Mechanisms and Functions of the RNA Polymerase II General Transcription Machinery during the Transcription Cycle
Abstract
1. Introduction
2. RNA Polymerase II
3. Preinitiation Complex (PIC) Formation and the GTFs
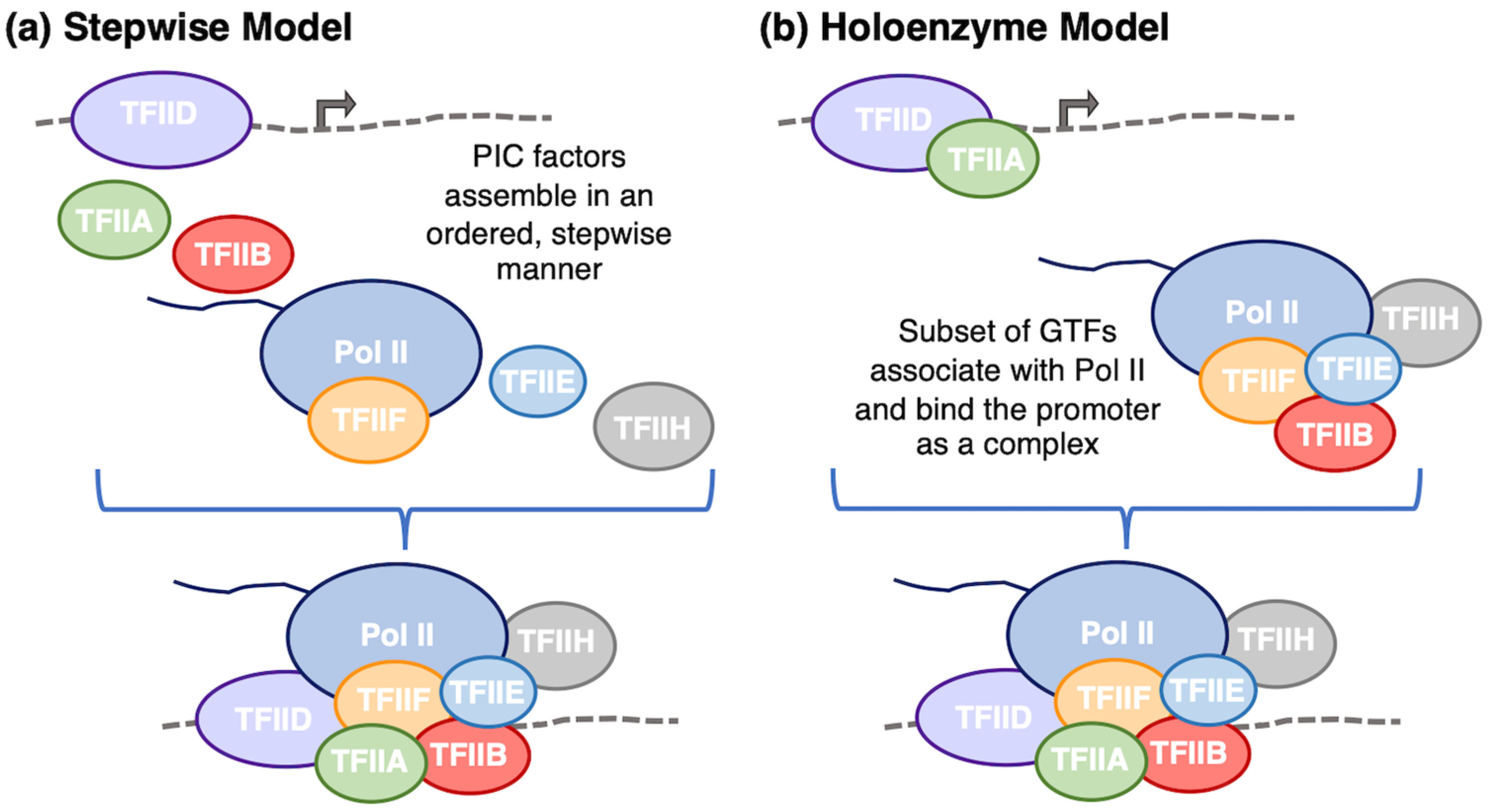
3.1. PIC Assembly Mechanisms
3.2. TFIID and the Core Promoter
3.3. TFIIA
3.4. TFIIB
3.5. TFIIF
3.6. TFIIE and TFIIH
4. Promoter Melting and Initiation
5. Early Transcription
5.1. Promoter Escape
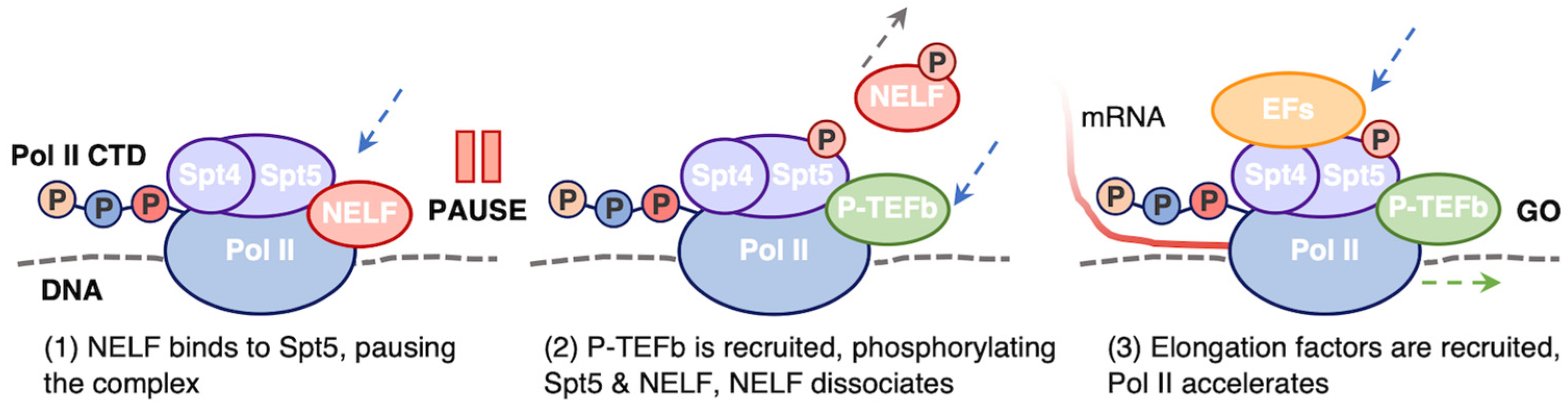
5.2. Promoter Proximal Pausing
6. Elongation
7. Termination
8. Summary and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Richter, W.F.; Nayak, S.; Iwasa, J.; Taatjes, D.J. The Mediator Complex as a Master Regulator of Transcription by RNA Polymerase II. Nat. Rev. Mol. Cell Biol. 2022, 23, 732–749. [Google Scholar] [CrossRef] [PubMed]
- Soutourina, J. Transcription Regulation by the Mediator Complex. Nat. Rev. Mol. Cell Biol. 2018, 19, 262–274. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Carey, M.; Workman, J.L. The Role of Chromatin during Transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Kujirai, T.; Kurumizaka, H. Transcription through the Nucleosome. Curr. Opin. Struct. Biol. 2020, 61, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.; Levine, M.S. Enhancer-Promoter Communication: Hubs or Loops? Curr. Opin. Genet. Dev. 2021, 67, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Kadonaga, J.T. Regulation of RNA Polymerase II Transcription by Sequence-Specific DNA Binding Factors. Cell 2004, 116, 247–257. [Google Scholar] [CrossRef]
- Malik, S.; Roeder, R.G. Regulation of the RNA Polymerase II Pre-Initiation Complex by Its Associated Coactivators. Nat. Rev. Genet. 2023, 24, 767–782. [Google Scholar] [CrossRef]
- Chen, H.; Pugh, B.F. What Do Transcription Factors Interact with? J. Mol. Biol. 2021, 433, 166883. [Google Scholar] [CrossRef]
- Cramer, P.; Armache, K.-J.; Baumli, S.; Benkert, S.; Brueckner, F.; Buchen, C.; Damsma, G.E.; Dengl, S.; Geiger, S.R.; Jasiak, A.J.; et al. Structure of Eukaryotic RNA Polymerases. Annu. Rev. Biophys. 2008, 37, 337–352. [Google Scholar] [CrossRef]
- Vannini, A.; Cramer, P. Conservation between the RNA Polymerase I, II, and III Transcription Initiation Machineries. Mol. Cell 2012, 45, 439–446. [Google Scholar] [CrossRef]
- Li, Y.; Huang, J.; Zhu, J.; Bao, L.; Wang, H.; Jiang, Y.; Tian, K.; Wang, R.; Zheng, H.; Duan, W.; et al. Targeted Protein Degradation Reveals RNA Pol II Heterogeneity and Functional Diversity. Mol. Cell 2022, 82, 3943–3959.e11. [Google Scholar] [CrossRef]
- Osman, S.; Cramer, P. Structural Biology of RNA Polymerase II Transcription: 20 Years on. Annu. Rev. Cell Dev. Biol. 2020, 36, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Schier, A.C.; Taatjes, D.J. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Genes Dev. 2020, 34, 465–488. [Google Scholar] [CrossRef]
- Aibara, S.; Schilbach, S.; Cramer, P. Structures of Mammalian RNA Polymerase II Pre-Initiation Complexes. Nature 2021, 594, 124–128. [Google Scholar] [CrossRef]
- Schier, A.C.; Taatjes, D.J. Everything at Once: Cryo-EM Yields Remarkable Insights into Human RNA Polymerase II Transcription. Nat. Struct. Mol. Biol. 2021, 28, 540–543. [Google Scholar] [CrossRef]
- Gnatt, A.L.; Cramer, P.; Fu, J.; Bushnell, D.A.; Kornberg, R.D. Structural Basis of Transcription: An RNA Polymerase II Elongation Complex at 3.3 Å Resolution. Science 2001, 292, 1876–1882. [Google Scholar] [CrossRef] [PubMed]
- Hahn, S. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Nat. Struct. Mol. Biol. 2004, 11, 394–403. [Google Scholar] [CrossRef]
- Armache, K.-J.; Kettenberger, H.; Cramer, P. Architecture of Initiation-Competent 12-Subunit RNA Polymerase II. Proc. Natl. Acad. Sci. USA 2003, 100, 6964–6968. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, D.A.; Westover, K.D.; Davis, R.E.; Kornberg, R.D. Structural Basis of Transcription: An RNA Polymerase II–TFIIB Cocrystal at 4.5 Angstroms. Science 2004, 303, 983–988. [Google Scholar] [CrossRef]
- Dienemann, C.; Schwalb, B.; Schilbach, S.; Cramer, P. Promoter Distortion and Opening in the RNA Polymerase II Cleft. Mol. Cell 2019, 73, 97–106.e4. [Google Scholar] [CrossRef]
- Bernecky, C.; Herzog, F.; Baumeister, W.; Plitzko, J.M.; Cramer, P. Structure of Transcribing Mammalian RNA Polymerase II. Nature 2016, 529, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Lyons, D.E.; McMahon, S.; Ott, M. A Combinatorial View of Old and New RNA Polymerase II Modifications. Transcription 2020, 11, 66–82. [Google Scholar] [CrossRef]
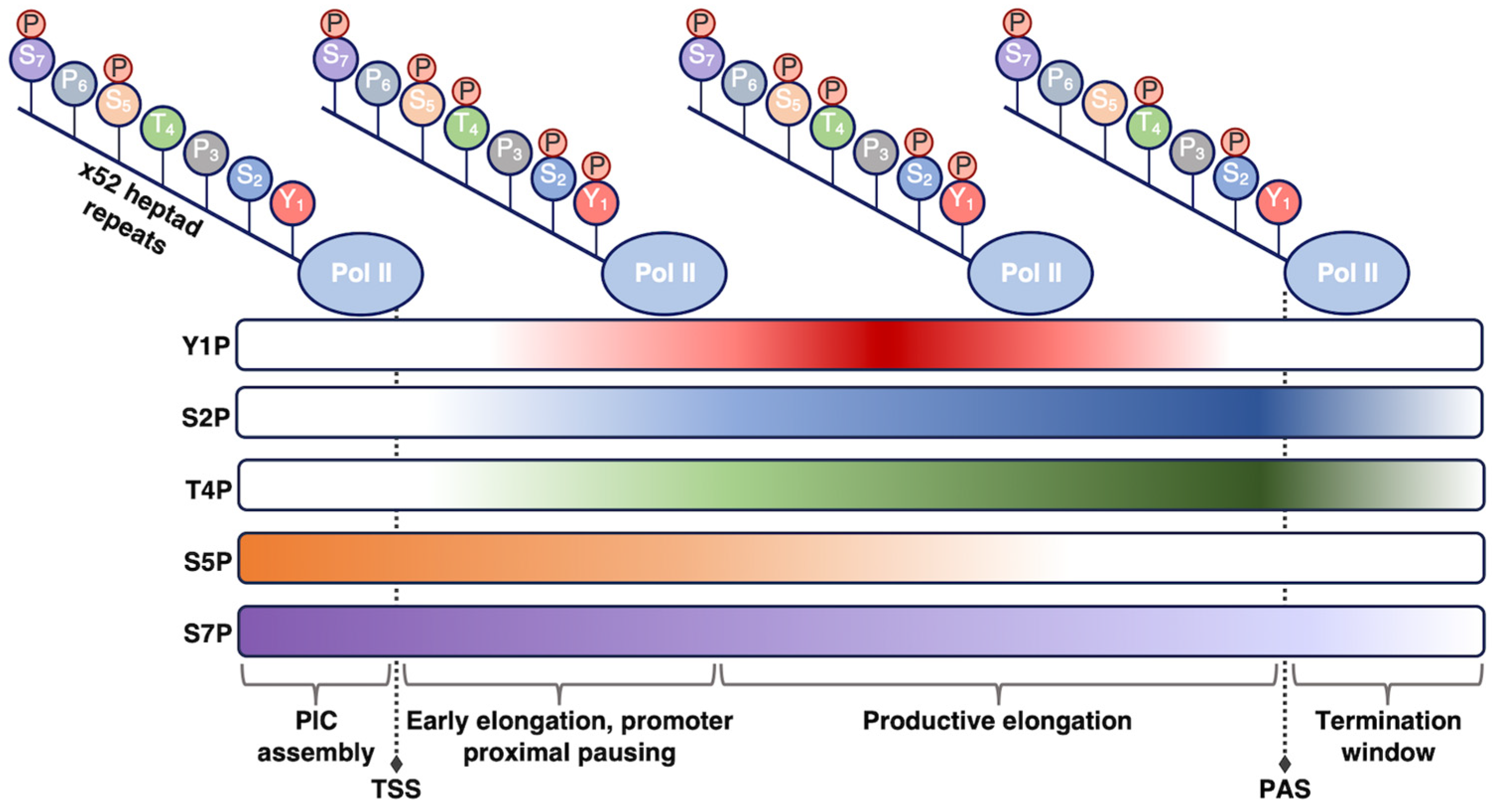
- Harlen, K.M.; Churchman, L.S. The Code and beyond: Transcription Regulation by the RNA Polymerase II Carboxy-Terminal Domain. Nat. Rev. Mol. Cell Biol. 2017, 18, 263–273. [Google Scholar] [CrossRef]
- Liu, P.; Kenney, J.M.; Stiller, J.W.; Greenleaf, A.L. Genetic Organization, Length Conservation, and Evolution of RNA Polymerase II Carboxyl-Terminal Domain. Mol. Biol. Evol. 2010, 27, 2628–2641. [Google Scholar] [CrossRef] [PubMed]
- Zehring, W.A.; Lee, J.M.; Weeks, J.R.; Jokerst, R.S.; Greenleaf, A.L. The C-Terminal Repeat Domain of RNA Polymerase II Largest Subunit Is Essential in Vivo but Is Not Required for Accurate Transcription Initiation in Vitro. Proc. Natl. Acad. Sci. USA 1988, 85, 3698–3702. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.Y.; Dahmus, M.E. The Major Late Promoter of Adenovirus-2 Is Accurately Transcribed by RNA Polymerases IIO, IIA, and IIB. J. Biol. Chem. 1989, 264, 3169–3176. [Google Scholar] [CrossRef] [PubMed]
- Gerber, H.-P.; Hagmann, M.; Seipel, K.; Georgiev, O.; West, M.A.L.; Litingtung, Y.; Schaffner, W.; Corden, J.L. RNA Polymerase II C-Terminal Domain Required for Enhancer-Driven Transcription. Nature 1995, 374, 660–662. [Google Scholar] [CrossRef] [PubMed]
- Yahia, Y.; Pigeot, A.; El Aabidine, A.Z.; Shah, N.; Karasu, N.; Forné, I.; Krebs, S.; Blum, H.; Esnault, C.; Sexton, T.; et al. RNA Polymerase II CTD Is Dispensable for Transcription and Required for Termination in Human Cells. EMBO Rep. 2023, 24, e56150. [Google Scholar] [CrossRef]
- Garg, A.; Sanchez, A.M.; Schwer, B.; Shuman, S. Transcriptional Profiling of Fission Yeast RNA Polymerase II CTD Mutants. RNA 2021, 27, 560–570. [Google Scholar] [CrossRef]
