.; He, Q.; Zhu, Y.; Wang, N.; Huang, L.; Lu, L.; Hu, T.; et al. Associations between common genetic variants in microRNAs and Hirschsprung disease susceptibility in Southern Chinese children. J. Gene Med. 2021, 23, e3301. [Google Scholar] [CrossRef] [PubMed]
- Pio, G.; Ceci, M.; D’Elia, D.; Loglisci, C.; Malerba, D. A novel biclustering algorithm for the discovery of meaningful biological correlations between microRNAs and their target genes. BMC Bioinform. 2013, 14, S8. [Google Scholar] [CrossRef]
- Pio, G.; Ceci, M.; Malerba, D.; D’Elia, D. ComiRNet: A web-based system for the analysis of miRNA-gene regulatory networks. BMC Bioinform. 2015, 16, S7. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Z.; Xiao, J.; Wu, L.; Chen, K.; Zhu, T.; Feng, C.; Zhuansun, D.; Meng, X.; Feng, J. Identification and validation of the common pathogenesis and hub biomarkers in Hirschsprung disease complicated with Crohn’s disease. Front. Immunol. 2022, 13, 961217. [Google Scholar] [CrossRef]
- Plotnikova, O.; Baranova, A.; Skoblov, M. Comprehensive Analysis of Human microRNA-mRNA Interactome. Front. Genet. 2019, 10, 933. [Google Scholar] [CrossRef]
- Ma, B.; Wang, S.; Wu, W.; Shan, P.; Chen, Y.; Meng, J.; Xing, L.; Yun, J.; Hao, L.; Wang, X.; et al. Mechanisms of circRNA/lncRNA-miRNA interactions and applications in disease and drug research. Biomed. Pharmacother. 2023, 162, 114672. [Google Scholar] [CrossRef]
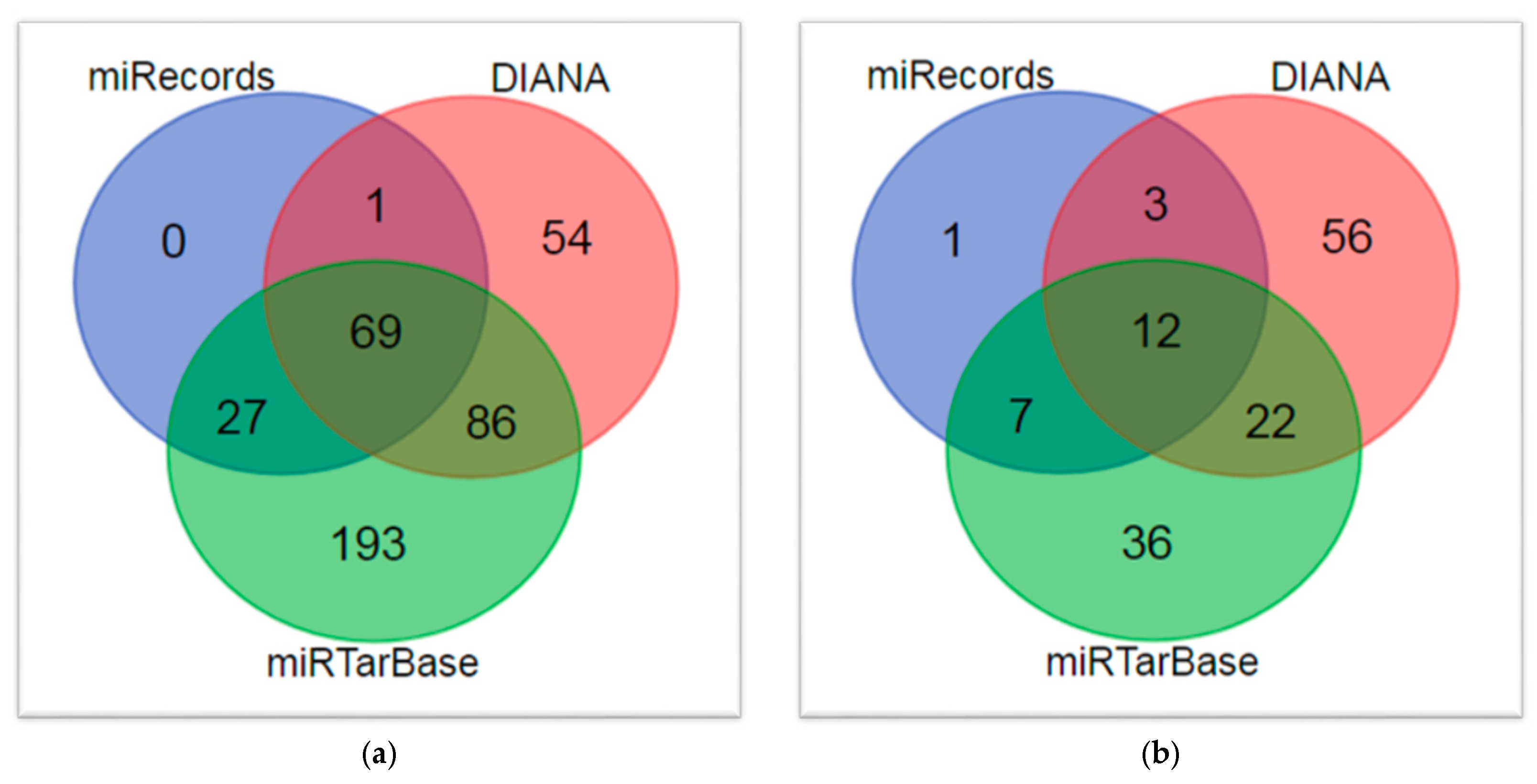
| Characteristic | PPI 1 Network |
|---|---|
| Number of edges 2 | 14,343 |
| Number of nodes 3 | 3806 |
| Diameter 4 | 11.00 |
| Average path length 5 | 3.83 |
| Clustering coefficient 6 | 0.05 |
| Modularity 7 | 0.42 |
| Number of self loops 8 | 269.00 |
| Average eccentricity 9 | 8.16 |
| Average eigenvector centrality 10 | 0.03 |
| Average number of neighbors 11 | 0.97 |
| Centralization betweenness 12 | 0.18 |
| Centralization degree 13 | 0.14 |
| Fit power law 14 | TRUE |
| Density | Total Cluster | Max Size | Avg. Size | Significant Cluster Count |
|---|---|---|---|---|
| 0.1 | 404 | 148 | 25.74 | 59 |
| 0.2 | 770 | 73 | 12.67 | 61 |
| 0.3 | 1213 | 47 | 8.2 | 257 |
| 0.4 | 1639 | 37 | 6.2 | 464 |
| 0.5 | 2093 | 27 | 4.94 | 563 |
| 0.6 | 3602 | 19 | 2.46 | 962 |
| 0.7 | 3796 | 17 | 2.49 | 1051 |
| 0.8 | 3851 | 12 | 2.39 | 1056 |
| 0.9 | 3861 | 8 | 2.38 | 1070 |
| Enriched Category | ID | Name | FDR 1 | Gene Count |
|---|---|---|---|---|
| GO: Molecular Function | GO:0050839 | Cell adhesion molecule binding | 6.15 × 10−21 | 43 |
| GO:0045296 | Cadherin binding | 4.14 × 10−19 | 33 | |
| GO:0019901 | Protein kinase binding | 2.34 × 10−15 | 43 | |
| GO:0019900 | Kinase binding | 1.50 × 10−14 | 44 | |
| GO:0044877 | Protein-containing complex binding | 1.27 × 10−11 | 54 | |
| GO:0019904 | Protein domain specific binding | 7.72 × 10−5 | 30 | |
| GO:0005102 | Signaling receptor binding 2 | 2.25 × 10−4 | 43 | |
| GO:0005158 | Insulin receptor binding | 7.29 × 10−4 | 7 | |
| GO:1990782 | Protein tyrosine kinase binding | 9.09 × 10−4 | 12 | |
| GO:0030527 | Structural constituent of chromatin | 2.01 × 10−3 | 10 | |
| GO: Biological Process | GO:0051094 | Positive regulation of developmental process | 2.14 × 10−12 | 57 |
| GO:0002009 | Morphogenesis of an epithelium | 4.09 × 10−12 | 42 | |
| GO:0000902 | Cell morphogenesis 2 | 1.05 × 10−10 | 48 | |
| GO:0048729 | Tissue morphogenesis | 1.05 × 10−10 | 43 | |
| GO:0007155 | Cell adhesion 2 | 1.25 × 10−10 | 53 | |
| GO:0022603 | Regulation of anatomical structure morphogenesis | 2.32 × 10−10 | 45 | |
| GO:0040011 | Locomotion | 7.88 × 10−10 | 53 | |
| GO:0043067 | Regulation of programmed cell death | 1.61 × 10−9 | 54 | |
| GO:0030155 | Regulation of cell adhesion 2 | 1.61 × 10−9 | 38 | |
| GO:0042981 | Regulation of apoptotic process | 2.35 × 10−9 | 53 | |
| GO: Cellular Component | GO:0031252 | Cell leading edge | 2.28 × 10−15 | 34 |
| GO:0070161 | Anchoring junction | 1.65 × 10−14 | 52 | |
| GO:0098590 | Plasma membrane region 2 | 1.18 × 10−13 | 52 | |
| GO:0045121 | Membrane raft 2 | 2.43 × 10−13 | 30 | |
| GO:0098857 | Membrane microdomain | 2.43 × 10−13 | 30 | |
| GO:0030027 | Lamellipodium | 3.46 × 10−13 | 23 | |
| GO:0005911 | Cell–cell junction | 5.31 × 10−13 | 33 | |
| GO:0043005 | Neuron projection | 1.67 × 10−11 | 53 | |
| GO:0030055 | Cell-substrate junction | 6.67 × 10−11 | 27 | |
| GO:0005925 | Focal adhesion | 2.74 × 10−10 | 26 | |
| KEGG Pathway | 83070 | Adherens junction | 4.01 × 10−9 | 15 |
| 782000 | Proteoglycans in cancer | 8.18 × 10−7 | 19 | |