- Singh, N.; Asalam, M.; Ansari, M.O.; Gerasimova, N.S.; Studitsky, V.M.; Akhtar, M.S. Transcription by RNA Polymerase II and the CTD-Chromatin Crosstalk. Biochem. Biophys. Res. Commun. 2022, 599, 81–86. [Google Scholar] [CrossRef]
- Maita, H.; Nakagawa, S. What Is the Switch for Coupling Transcription and Splicing? RNA Polymerase II C-terminal Domain Phosphorylation, Phase Separation and Beyond. WIREs RNA 2020, 11, e1574. [Google Scholar] [CrossRef]
- Jeronimo, C.; Bataille, A.R.; Robert, F. The Writers, Readers, and Functions of the RNA Polymerase II C-Terminal Domain Code. Chem. Rev. 2013, 113, 8491–8522. [Google Scholar] [CrossRef] [PubMed]
- Davidson, L.; Muniz, L.; West, S. 3′ End Formation of Pre-mRNA and Phosphorylation of Ser2 on the RNA Polymerase II CTD Are Reciprocally Coupled in Human Cells. Genes Dev. 2014, 28, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Nojima, T.; Gomes, T.; Grosso, A.R.F.; Kimura, H.; Dye, M.J.; Dhir, S.; Carmo-Fonseca, M.; Proudfoot, N.J. Mammalian NET-Seq Reveals Genome-Wide Nascent Transcription Coupled to RNA Processing. Cell 2015, 161, 526–540. [Google Scholar] [CrossRef]
- Ahn, S.H.; Kim, M.; Buratowski, S. Phosphorylation of Serine 2 within the RNA Polymerase II C-Terminal Domain Couples Transcription and 3′ End Processing. Mol. Cell 2004, 13, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Eick, D.; Bensaude, O. CTD Serine-2 Plays a Critical Role in Splicing and Termination Factor Recruitment to RNA Polymerase II in Vivo. Nucleic Acids Res. 2013, 41, 1591–1603. [Google Scholar] [CrossRef]
- Heidemann, M.; Eick, D. Tyrosine-1 and Threonine-4 Phosphorylation Marks Complete the RNA Polymerase II CTD Phospho-Code. RNA Biol. 2012, 9, 1144–1146. [Google Scholar] [CrossRef][Green Version]
- Yurko, N.M.; Manley, J.L. The RNA Polymerase II CTD “Orphan” Residues: Emerging Insights into the Functions of Tyr-1, Thr-4, and Ser-7. Transcription 2018, 9, 30–40. [Google Scholar] [CrossRef]
- Thomas, M.C.; Chiang, C.-M. The General Transcription Machinery and General Cofactors. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 105–178. [Google Scholar] [CrossRef]
- Luse, D.S. The RNA Polymerase II Preinitiation Complex: Through What Pathway Is the Complex Assembled? Transcription 2014, 5, e27050. [Google Scholar] [CrossRef]
- Flores, O.; Lu, H.; Reinberg, D. Factors Involved in Specific Transcription by Mammalian RNA Polymerase II. Identification and Characterization of Factor IIH. J. Biol. Chem. 1992, 267, 2786–2793. [Google Scholar] [CrossRef] [PubMed]
- Buratowski, S.; Hahn, S.; Guarente, L.; Sharp, P.A. Five Intermediate Complexes in Transcription Initiation by RNA Polymerase II. Cell 1989, 56, 549–561. [Google Scholar] [CrossRef]
- Ossipow, V.; Tassan, J.-P.; Nigg, E.A.; Schibler, U. A Mammalian RNA Polymerase II Holoenzyme Containing All Components Required for Promoter-Specific Transcription Initiation. Cell 1995, 83, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-Y.; Chiang, C.-M. Properties of PC4 and an RNA Polymerase II Complex in Directing Activated and Basal Transcription in Vitro. J. Biol. Chem. 1998, 273, 12492–12498. [Google Scholar] [CrossRef] [PubMed]
- Cramer, P. Organization and Regulation of Gene Transcription. Nature 2019, 573, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Hantsche, M.; Cramer, P. Conserved RNA Polymerase II Initiation Complex Structure. Curr. Opin. Struct. Biol. 2017, 47, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.-W.; Maekiniemi, A.; Singer, R.H.; Sato, H. Real-Time Single-Molecule Imaging of Transcriptional Regulatory Networks in Living Cells. Nat. Rev. Genet. 2024. [Google Scholar] [CrossRef]
- Wagh, K.; Stavreva, D.A.; Upadhyaya, A.; Hager, G.L. Transcription Factor Dynamics: One Molecule at a Time. Annu. Rev. Cell Dev. Biol. 2023, 39, 277–305. [Google Scholar] [CrossRef]
- Wang, Z.; Deng, W. Dynamic Transcription Regulation at the Single-Molecule Level. Dev. Biol. 2022, 482, 67–81. [Google Scholar] [CrossRef]
- Lionnet, T.; Wu, C. Single-Molecule Tracking of Transcription Protein Dynamics in Living Cells: Seeing Is Believing, but What Are We Seeing? Curr. Opin. Genet. Dev. 2021, 67, 94–102. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Z.; Li, L.; Chen, B.-C.; Revyakin, A.; Hajj, B.; Legant, W.; Dahan, M.; Lionnet, T.; Betzig, E.; et al. Single-Molecule Dynamics of Enhanceosome Assembly in Embryonic Stem Cells. Cell 2014, 156, 1274–1285. [Google Scholar] [CrossRef]
- Hipp, L.; Beer, J.; Kuchler, O.; Reisser, M.; Sinske, D.; Michaelis, J.; Gebhardt, J.C.M.; Knöll, B. Single-Molecule Imaging of the Transcription Factor SRF Reveals Prolonged Chromatin-Binding Kinetics upon Cell Stimulation. Proc. Natl. Acad. Sci. USA 2019, 116, 880–889. [Google Scholar] [CrossRef]
- Hansen, A.S.; Amitai, A.; Cattoglio, C.; Tjian, R.; Darzacq, X. Guided Nuclear Exploration Increases CTCF Target Search Efficiency. Nat. Chem. Biol. 2020, 16, 257–266. [Google Scholar] [CrossRef]
- Izeddin, I.; Récamier, V.; Bosanac, L.; Cissé, I.I.; Boudarene, L.; Dugast-Darzacq, C.; Proux, F.; Bénichou, O.; Voituriez, R.; Bensaude, O.; et al. Single-Molecule Tracking in Live Cells Reveals Distinct Target-Search Strategies of Transcription Factors in the Nucleus. eLife 2014, 3, e02230. [Google Scholar] [CrossRef]
- Paakinaho, V.; Presman, D.M.; Ball, D.A.; Johnson, T.A.; Schiltz, R.L.; Levitt, P.; Mazza, D.; Morisaki, T.; Karpova, T.S.; Hager, G.L. Single-Molecule Analysis of Steroid Receptor and Cofactor Action in Living Cells. Nat. Commun. 2017, 8, 15896. [Google Scholar] [CrossRef]
- Zhang, Z.; English, B.P.; Grimm, J.B.; Kazane, S.A.; Hu, W.; Tsai, A.; Inouye, C.; You, C.; Piehler, J.; Schultz, P.G.; et al. Rapid Dynamics of General Transcription Factor TFIIB Binding during Preinitiation Complex Assembly Revealed by Single-Molecule Analysis. Genes Dev. 2016, 30, 2106–2118. [Google Scholar] [CrossRef]
- Ly, E.; Powell, A.E.; Goodrich, J.A.; Kugel, J.F. Release of Human TFIIB from Actively Transcribing Complexes Is Triggered upon Synthesis of 7- and 9-Nt RNAs. J. Mol. Biol. 2020, 432, 4049–4060. [Google Scholar] [CrossRef]
- Baek, I.; Friedman, L.J.; Gelles, J.; Buratowski, S. Single-Molecule Studies Reveal Branched Pathways for Activator-Dependent Assembly of RNA Polymerase II Pre-Initiation Complexes. Mol. Cell 2021, 81, 3576–3588.e6. [Google Scholar] [CrossRef]