| 585563 | Alcoholism | 4.68 × 10−5 | 16 | |
| 83083 | Leukocyte transendothelial migration | 7.43 × 10−5 | 13 | |
| 868086 | Rap1 signaling pathway | 2.66 × 10−4 | 16 | |
| 83109 | Endometrial cancer | 3.84 × 10−4 | 9 | |
| 83067 | Focal adhesion | 6.72 × 10−4 | 15 | |
| 101143 | Neurotrophin signaling pathway | 7.67 × 10−4 | 12 | |
| 946598 | Thyroid hormone signaling pathway | 3.57 × 10−3 | 11 | |
| 117293 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 4.35 × 10−3 | 9 | |
| Reactome Pathway | 1269326 | Interleukin-7 signaling | 2.00 × 10−7 | 10 |
| 1269507 | Signaling by Rho GTPases | 3.61 × 10−7 | 27 | |
| 1269512 | RHO GTPases activate PKNs | 1.06 × 10−6 | 14 | |
| 1269509 | RHO GTPase effectors | 1.16 × 10−6 | 22 | |
| 1270302 | Developmental biology 2 | 2.33 × 10−6 | 41 | |
| 1269340 | Hemostasis | 1.14 × 10−5 | 30 | |
| 1269811 | Mitotic prophase | 1.65 × 10−5 | 15 | |
| 1270437 | HDMs demethylate histones | 4.72 × 10−5 | 10 | |
| 1269318 | Signaling by interleukins 2 | 6.46 × 10−5 | 26 | |
| 1269602 | Formation of the beta-catenin:TCF transactivating complex | 7.43 × 10−5 | 12 | |
| PID Pathway | 138071 | PDGFR-beta signaling pathway | 6.46 × 10−5 | 10 |
| 137930 | Signaling events mediated by hepatocyte growth factor receptor (c-Met) | 7.20 × 10−5 | 11 | |
| 137940 | Signaling events mediated by VEGFR1 and VEGFR2 | 2.35 × 10−4 | 10 | |
| 137919 | N-cadherin signaling events | 2.58 × 10−4 | 8 | |
| 137970 | EGF receptor (ErbB1) signaling pathway | 6.17 × 10−4 | 7 | |
| 169348 | Signaling events mediated by focal adhesion kinase | 6.52 × 10−4 | 9 | |
| 137989 | FGF signaling pathway | 2.31 × 10−3 | 8 | |
| 138017 | Signaling events mediated by PTP1B | 3.37 × 10−3 | 8 | |
| 137915 | Signaling events regulated by Ret tyrosine kinase 2 | 6.52 × 10−3 | 7 | |
| 137977 | Neurotrophic factor-mediated Trk receptor signaling | 6.83 × 10−3 | 8 |
| MTI Network 1 | MRM 2 | Sub-MRM 3 | miRNA 4 | Gene 5 |
|---|---|---|---|---|
| DIANA | 636 | 266 | 98 | 1160 |
| miRTarbase | 58 | 23 | 81 | 407 |
| miRecords | 16 | 13 | 24 | 143 |
| Category | Subcategory | Pathway ID | Adjusted p-Value | Count 1 |
|---|---|---|---|---|
| Reactome (miRPathDB) | Cellular senescence | R-HSA-2559583 | 3.73 × 10−34 | 71 |
| Signal transduction | R-HSA-162582 | 5.54 × 10−33 | 66 | |
| Cellular responses to stress | R-HSA-2262752 | 1.95 × 10−30 | 71 | |
| PIP3 activates AKT signaling | R-HSA-1257604 | 5.59 × 10−26 | 57 | |
| Cytokine signaling in immune system | R-HSA-1280215 | 1.30 × 10−24 | 53 | |
| Signaling by interleukins | R-HSA-449147 | 3.74 × 10−22 | 51 | |
| Disease | R-HSA-1643685 | 3.79 × 10−21 | 45 | |
| Diseases of signal transduction | R-HSA-5663202 | 2.37 × 10−20 | 46 | |
| Immune system | R-HSA-168256 | 5.19 × 10−20 | 45 | |
| Oxidative stress-induced senescence | R-HSA-2559580 | 1.73 × 10−18 | 51 | |
| Developmental biology | R-HSA-1266738 | 5.94 × 10−16 | 41 | |
| Signaling by PTK6 | R-HSA-8848021 | 3.90 × 10−14 | 32 | |
| Post-translational protein modification | R-HSA-597592 | 1.37 × 10−12 | 34 | |
| Signaling by ERBB2 | R-HSA-1227986 | 2.24 × 10−11 | 26 | |
| Metabolism of proteins | R-HSA-392499 | 2.49 × 10−11 | 31 | |
| VEGFA-VEGFR2 pathway | R-HSA-4420097 | 1.15 × 10−9 | 20 | |
| SUMOylation | R-HSA-2990846 | 2.08 × 10−9 | 24 | |
| Signaling by EGFR | R-HSA-177929 | 7.80 × 10−9 | 15 | |
| SUMO E3 ligases SUMOylate target proteins | R-HSA-3108232 | 8.52 × 10−9 | 23 | |
| Downregulation of ERBB2 signaling | R-HSA-8863795 | 4.76 × 10−8 | 18 | |
| Adaptive immune system | R-HSA-1280218 | 6.79 × 10−8 | 19 | |
| Signaling by VEGF | R-HSA-194138 | 6.79 × 10−8 | 19 | |
| MAPK family signaling cascades | R-HSA-5683057 | 8.15 × 10−8 | 23 | |
| SHC1 events in ERBB2 signaling | R-HSA-1250196 | 3.96 × 10−7 | 15 | |
| Axon guidance | R-HSA-422475 | 1.11 × 10−6 | 22 | |
| SHC1 events in EGFR signaling | R-HSA-180336 | 8.22 × 10−6 | 12 | |
| SHC1 events in ERBB4 signaling | R-HSA-1250347 | 1.86 × 10−5 | 10 | |
| RET signaling | R-HSA-8853659 | 3.17 × 10−5 | 8 | |
| GRB2 events in ERBB2 signaling | R-HSA-1963640 | 5.86 × 10−5 | 10 | |
| Signaling to RAS | R-HSA-167044 | 6.64 × 10−5 | 8 | |
| Signaling by ERBB4 | R-HSA-1236394 | 8.88 × 10−5 | 13 | |
| Downstream signal transduction | R-HSA-186763 | 1.13 × 10−4 | 12 | |
| SOS-mediated signaling | R-HSA-112412 | 1.53 × 10−4 | 9 | |
| Innate immune system | R-HSA-168249 | 1.64 × 10−4 | 13 | |
| ERBB2 activates PTK6 signaling | R-HSA-8847993 | 3.34 × 10−4 | 7 | |
| ERBB2 regulates cell motility | R-HSA-6785631 | 3.83 × 10−4 | 9 | |