- Hou, T.Y.; Kraus, W.L. Come One, Come All? Re-Evaluating RNA Polymerase II Pre-Initiation Complex Assembly Using Single-Molecule Microscopy. Mol. Cell 2021, 81, 3443–3445. [Google Scholar] [CrossRef]
- Nguyen, V.Q.; Ranjan, A.; Liu, S.; Tang, X.; Ling, Y.H.; Wisniewski, J.; Mizuguchi, G.; Li, K.Y.; Jou, V.; Zheng, Q.; et al. Spatiotemporal Coordination of Transcription Preinitiation Complex Assembly in Live Cells. Mol. Cell 2021, 81, 3560–3575.e6. [Google Scholar] [CrossRef]
- Li, J.; Dong, A.; Saydaminova, K.; Chang, H.; Wang, G.; Ochiai, H.; Yamamoto, T.; Pertsinidis, A. Single-Molecule Nanoscopy Elucidates RNA Polymerase II Transcription at Single Genes in Live Cells. Cell 2019, 178, 491–506.e28. [Google Scholar] [CrossRef]
- Stasevich, T.J.; Hayashi-Takanaka, Y.; Sato, Y.; Maehara, K.; Ohkawa, Y.; Sakata-Sogawa, K.; Tokunaga, M.; Nagase, T.; Nozaki, N.; McNally, J.G.; et al. Regulation of RNA Polymerase II Activation by Histone Acetylation in Single Living Cells. Nature 2014, 516, 272–275. [Google Scholar] [CrossRef]
- Steurer, B.; Janssens, R.C.; Geverts, B.; Geijer, M.E.; Wienholz, F.; Theil, A.F.; Chang, J.; Dealy, S.; Pothof, J.; Van Cappellen, W.A.; et al. Live-Cell Analysis of Endogenous GFP-RPB1 Uncovers Rapid Turnover of Initiating and Promoter-Paused RNA Polymerase II. Proc. Natl. Acad. Sci. USA 2018, 115, E4368–E4376. [Google Scholar] [CrossRef]
- Tunnacliffe, E.; Chubb, J.R. What Is a Transcriptional Burst? Trends Genet. 2020, 36, 288–297. [Google Scholar] [CrossRef]
- Cisse, I.I.; Izeddin, I.; Causse, S.Z.; Boudarene, L.; Senecal, A.; Muresan, L.; Dugast-Darzacq, C.; Hajj, B.; Dahan, M.; Darzacq, X. Real-Time Dynamics of RNA Polymerase II Clustering in Live Human Cells. Science 2013, 341, 664–667. [Google Scholar] [CrossRef]
- Tantale, K.; Mueller, F.; Kozulic-Pirher, A.; Lesne, A.; Victor, J.-M.; Robert, M.-C.; Capozi, S.; Chouaib, R.; Bäcker, V.; Mateos-Langerak, J.; et al. A Single-Molecule View of Transcription Reveals Convoys of RNA Polymerases and Multi-Scale Bursting. Nat. Commun. 2016, 7, 12248. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator Condensation at Super-Enhancers Links Phase Separation and Gene Control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef]
- Cho, W.-K.; Spille, J.-H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA Polymerase II Clusters Associate in Transcription-Dependent Condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef]
- Du, M.; Stitzinger, S.H.; Spille, J.-H.; Cho, W.-K.; Lee, C.; Hijaz, M.; Quintana, A.; Cissé, I.I. Direct Observation of a Condensate Effect on Super-Enhancer Controlled Gene Bursting. Cell 2024, 187, 331–344.e17. [Google Scholar] [CrossRef]
- Palacio, M.; Taatjes, D.J. Merging Established Mechanisms with New Insights: Condensates, Hubs, and the Regulation of RNA Polymerase II Transcription. J. Mol. Biol. 2022, 434, 167216. [Google Scholar] [CrossRef]
- Kimura, H.; Sato, Y. Imaging Transcription Elongation Dynamics by New Technologies Unveils the Organization of Initiation and Elongation in Transcription Factories. Curr. Opin. Cell Biol. 2022, 74, 71–79. [Google Scholar] [CrossRef]
- Luse, D.S.; Parida, M.; Spector, B.M.; Nilson, K.A.; Price, D.H. A Unified View of the Sequence and Functional Organization of the Human RNA Polymerase II Promoter. Nucleic Acids Res. 2020, 48, 7767–7785. [Google Scholar] [CrossRef]
- Starr, D.B.; Hawley, D.K. TFIID Binds in the Minor Groove of the TATA Box. Cell 1991, 67, 1231–1240. [Google Scholar] [CrossRef]
- Santana, J.F.; Collins, G.S.; Parida, M.; Luse, D.S.; Price, D.H. Differential Dependencies of Human RNA Polymerase II Promoters on TBP, TAF1, TFIIB and XPB. Nucleic Acids Res. 2022, 50, 9127–9148. [Google Scholar] [CrossRef]
- Nikolov, D.B.; Chen, H.; Halay, E.D.; Hoffman, A.; Roeder, R.G.; Burley, S.K. Crystal Structure of a Human TATA Box-Binding Protein/TATA Element Complex. Proc. Natl. Acad. Sci. USA 1996, 93, 4862–4867. [Google Scholar] [CrossRef]
- Starr, B.D.; Hoopes, B.C.; Hawley, D.K. DNA Bending Is an Important Component of Site-Specific Recognition by the TATA Binding Protein. J. Mol. Biol. 1995, 250, 434–446. [Google Scholar] [CrossRef]
- White, R.; Jackson, S. The TATA-Binding Protein: A Central Role in Transcription by RNA Polymerases I, II and III. Trends Genet. 1992, 8, 284–288. [Google Scholar] [CrossRef]
- Chen, X.; Qi, Y.; Wu, Z.; Wang, X.; Li, J.; Zhao, D.; Hou, H.; Li, Y.; Yu, Z.; Liu, W.; et al. Structural Insights into Preinitiation Complex Assembly on Core Promoters. Science 2021, 372, eaba8490. [Google Scholar] [CrossRef]
- Gershenzon, N.I.; Ioshikhes, I.P. Synergy of Human Pol II Core Promoter Elements Revealed by Statistical Sequence Analysis. Bioinformatics 2005, 21, 1295–1300. [Google Scholar] [CrossRef]
- Yang, C.; Bolotin, E.; Jiang, T.; Sladek, F.M.; Martinez, E. Prevalence of the Initiator over the TATA Box in Human and Yeast Genes and Identification of DNA Motifs Enriched in Human TATA-Less Core Promoters. Gene 2007, 389, 52–65. [Google Scholar] [CrossRef]
- Suzuki, Y.; Tsunoda, T.; Sese, J.; Taira, H.; Mizushima-Sugano, J.; Hata, H.; Ota, T.; Isogai, T.; Tanaka, T.; Nakamura, Y.; et al. Identification and Characterization of the Potential Promoter Regions of 1031 Kinds of Human Genes. Genome Res. 2001, 11, 677–684. [Google Scholar] [CrossRef]
- Vo Ngoc, L.; Cassidy, C.J.; Huang, C.Y.; Duttke, S.H.C.; Kadonaga, J.T. The Human Initiator Is a Distinct and Abundant Element That Is Precisely Positioned in Focused Core Promoters. Genes Dev. 2017, 31, 6–11. [Google Scholar] [CrossRef]
- Zhang, M.Q. Identification of Human Gene Core Promoters in Silico. Genome Res. 1998, 8, 319–326. [Google Scholar] [CrossRef]
- Vo Ngoc, L.; Wang, Y.-L.; Kassavetis, G.A.; Kadonaga, J.T. The Punctilious RNA Polymerase II Core Promoter. Genes Dev. 2017, 31, 1289–1301. [Google Scholar] [CrossRef]
- Chalkley, G.E. DNA Binding Site Selection by RNA Polymerase II TAFs: A TAFII250-TAFII150 Complex Recognizes the Initiator. EMBO J. 1999, 18, 4835–4845. [Google Scholar] [CrossRef]
- Dreos, R.; Sloutskin, A.; Malachi, N.; Ideses, D.; Bucher, P.; Juven-Gershon, T. Computational Identification and Experimental Characterization of Preferred Downstream Positions in Human Core Promoters. PLOS Comput. Biol. 2021, 17, e1009256. [Google Scholar] [CrossRef]
- Vo Ngoc, L.; Huang, C.Y.; Cassidy, C.J.; Medrano, C.; Kadonaga, J.T. Identification of the Human DPR Core Promoter Element Using Machine Learning. Nature 2020, 585, 459–463. [Google Scholar] [CrossRef]
- Theisen, J.W.M.; Lim, C.Y.; Kadonaga, J.T. Three Key Subregions Contribute to the Function of the Downstream RNA Polymerase II Core Promoter. Mol. Cell. Biol. 2010, 30, 3471–3479. [Google Scholar] [CrossRef]
- Louder, R.K.; He, Y.; López-Blanco, J.R.; Fang, J.; Chacón, P.; Nogales, E. Structure of Promoter-Bound TFIID and Model of Human Pre-Initiation Complex Assembly. Nature 2016, 531, 604–609. [Google Scholar] [CrossRef]
- Joo, Y.J.; Ficarro, S.B.; Soares, L.M.; Chun, Y.; Marto, J.A.; Buratowski, S. Downstream Promoter Interactions of TFIID TAFs Facilitate Transcription Reinitiation. Genes Dev. 2017, 31, 2162–2174. [Google Scholar] [CrossRef]
- Nogales, E.; Louder, R.K.; He, Y. Structural Insights into the Eukaryotic Transcription Initiation Machinery. Annu. Rev. Biophys. 2017, 46, 59–83. [Google Scholar] [CrossRef] [PubMed]
- Fant, C.B.; Levandowski, C.B.; Gupta, K.; Maas, Z.L.; Moir, J.; Rubin, J.D.; Sawyer, A.; Esbin, M.N.; Rimel, J.K.; Luyties, O.; et al. TFIID Enables RNA Polymerase II Promoter-Proximal Pausing. Mol. Cell 2020, 78, 785–793.e8. [Google Scholar] [CrossRef] [PubMed]
- Sloutskin, A.; Shir-Shapira, H.; Freiman, R.N.; Juven-Gershon, T. The Core Promoter Is a Regulatory Hub for Developmental Gene Expression. Front. Cell Dev. Biol. 2021, 9, 666508. [Google Scholar] [CrossRef] [PubMed]
- Haberle, V.; Stark, A. Eukaryotic Core Promoters and the Functional Basis of Transcription Initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, A.; Hollinger, C.; Willgenss, D.; Müller, F.; Devys, D.; Tora, L. Transcription Factor IID Parks and Drives Preinitiation Complexes at Sharp or Broad Promoters. Trends Biochem. Sci. 2023, 48, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Serebreni, L.; Pleyer, L.; Haberle, V.; Hendy, O.; Vlasova, A.; Loubiere, V.; Nemčko, F.; Bergauer, K.; Roitinger, E.; Mechtler, K.; et al. Functionally Distinct Promoter Classes Initiate Transcription via Different Mechanisms Reflected in Focused versus Dispersed Initiation Patterns. EMBO J. 2023, 42, e113519. [Google Scholar] [CrossRef] [PubMed]
- Grzechnik, P.; Tan-Wong, S.M.; Proudfoot, N.J. Terminate and Make a Loop: Regulation of Transcriptional Directionality. Trends Biochem. Sci. 2014, 39, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, D.N.; Iyer, V.R. The Determinants of Directionality in Transcriptional Initiation. Trends Genet. 2016, 32, 322–333. [Google Scholar] [CrossRef]
- Seila, A.C.; Calabrese, J.M.; Levine, S.S.; Yeo, G.W.; Rahl, P.B.; Flynn, R.A.; Young, R.A.; Sharp, P.A. Divergent Transcription from Active Promoters. Science 2008, 322, 1849–1851. [Google Scholar] [CrossRef]
- Preker, P.; Nielsen, J.; Kammler, S.; Lykke-Andersen, S.; Christensen, M.S.; Mapendano, C.K.; Schierup, M.H.; Jensen, T.H. RNA Exosome Depletion Reveals Transcription Upstream of Active Human Promoters. Science 2008, 322, 1851–1854. [Google Scholar] [CrossRef]
- Core, L.J.; Waterfall, J.J.; Lis, J.T. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science 2008, 322, 1845–1848. [Google Scholar] [CrossRef] [PubMed]
- Andersson, R.; Gebhard, C.; Miguel-Escalada, I.; Hoof, I.; Bornholdt, J.; Boyd, M.; Chen, Y.; Zhao, X.; Schmidl, C.; Suzuki, T.; et al. An Atlas of Active Enhancers across Human Cell Types and Tissues. Nature 2014, 507, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-K.; Hemberg, M.; Gray, J.M.; Costa, A.M.; Bear, D.M.; Wu, J.; Harmin, D.A.; Laptewicz, M.; Barbara-Haley, K.; Kuersten, S.; et al. Widespread Transcription at Neuronal Activity-Regulated Enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef]
- He, Y.; Yan, C.; Fang, J.; Inouye, C.; Tjian, R.; Ivanov, I.; Nogales, E. Near-Atomic Resolution Visualization of Human Transcription Promoter Opening. Nature 2016, 533, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Høiby, T.; Zhou, H.; Mitsiou, D.J.; Stunnenberg, H.G. A Facelift for the General Transcription Factor TFIIA. Biochim. Biophys. Acta BBA Gene Struct. Expr. 2007, 1769, 429–436. [Google Scholar] [CrossRef]
- Wang, W.; Gralla, J.D.; Carey, M. The Acidic Activator GAL4-AH Can Stimulate Polymerase II Transcription by Promoting Assembly of a Closed Complex Requiring TFIID and TFIIA. Genes Dev. 1992, 6, 1716–1727. [Google Scholar] [CrossRef][Green Version]
- Lee, D.K.; DeJong, J.; Hashimoto, S.; Horikoshi, M.; Roeder, R.G. TFIIA Induces Conformational Changes in TFIID via Interactions with the Basic Repeat. Mol. Cell. Biol. 1992, 12, 5189–5196. [Google Scholar] [CrossRef] [PubMed]
- Imbalzano, A.N.; Zaret, K.S.; Kingston, R.E. Transcription Factor (TF) IIB and TFIIA Can Independently Increase the Affinity of the TATA-Binding Protein for DNA. J. Biol. Chem. 1994, 269, 8280–8286. [Google Scholar] [CrossRef]
- Coleman, R.A.; Taggart, A.K.P.; Burma, S.; Chicca, J.J.; Pugh, B.F. TFIIA Regulates TBP and TFIID Dimers. Mol. Cell 1999, 4, 451–457. [Google Scholar] [CrossRef]
- Mitsiou, D.J.; Stunnenberg, H.G. TAC, a TBP-sans-TAFs Complex Containing the Unprocessed TFIIAαβ Precursor and the TFIIAγ Subunit. Mol. Cell 2000, 6, 527–537. [Google Scholar] [CrossRef]
- Kang, J.J.; Auble, D.T.; Ranish, J.A.; Hahn, S. Analysis of the Yeast Transcription Factor TFIIA: Distinct Functional Regions and a Polymerase II-Specific Role in Basal and Activated Transcription. Mol. Cell. Biol. 1995, 15, 1234–1243. [Google Scholar] [CrossRef][Green Version]
- Liu, Q.; Gabriel, S.E.; Roinick, K.L.; Ward, R.D.; Arndt, K.M. Analysis of TFIIA Function In Vivo: Evidence for a Role in TATA-Binding Protein Recruitment and Gene-Specific Activation. Mol. Cell. Biol. 1999, 19, 8673–8685. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Deng, W.; Roberts, S.G.E. TFIIB and the Regulation of Transcription by RNA Polymerase II. Chromosoma 2007, 116, 417–429. [Google Scholar] [CrossRef]
- Deng, W.; Roberts, S.G.E. A Core Promoter Element Downstream of the TATA Box That Is Recognized by TFIIB. Genes Dev. 2005, 19, 2418–2423. [Google Scholar] [CrossRef]
- Lagrange, T.; Kapanidis, A.N.; Tang, H.; Reinberg, D.; Ebright, R.H. New Core Promoter Element in RNA Polymerase II-Dependent Transcription: Sequence-Specific DNA Binding by Transcription Factor IIB. Genes Dev. 1998, 12, 34–44. [Google Scholar] [CrossRef]
- Nikolov, D.B.; Chen, H.; Halay, E.D.; Usheva, A.A.; Hisatake, K.; Lee, D.K.; Roeder, R.G.; Burley, S.K. Crystal Structure of a TFIIB–TBP–TATA-Element Ternary Complex. Nature 1995, 377, 119–128. [Google Scholar] [CrossRef]
- Orphanides, G.; Lagrange, T.; Reinberg, D. The General Transcription Factors of RNA Polymerase II. Genes Dev. 1996, 10, 2657–2683. [Google Scholar] [CrossRef]
- Pardee, T.S.; Bangur, C.S.; Ponticelli, A.S. The N-Terminal Region of Yeast TFIIB Contains Two Adjacent Functional Domains Involved in Stable RNA Polymerase II Binding and Transcription Start Site Selection. J. Biol. Chem. 1998, 273, 17859–17864. [Google Scholar] [CrossRef]
- Elsby, L.M.; Roberts, S.G.E. The Role of TFIIB Conformation in Transcriptional Regulation. Biochem. Soc. Trans. 2004, 32, 1098–1099. [Google Scholar] [CrossRef]
- Sainsbury, S.; Niesser, J.; Cramer, P. Structure and Function of the Initially Transcribing RNA Polymerase II–TFIIB Complex. Nature 2013, 493, 437–440. [Google Scholar] [CrossRef]
- Flores, O.; Maldonado, E.; Reinberg, D. Factors Involved in Specific Transcription by Mammalian RNA Polymerase II. Factors IIE and IIF Independently Interact with RNA Polymerase II. J. Biol. Chem. 1989, 264, 8913–8921. [Google Scholar] [CrossRef] [PubMed]
- Flores, O.; Lu, H.; Killeen, M.; Greenblatt, J.; Burton, Z.F.; Reinberg, D. The Small Subunit of Transcription Factor IIF Recruits RNA Polymerase II into the Preinitiation Complex. Proc. Natl. Acad. Sci. USA 1991, 88, 9999–10003. [Google Scholar] [CrossRef]
- Price, D.H.; Sluder, A.E.; Greenleaf, A.L. Dynamic Interaction between a Drosophila Transcription Factor and RNA Polymerase II. Mol. Cell. Biol. 1989, 9, 1465–1475. [Google Scholar] [CrossRef] [PubMed]
- Killeen, M.T.; Greenblatt, J.F. The General Transcription Factor RAP30 Binds to RNA Polymerase II and Prevents It from Binding Nonspecifically to DNA. Mol. Cell Biol. 1992, 12, 30–37. [Google Scholar] [CrossRef]
- Robert, F.; Forget, D.; Li, J.; Greenblatt, J.; Coulombe, B. Localization of Subunits of Transcription Factors IIE and IIF Immediately Upstream of the Transcriptional Initiation Site of the Adenovirus Major Late Promoter. J. Biol. Chem. 1996, 271, 8517–8520. [Google Scholar] [CrossRef]
- He, Y.; Fang, J.; Taatjes, D.J.; Nogales, E. Structural Visualization of Key Steps in Human Transcription Initiation. Nature 2013, 495, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Groft, C.M.; Uljon, S.N.; Wang, R.; Werner, M.H. Structural Homology between the Rap30 DNA-Binding Domain and Linker Histone H5: Implications for Preinitiation Complex Assembly. Proc. Natl. Acad. Sci. USA 1998, 95, 9117–9122. [Google Scholar] [CrossRef]
- Robert, F.; Douziech, M.; Forget, D.; Egly, J.-M.; Greenblatt, J.; Burton, Z.F.; Coulombe, B. Wrapping of Promoter DNA around the RNA Polymerase II Initiation Complex Induced by TFIIF. Mol. Cell 1998, 2, 341–351. [Google Scholar] [CrossRef]
- Shilatifard, A.; Conaway, R.C.; Conaway, J.W. The RNA Polymerase II Elongation Complex. Annu. Rev. Biochem. 2003, 72, 693–715. [Google Scholar] [CrossRef]
- Yan, Q.; Moreland, R.J.; Conaway, J.W.; Conaway, R.C. Dual Roles for Transcription Factor IIF in Promoter Escape by RNA Polymerase II. J. Biol. Chem. 1999, 274, 35668–35675. [Google Scholar] [CrossRef][Green Version]
- Elmendorf, B.J.; Shilatifard, A.; Yan, Q.; Conaway, J.W.; Conaway, R.C. Transcription Factors TFIIF, ELL, and Elongin Negatively Regulate SII-Induced Nascent Transcript Cleavage by Non-Arrested RNA Polymerase II Elongation Intermediates. J. Biol. Chem. 2001, 276, 23109–23114. [Google Scholar] [CrossRef] [PubMed]
- Joo, Y.J.; Ficarro, S.B.; Chun, Y.; Marto, J.A.; Buratowski, S. In Vitro Analysis of RNA Polymerase II Elongation Complex Dynamics. Genes Dev. 2019, 33, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Zawel, L.; Kumar, K.P.; Reinberg, D. Recycling of the General Transcription Factors during RNA Polymerase II Transcription. Genes Dev. 1995, 9, 1479–1490. [Google Scholar] [CrossRef]
- Cojocaru, M.; Jeronimo, C.; Forget, D.; Bouchard, A.; Bergeron, D.; Côte, P.; Poirier, G.G.; Greenblatt, J.; Coulombe, B. Genomic Location of the Human RNA Polymerase II General Machinery: Evidence for a Role of TFIIF and Rpb7 at Both Early and Late Stages of Transcription. Biochem. J. 2008, 409, 139–147. [Google Scholar] [CrossRef]
- Chen, Y.; Kokic, G.; Dienemann, C.; Dybkov, O.; Urlaub, H.; Cramer, P. Structure of the Transcribing RNA Polymerase II–Elongin Complex. Nat. Struct. Mol. Biol. 2023, 30, 1925–1935. [Google Scholar] [CrossRef]
- Chen, Y.; Cramer, P. RNA Polymerase II Elongation Factors Use Conserved Regulatory Mechanisms. Curr. Opin. Struct. Biol. 2024, 84, 102766. [Google Scholar] [CrossRef]
- Ohkuma, Y.; Hashimoto, S.; Wang, C.K.; Horikoshi, M.; Roeder, R.G. Analysis of the Role of TFIIE in Basal Transcription and TFIIH-Mediated Carboxy-Terminal Domain Phosphorylation through Structure-Function Studies of TFIIE-α. Mol. Cell. Biol. 1995, 15, 4856–4866. [Google Scholar] [CrossRef]
- Ohkuma, Y.; Roeder, R.G. Regulation of TFIIH ATPase and Kinase Activities by TFIIE during Active Initiation Complex Formation. Nature 1994, 368, 160–163. [Google Scholar] [CrossRef]
- Okamoto, T.; Yamamoto, S.; Watanabe, Y.; Ohta, T.; Hanaoka, F.; Roeder, R.G.; Ohkuma, Y. Analysis of the Role of TFIIE in Transcriptional Regulation through Structure-Function Studies of the TFIIEβ Subunit. J. Biol. Chem. 1998, 273, 19866–19876. [Google Scholar] [CrossRef]
- Serizawa, H.; Conaway, J.W.; Conaway, R.C. An Oligomeric Form of the Large Subunit of Transcription Factor (TF) IIE Activates Phosphorylation of the RNA Polymerase II Carboxyl-Terminal Domain by TFIIH. J. Biol. Chem. 1994, 269, 20750–20756. [Google Scholar] [CrossRef]
- Kuldell, N.H.; Buratowski, S. Genetic Analysis of the Large Subunit of Yeast Transcription Factor IIE Reveals Two Regions with Distinct Functions. Mol. Cell. Biol. 1997, 17, 5288–5298. [Google Scholar] [CrossRef] [PubMed]
- Miwa, K.; Kojima, R.; Obita, T.; Ohkuma, Y.; Tamura, Y.; Mizuguchi, M. Crystal Structure of Human General Transcription Factor TFIIE at Atomic Resolution. J. Mol. Biol. 2016, 428, 4258–4266. [Google Scholar] [CrossRef] [PubMed]
- Okuda, M.; Tanaka, A.; Arai, Y.; Satoh, M.; Okamura, H.; Nagadoi, A.; Hanaoka, F.; Ohkuma, Y.; Nishimura, Y. A Novel Zinc Finger Structure in the Large Subunit of Human General Transcription Factor TFIIE. J. Biol. Chem. 2004, 279, 51395–51403. [Google Scholar] [CrossRef]
- Yokomori, K.; Verrijzer, C.P.; Tjian, R. An Interplay between TATA Box-Binding Protein and Transcription Factors IIE and IIA Modulates DNA Binding and Transcription. Proc. Natl. Acad. Sci. USA 1998, 95, 6722–6727. [Google Scholar] [CrossRef]
- Okuda, M.; Watanabe, Y.; Okamura, H.; Hanaoka, F.; Ohkuma, Y.; Nishimura, Y. Structure of the Central Core Domain of TFIIEβ with a Novel Double-Stranded DNA-Binding Surface. EMBO J. 2000, 19, 1346–1356. [Google Scholar] [CrossRef]
- Forget, D.; Langelier, M.-F.; Thérien, C.; Trinh, V.; Coulombe, B. Photo-Cross-Linking of a Purified Preinitiation Complex Reveals Central Roles for the RNA Polymerase II Mobile Clamp and TFIIE in Initiation Mechanisms. Mol. Cell. Biol. 2004, 24, 1122–1131. [Google Scholar] [CrossRef]
- Compe, E.; Genes, C.M.; Braun, C.; Coin, F.; Egly, J.-M. TFIIE Orchestrates the Recruitment of the TFIIH Kinase Module at Promoter before Release during Transcription. Nat. Commun. 2019, 10, 2084. [Google Scholar] [CrossRef]
- Holstege, F.C.; van der Vliet, P.C.; Timmers, H.T. Opening of an RNA Polymerase II Promoter Occurs in Two Distinct Steps and Requires the Basal Transcription Factors IIE and IIH. EMBO J. 1996, 15, 1666–1677. [Google Scholar] [CrossRef]
- Maxon, M.E.; Goodrich, J.A.; Tjian, R. Transcription Factor IIE Binds Preferentially to RNA Polymerase IIa and Recruits TFIIH: A Model for Promoter Clearance. Genes Dev. 1994, 8, 515–524. [Google Scholar] [CrossRef][Green Version]
- Watanabe, T.; Hayashi, K.; Tanaka, A.; Furumoto, T.; Hanaoka, F.; Ohkuma, Y. The Carboxy Terminus of the Small Subunit of TFIIE Regulates the Transition from Transcription Initiation to Elongation by RNA Polymerase II. Mol. Cell. Biol. 2003, 23, 2914–2926. [Google Scholar] [CrossRef][Green Version]
- Nogales, E.; Greber, B.J. High-Resolution Cryo-EM Structures of TFIIH and Their Functional Implications. Curr. Opin. Struct. Biol. 2019, 59, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Rimel, J.K.; Taatjes, D.J. The Essential and Multifunctional TFIIH Complex. Protein Sci. 2018, 27, 1018–1037. [Google Scholar] [CrossRef] [PubMed]
- Compe, E.; Egly, J.-M. TFIIH: When Transcription Met DNA Repair. Nat. Rev. Mol. Cell Biol. 2012, 13, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, L.; Roy, R.; Humbert, S.; Moncollin, V.; Vermeulen, W.; Hoeijmakers, J.H.J.; Chambon, P.; Egly, J.-M. DNA Repair Helicase: A Component of BTF2 (TFIIH) Basic Transcription Factor. Science 1993, 260, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Svejstrup, J.Q.; Vichi, P.; Egly, J.M. The Multiple Roles of Transcription/Repair Factor TFIIH. Trends Biochem. Sci. 1996, 21, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Coin, F.; Oksenych, V.; Mocquet, V.; Groh, S.; Blattner, C.; Egly, J.M. Nucleotide Excision Repair Driven by the Dissociation of CAK from TFIIH. Mol. Cell 2008, 31, 9–20. [Google Scholar] [CrossRef]
- Wong, K.H.; Jin, Y.; Struhl, K. TFIIH Phosphorylation of the Pol II CTD Stimulates Mediator Dissociation from the Preinitiation Complex and Promoter Escape. Mol. Cell 2014, 54, 601–612. [Google Scholar] [CrossRef]
- Fishburn, J.; Tomko, E.; Galburt, E.; Hahn, S. Double-Stranded DNA Translocase Activity of Transcription Factor TFIIH and the Mechanism of RNA Polymerase II Open Complex Formation. Proc. Natl. Acad. Sci. USA 2015, 112, 3961–3966. [Google Scholar] [CrossRef]
- Plaschka, C.; Hantsche, M.; Dienemann, C.; Burzinski, C.; Plitzko, J.; Cramer, P. Transcription Initiation Complex Structures Elucidate DNA Opening. Nature 2016, 533, 353–358. [Google Scholar] [CrossRef]
- Cramer, P.; Bushnell, D.A.; Kornberg, R.D. Structural Basis of Transcription: RNA Polymerase II at 2.8 Ångstrom Resolution. Science 2001, 292, 1863–1876. [Google Scholar] [CrossRef]
- Unarta, I.C.; Goonetilleke, E.C.; Wang, D.; Huang, X. Nucleotide Addition and Cleavage by RNA Polymerase II: Coordination of Two Catalytic Reactions Using a Single Active Site. J. Biol. Chem. 2023, 299, 102844. [Google Scholar] [CrossRef]
- Brueckner, F.; Cramer, P. Structural Basis of Transcription Inhibition by α-Amanitin and Implications for RNA Polymerase II Translocation. Nat. Struct. Mol. Biol. 2008, 15, 811–818. [Google Scholar] [CrossRef]
- Robinson, P.J.; Trnka, M.J.; Bushnell, D.A.; Davis, R.E.; Mattei, P.-J.; Burlingame, A.L.; Kornberg, R.D. Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell 2016, 166, 1411–1422.e16. [Google Scholar] [CrossRef]
- Cho, E.-J.; Takagi, T.; Moore, C.R.; Buratowski, S. mRNA Capping Enzyme Is Recruited to the Transcription Complex by Phosphorylation of the RNA Polymerase II Carboxy-Terminal Domain. Genes Dev. 1997, 11, 3319–3326. [Google Scholar] [CrossRef]
- Komarnitsky, P.; Cho, E.-J.; Buratowski, S. Different Phosphorylated Forms of RNA Polymerase II and Associated mRNA Processing Factors during Transcription. Genes Dev. 2000, 14, 2452–2460. [Google Scholar] [CrossRef]
- Pei, Y.; Hausmann, S.; Ho, C.K.; Schwer, B.; Shuman, S. The Length, Phosphorylation State, and Primary Structure of the RNA Polymerase II Carboxyl-Terminal Domain Dictate Interactions with mRNA Capping Enzymes. J. Biol. Chem. 2001, 276, 28075–28082. [Google Scholar] [CrossRef]
- Rimel, J.K.; Poss, Z.C.; Erickson, B.; Maas, Z.L.; Ebmeier, C.C.; Johnson, J.L.; Decker, T.-M.; Yaron, T.M.; Bradley, M.J.; Hamman, K.B.; et al. Selective Inhibition of CDK7 Reveals High-Confidence Targets and New Models for TFIIH Function in Transcription. Genes Dev. 2020, 34, 1452–1473. [Google Scholar] [CrossRef] [PubMed]
- Hieb, A.R.; Baran, S.; Goodrich, J.A.; Kugel, J.F. An 8 Nt RNA Triggers a Rate-Limiting Shift of RNA Polymerase II Complexes into Elongation. EMBO J. 2006, 25, 3100–3109. [Google Scholar] [CrossRef] [PubMed]
- Ebmeier, C.C.; Erickson, B.; Allen, B.L.; Allen, M.A.; Kim, H.; Fong, N.; Jacobsen, J.R.; Liang, K.; Shilatifard, A.; Dowell, R.D.; et al. Human TFIIH Kinase CDK7 Regulates Transcription-Associated Chromatin Modifications. Cell Rep. 2017, 20, 1173–1186. [Google Scholar] [CrossRef]
- Glover-Cutter, K.; Larochelle, S.; Erickson, B.; Zhang, C.; Shokat, K.; Fisher, R.P.; Bentley, D.L. TFIIH-Associated Cdk7 Kinase Functions in Phosphorylation of C-Terminal Domain Ser7 Residues, Promoter-Proximal Pausing, and Termination by RNA Polymerase II. Mol. Cell. Biol. 2009, 29, 5455–5464. [Google Scholar] [CrossRef] [PubMed]
- Luse, D.S. Promoter Clearance by RNA Polymerase II. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2013, 1829, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Saunders, A.; Core, L.J.; Lis, J.T. Breaking Barriers to Transcription Elongation. Nat. Rev. Mol. Cell Biol. 2006, 7, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Pal, M.; Ponticelli, A.S.; Luse, D.S. The Role of the Transcription Bubble and TFIIB in Promoter Clearance by RNA Polymerase II. Mol. Cell 2005, 19, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Kugel, J.F.; Goodrich, J.A. Promoter Escape Limits the Rate of RNA Polymerase II Transcription and Is Enhanced by TFIIE, TFIIH, and ATP on Negatively Supercoiled DNA. Proc. Natl. Acad. Sci. USA 1998, 95, 9232–9237. [Google Scholar] [CrossRef] [PubMed]
- Horn, A.E.; Kugel, J.F.; Goodrich, J.A. Single Molecule Microscopy Reveals Mechanistic Insight into RNA Polymerase II Preinitiation Complex Assembly and Transcriptional Activity. Nucleic Acids Res. 2016, 44, 7132–7143. [Google Scholar] [CrossRef] [PubMed]
- Kamakaka, R.T.; Tyree, C.M.; Kadonaga, J.T. Accurate and Efficient RNA Polymerase II Transcription with a Soluble Nuclear Fraction Derived from Drosophila Embryos. Proc. Natl. Acad. Sci. USA 1991, 88, 1024–1028. [Google Scholar] [CrossRef]
- Darzacq, X.; Shav-Tal, Y.; De Turris, V.; Brody, Y.; Shenoy, S.M.; Phair, R.D.; Singer, R.H. In Vivo Dynamics of RNA Polymerase II Transcription. Nat. Struct. Mol. Biol. 2007, 14, 796–806. [Google Scholar] [CrossRef]
- Forero-Quintero, L.S.; Raymond, W.; Handa, T.; Saxton, M.N.; Morisaki, T.; Kimura, H.; Bertrand, E.; Munsky, B.; Stasevich, T.J. Live-Cell Imaging Reveals the Spatiotemporal Organization of Endogenous RNA Polymerase II Phosphorylation at a Single Gene. Nat. Commun. 2021, 12, 3158. [Google Scholar] [CrossRef]
- Pal, S.; Biswas, D. Promoter-Proximal Regulation of Gene Transcription: Key Factors Involved and Emerging Role of General Transcription Factors in Assisting Productive Elongation. Gene 2023, 878, 147571. [Google Scholar] [CrossRef]
- Yudkovsky, N.; Ranish, J.A.; Hahn, S. A Transcription Reinitiation Intermediate That Is Stabilized by Activator. Nature 2000, 408, 225–229. [Google Scholar] [CrossRef]
- Christova, R.; Oelgeschläger, T. Association of Human TFIID–Promoter Complexes with Silenced Mitotic Chromatin in Vivo. Nat. Cell Biol. 2002, 4, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Dollinger, R.; Gilmour, D.S. Regulation of Promoter Proximal Pausing of RNA Polymerase II in Metazoans. J. Mol. Biol. 2021, 433, 166897. [Google Scholar] [CrossRef] [PubMed]
- Core, L.; Adelman, K. Promoter-Proximal Pausing of RNA Polymerase II: A Nexus of Gene Regulation. Genes Dev. 2019, 33, 960–982. [Google Scholar] [CrossRef] [PubMed]
- Adelman, K.; Lis, J.T. Promoter-Proximal Pausing of RNA Polymerase II: Emerging Roles in Metazoans. Nat. Rev. Genet. 2012, 13, 720–731. [Google Scholar] [CrossRef]
- Adelman, K.; Kennedy, M.A.; Nechaev, S.; Gilchrist, D.A.; Muse, G.W.; Chinenov, Y.; Rogatsky, I. Immediate Mediators of the Inflammatory Response Are Poised for Gene Activation through RNA Polymerase II Stalling. Proc. Natl. Acad. Sci. USA 2009, 106, 18207–18212. [Google Scholar] [CrossRef]
- Gupte, R.; Muse, G.W.; Chinenov, Y.; Adelman, K.; Rogatsky, I. Glucocorticoid Receptor Represses Proinflammatory Genes at Distinct Steps of the Transcription Cycle. Proc. Natl. Acad. Sci. USA 2013, 110, 14616–14621. [Google Scholar] [CrossRef]
- Hah, N.; Danko, C.G.; Core, L.; Waterfall, J.J.; Siepel, A.; Lis, J.T.; Kraus, W.L. A Rapid, Extensive, and Transient Transcriptional Response to Estrogen Signaling in Breast Cancer Cells. Cell 2011, 145, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Lagha, M.; Bothma, J.P.; Esposito, E.; Ng, S.; Stefanik, L.; Tsui, C.; Johnston, J.; Chen, K.; Gilmour, D.S.; Zeitlinger, J.; et al. Paused Pol II Coordinates Tissue Morphogenesis in the Drosophila Embryo. Cell 2013, 153, 976–987. [Google Scholar] [CrossRef]
- Williams, L.H.; Fromm, G.; Gokey, N.G.; Henriques, T.; Muse, G.W.; Burkholder, A.; Fargo, D.C.; Hu, G.; Adelman, K. Pausing of RNA Polymerase II Regulates Mammalian Developmental Potential through Control of Signaling Networks. Mol. Cell 2015, 58, 311–322. [Google Scholar] [CrossRef]
- Vos, S.M.; Farnung, L.; Urlaub, H.; Cramer, P. Structure of Paused Transcription Complex Pol II–DSIF–NELF. Nature 2018, 560, 601–606. [Google Scholar] [CrossRef]
- Pei, Y.; Shuman, S. Interactions between Fission Yeast mRNA Capping Enzymes and Elongation Factor Spt5. J. Biol. Chem. 2002, 277, 19639–19648. [Google Scholar] [CrossRef]
- Wen, Y.; Shatkin, A.J. Transcription Elongation Factor hSPT5 Stimulates mRNA Capping. Genes Dev. 1999, 13, 1774–1779. [Google Scholar] [CrossRef]
- Schulz, S.; Gietl, A.; Smollett, K.; Tinnefeld, P.; Werner, F.; Grohmann, D. TFE and Spt4/5 Open and Close the RNA Polymerase Clamp during the Transcription Cycle. Proc. Natl. Acad. Sci. USA 2016, 113, E1816–E1825. [Google Scholar] [CrossRef]
- Larochelle, S.; Amat, R.; Glover-Cutter, K.; Sansó, M.; Zhang, C.; Allen, J.J.; Shokat, K.M.; Bentley, D.L.; Fisher, R.P. Cyclin-Dependent Kinase Control of the Initiation-to-Elongation Switch of RNA Polymerase II. Nat. Struct. Mol. Biol. 2012, 19, 1108–1115. [Google Scholar] [CrossRef]
- Li, J.; Liu, Y.; Rhee, H.S.; Ghosh, S.K.B.; Bai, L.; Pugh, B.F.; Gilmour, D.S. Kinetic Competition between Elongation Rate and Binding of NELF Controls Promoter-Proximal Pausing. Mol. Cell 2013, 50, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Price, D.H. Properties of RNA Polymerase II Elongation Complexes Before and After the P-TEFb-Mediated Transition into Productive Elongation. J. Biol. Chem. 2007, 282, 21901–21912. [Google Scholar] [CrossRef] [PubMed]
- Henriques, T.; Gilchrist, D.A.; Nechaev, S.; Bern, M.; Muse, G.W.; Burkholder, A.; Fargo, D.C.; Adelman, K. Stable Pausing by RNA Polymerase II Provides an Opportunity to Target and Integrate Regulatory Signals. Mol. Cell 2013, 52, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Egloff, S. CDK9 Keeps RNA Polymerase II on Track. Cell. Mol. Life Sci. 2021, 78, 5543–5567. [Google Scholar] [CrossRef] [PubMed]
- Sansó, M.; Levin, R.S.; Lipp, J.J.; Wang, V.Y.-F.; Greifenberg, A.K.; Quezada, E.M.; Ali, A.; Ghosh, A.; Larochelle, S.; Rana, T.M.; et al. P-TEFb Regulation of Transcription Termination Factor Xrn2 Revealed by a Chemical Genetic Screen for Cdk9 Substrates. Genes Dev. 2016, 30, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Decker, T.-M.; Forné, I.; Straub, T.; Elsaman, H.; Ma, G.; Shah, N.; Imhof, A.; Eick, D. Analog-Sensitive Cell Line Identifies Cellular Substrates of CDK9. Oncotarget 2019, 10, 6934–6943. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.J.; Tong, L.; Adelman, K. Integrator Is a Global Promoter-Proximal Termination Complex. Mol. Cell 2023, 83, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Jonkers, I.; Kwak, H.; Lis, J.T. Genome-Wide Dynamics of Pol II Elongation and Its Interplay with Promoter Proximal Pausing, Chromatin, and Exons. eLife 2014, 3, e02407. [Google Scholar] [CrossRef] [PubMed]
- Chao, S.-H.; Price, D.H. Flavopiridol Inactivates P-TEFb and Blocks Most RNA Polymerase II Transcription in Vivo. J. Biol. Chem. 2001, 276, 31793–31799. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.X.; Smith, E.R.; Shilatifard, A. Born to Run: Control of Transcription Elongation by RNA Polymerase II. Nat. Rev. Mol. Cell Biol. 2018, 19, 464–478. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Li, T.; Price, D.H. RNA Polymerase II Elongation Control. Annu. Rev. Biochem. 2012, 81, 119–143. [Google Scholar] [CrossRef] [PubMed]
- Izban, M.G.; Luse, D.S. Factor-Stimulated RNA Polymerase II Transcribes at Physiological Elongation Rates on Naked DNA but Very Poorly on Chromatin Templates. J. Biol. Chem. 1992, 267, 13647–13655. [Google Scholar] [CrossRef] [PubMed]
- Conaway, R.C.; Conaway, J.W. The Hunt for RNA Polymerase II Elongation Factors: A Historical Perspective. Nat. Struct. Mol. Biol. 2019, 26, 771–776. [Google Scholar] [CrossRef]
- Luo, Z.; Lin, C.; Shilatifard, A. The Super Elongation Complex (SEC) Family in Transcriptional Control. Nat. Rev. Mol. Cell Biol. 2012, 13, 543–547. [Google Scholar] [CrossRef]
- Chen, Y.; Vos, S.M.; Dienemann, C.; Ninov, M.; Urlaub, H.; Cramer, P. Allosteric Transcription Stimulation by RNA Polymerase II Super Elongation Complex. Mol. Cell 2021, 81, 3386–3399.e10. [Google Scholar] [CrossRef]
- Van Oss, S.B.; Cucinotta, C.E.; Arndt, K.M. Emerging Insights into the Roles of the Paf1 Complex in Gene Regulation. Trends Biochem. Sci. 2017, 42, 788–798. [Google Scholar] [CrossRef]
- Francette, A.M.; Tripplehorn, S.A.; Arndt, K.M. The Paf1 Complex: A Keystone of Nuclear Regulation Operating at the Interface of Transcription and Chromatin. J. Mol. Biol. 2021, 433, 166979. [Google Scholar] [CrossRef]
- Mbogning, J.; Nagy, S.; Pagé, V.; Schwer, B.; Shuman, S.; Fisher, R.P.; Tanny, J.C. The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast. PLoS Genet. 2013, 9, e1004029. [Google Scholar] [CrossRef]
- Qiu, H.; Hu, C.; Gaur, N.A.; Hinnebusch, A.G. Pol II CTD Kinases Bur1 and Kin28 Promote Spt5 CTR-Independent Recruitment of Paf1 Complex: Dual Pathway of Paf1C Recruitment. EMBO J. 2012, 31, 3494–3505. [Google Scholar] [CrossRef]
- Wier, A.D.; Mayekar, M.K.; Héroux, A.; Arndt, K.M.; VanDemark, A.P. Structural Basis for Spt5-Mediated Recruitment of the Paf1 Complex to Chromatin. Proc. Natl. Acad. Sci. USA 2013, 110, 17290–17295. [Google Scholar] [CrossRef]
- Liu, Y.; Warfield, L.; Zhang, C.; Luo, J.; Allen, J.; Lang, W.H.; Ranish, J.; Shokat, K.M.; Hahn, S. Phosphorylation of the Transcription Elongation Factor Spt5 by Yeast Bur1 Kinase Stimulates Recruitment of the PAF Complex. Mol. Cell. Biol. 2009, 29, 4852–4863. [Google Scholar] [CrossRef]
- Yu, M.; Yang, W.; Ni, T.; Tang, Z.; Nakadai, T.; Zhu, J.; Roeder, R.G. RNA Polymerase II–Associated Factor 1 Regulates the Release and Phosphorylation of Paused RNA Polymerase II. Science 2015, 350, 1383–1386. [Google Scholar] [CrossRef]
- Lu, X.; Zhu, X.; Li, Y.; Liu, M.; Yu, B.; Wang, Y.; Rao, M.; Yang, H.; Zhou, K.; Wang, Y.; et al. Multiple P-TEFbs Cooperatively Regulate the Release of Promoter-Proximally Paused RNA Polymerase II. Nucleic Acids Res. 2016, 44, 6853–6867. [Google Scholar] [CrossRef]
- Chen, F.X.; Woodfin, A.R.; Gardini, A.; Rickels, R.A.; Marshall, S.A.; Smith, E.R.; Shiekhattar, R.; Shilatifard, A. PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell 2015, 162, 1003–1015. [Google Scholar] [CrossRef]
- Chen, F.X.; Xie, P.; Collings, C.K.; Cao, K.; Aoi, Y.; Marshall, S.A.; Rendleman, E.J.; Ugarenko, M.; Ozark, P.A.; Zhang, A.; et al. PAF1 Regulation of Promoter-Proximal Pause Release via Enhancer Activation. Science 2017, 357, 1294–1298. [Google Scholar] [CrossRef]
- Mosimann, C.; Hausmann, G.; Basler, K. Parafibromin/Hyrax Activates Wnt/Wg Target Gene Transcription by Direct Association with β-Catenin/Armadillo. Cell 2006, 125, 327–341. [Google Scholar] [CrossRef]
- Kikuchi, I.; Takahashi-Kanemitsu, A.; Sakiyama, N.; Tang, C.; Tang, P.-J.; Noda, S.; Nakao, K.; Kassai, H.; Sato, T.; Aiba, A.; et al. Dephosphorylated Parafibromin Is a Transcriptional Coactivator of the Wnt/Hedgehog/Notch Pathways. Nat. Commun. 2016, 7, 12887. [Google Scholar] [CrossRef]
- Skene, P.J.; Hernandez, A.E.; Groudine, M.; Henikoff, S. The Nucleosomal Barrier to Promoter Escape by RNA Polymerase II Is Overcome by the Chromatin Remodeler Chd1. eLife 2014, 3, e02042. [Google Scholar] [CrossRef]
- Farnung, L.; Ochmann, M.; Engeholm, M.; Cramer, P. Structural Basis of Nucleosome Transcription Mediated by Chd1 and FACT. Nat. Struct. Mol. Biol. 2021, 28, 382–387. [Google Scholar] [CrossRef]
- Kulaeva, O.I.; Hsieh, F.-K.; Chang, H.-W.; Luse, D.S.; Studitsky, V.M. Mechanism of Transcription through a Nucleosome by RNA Polymerase II. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2013, 1829, 76–83. [Google Scholar] [CrossRef]
- Filipovski, M.; Soffers, J.H.M.; Vos, S.M.; Farnung, L. Structural Basis of