| GRB2 events in EGFR signaling | R-HSA-179812 | 3.83 × 10−4 | 9 | |
| PI3K events in ERBB2 signaling | R-HSA-1963642 | 6.03 × 10−4 | 7 | |
| PI3K events in ERBB4 signaling | R-HSA-1250342 | 6.03 × 10−4 | 4 | |
| GRB7 events in ERBB2 signaling | R-HSA-1306955 | 9.79 × 10−4 | 5 | |
| Signaling by SCF-KIT | R-HSA-1433557 | 1.26 × 10−3 | 10 | |
| Nuclear signaling by ERBB4 | R-HSA-1251985 | 1.57 × 10−3 | 6 | |
| Constitutive signaling by aberrant PI3K in cancer | R-HSA-2219530 | 1.65 × 10−3 | 10 | |
| rRNA processing | R-HSA-72312 | 1.65 × 10−3 | 10 | |
| FCERI mediated MAPK activation | R-HSA-2871796 | 1.83 × 10−3 | 12 | |
| Gastrin-CREB signaling pathway via PKC and MAPK | R-HSA-881907 | 2.12 × 10−3 | 9 | |
| Signaling by PDGF | R-HSA-186797 | 6.39 × 10−3 | 7 | |
| Signaling to ERKs | R-HSA-187687 | 8.02 × 10−3 | 10 | |
| IGF1R signaling cascade | R-HSA-2428924 | 1.86 × 10−2 | 8 | |
| IRS-related events triggered by IGF1R | R-HSA-2428928 | 1.86 × 10−2 | 8 | |
| VEGFR2 mediated cell proliferation | R-HSA-5218921 | 2.22 × 10−2 | 8 | |
| Signaling by leptin | R-HSA-2586552 | 2.25 × 10−2 | 3 | |
| Interleukin receptor SHC signaling | R-HSA-912526 | 2.43 × 10−2 | 2 | |
| DAP12 signaling | R-HSA-2424491 | 3.44 × 10−2 | 6 | |
| rRNA processing in the nucleus and cytosol | R-HSA-8868773 | 3.50 × 10−2 | 7 | |
| SUMOylation of DNA damage response and repair proteins | R-HSA-3108214 | 4.08 × 10−2 | 6 | |
| SUMOylation of RNA-binding proteins | R-HSA-4570464 | 4.08 × 10−2 | 7 | |
| Signaling by insulin receptor | R-HSA-74752 | 4.08 × 10−2 | 7 | |
| KEGG (miRPathDB) | MicroRNAs in cancer | hsa05206 | 1.68 × 10−42 | 98 |
| Pathways in cancer | hsa05200 | 6.85 × 10−28 | 78 | |
| ErbB signaling pathway | hsa04012 | 4.68 × 10−13 | 40 | |
| Thyroid cancer | hsa05216 | 1.29 × 10−11 | 33 | |
| Transcriptional misregulation in cancer | hsa05202 | 1.47 × 10−7 | 34 | |
| Endocytosis | hsa04144 | 4.77 × 10−6 | 22 | |
| Melanogenesis | hsa04916 | 3.44 × 10−4 | 12 |
| Subcategory | Adjusted p-Value | Count 1 |
|---|---|---|
| Gene silencing by miRNA GO:0035195 | 1.19 × 10−10 | 98 |
| mRNA binding involved in post-transcriptional gene silencing GO:1903231 | 6.99 × 10−9 | 97 |
| MiRNA mediated inhibition of translation GO:0035278 | 5.86 × 10−6 | 39 |
| Negative regulation of cell population proliferation GO:0008285 2 | 7.40 × 10−4 | 17 |
| Negative regulation of gene expression GO:0010629 2 | 7.40 × 10−4 | 21 |
| Positive regulation of ERK1 and ERK2 cascade GO:0070374 2 | 7.40 × 10−4 | 9 |
| Extracellular space GO:00056152 | 8.34 × 10−4 | 84 |
| Positive regulation of connective tissue replacement GO:1905205 2 | 1.99 × 10−3 | 8 |
| Extracellular exosome GO:0070062 2 | 3.66 × 10−3 | 15 |
| Negative regulation of cardiac muscle cell apoptotic process GO:0010667 2 | 4.25 × 10−3 | 10 |
| Positive regulation of vascular smooth muscle cell proliferation GO:1904707 | 4.25 × 10−3 | 12 |
| Negative regulation of apoptotic process GO:0043066 2 | 4.96 × 10−3 | 7 |
| Negative regulation of angiogenesis GO:0016525 | 7.43 × 10−3 | 19 |
| Positive regulation of angiogenesis GO:0045766 | 7.47 × 10−3 | 13 |
| Negative regulation of sprouting angiogenesis GO:1903671 | 8.07 × 10−3 | 12 |
| Negative regulation of cell migration GO:0030336 2 | 9.15 × 10−3 | 15 |
| Nucleus GO:0005634 2 | 1.30 × 10−2 | 6 |
| Positive regulation of apoptotic process GO:0043065 2 | 1.82 × 10−2 | 11 |
| Cytoplasm GO:0005737 2 | 2.09 × 10−2 | 8 |
| Positive regulation of protein kinase B signaling GO:0051897 2 | 2.09 × 10−2 | 8 |
| Cellular response to vascular endothelial growth factor stimulus GO:0035924 | 3.53 × 10−2 | 5 |
| Negative regulation of I-kappaB kinase/NF-kappaB signaling GO:0043124 | 3.53 × 10−2 | 5 |
| Positive regulation of metalloendopeptidase activity GO:1904685 | 3.53 × 10−2 | 5 |
| Negative regulation of protein kinase B signaling GO:0051898 | 3.60 × 10−2 | 10 |
| Negative regulation of cholesterol efflux GO:0090370 2 | 4.11 × 10−2 | 9 |
| Negative regulation of low-density lipoprotein particle clearance GO:0010989 | 4.40 × 10−2 | 6 |
| Positive regulation of cardiac muscle hypertrophy in response to stress GO:1903244 | 4.40 × 10−2 | 6 |
| Negative regulation of G1S transition of mitotic cell cycle GO:2000134 | 4.85 × 10−2 | 11 |
| Negative regulation of inflammatory response GO:0050728 | 4.85 × 10−2 | 15 |
| Predicted HSCR-Related miRNAs in This Study | miRNA Symbol | NSR Value | p-Value of NSR | TFP Value | p-Value of TFP |
|---|---|---|---|---|---|
| hsa-let-7a-5p | 113 | 1.88 × 10−23 | 218 | 1.16 × 107 | |
| hsa-miR-200b-3p | 114 | 1.36 × 10−22 | 151 | 1.40 × 1010 | |
| hsa-miR-107 | 160 | 2.30 × 10−26 | 244 | 2.33 × 106 | |
| hsa-miR-30a-5p | 116 | 5.87 × 10−24 | 174 | 8.29 × 107 | |
| hsa-miR-141-3p | 6 | 5.83 × 108 | 103 | 1.41 × 10−3 | |
| hsa-miR-195-5p | 107 | 8.01 × 10−23 | 194 | 7.92 × 106 | |
| hsa-miR-148a-3p | 41 | 6.11 × 10−4 | 126 | 1.75 × 1010 | |
| hsa-miR-218-5p | 69 | 1.62 × 10−13 | 137 | 6.68 × 109 | |
| hsa-miR-429 | 123 | 2.07 × 10−23 | 120 | 1.59 × 1012 | |
| hsa-miR-128-3p | 186 | 1.15 × 10−25 | 200 | 4.42 × 106 | |
| hsa-miR-214-3p | 60 | 3.34 × 10−12 | 93 | 1.85 × 10−2 | |
| hsa-miR-24-3p | 98 | 2.58 × 10−21 | 159 | 6.91× 108 | |
| DE miRNAs from GSE77296 | miRNA symbol | NSR value | p-value of NSR | TFP value | p-value of TFP |
| hsa-miR-146a-5p | 956 | 4.66 × 106 | 259 | 1.95 × 10−3 | |
| hsa-miR-200b-3p | 114 | 2.16 × 1010 | 151 | 3.71 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lucena-Padros, H.; Bravo-Gil, N.; Tous, C.; Rojano, E.; Seoane-Zonjic, P.; Fernández, R.M.; Ranea, J.A.G.; Antiñolo, G.; Borrego, S. Bioinformatics Prediction for Network-Based Integrative Multi-Omics Expression Data Analysis in Hirschsprung Disease. Biomolecules 2024, 14, 164. https://doi.org/10.3390/biom14020164
Lucena-Padros H, Bravo-Gil N, Tous C, Rojano E, Seoane-Zonjic P, Fernández RM, Ranea JAG, Antiñolo G, Borrego S. Bioinformatics Prediction for Network-Based Integrative Multi-Omics Expression Data Analysis in Hirschsprung Disease. Biomolecules. 2024; 14(2):164. https://doi.org/10.3390/biom14020164
Chicago/Turabian StyleLucena-Padros, Helena, Nereida Bravo-Gil, Cristina Tous, Elena Rojano, Pedro Seoane-Zonjic, Raquel María Fernández, Juan A. G. Ranea, Guillermo Antiñolo, and Salud Borrego. 2024. "Bioinformatics Prediction for Network-Based Integrative Multi-Omics Expression Data Analysis in Hirschsprung Disease" Biomolecules 14, no. 2: 164. https://doi.org/10.3390/biom14020164
APA StyleLucena-Padros, H., Bravo-Gil, N., Tous, C., Rojano, E., Seoane-Zonjic, P., Fernández, R. M., Ranea, J. A. G., Antiñolo, G., & Borrego, S. (2024). Bioinformatics Prediction for Network-Based Integrative Multi-Omics Expression Data Analysis in Hirschsprung Disease. Biomolecules, 14(2), 164. https://doi.org/10.3390/biom14020164







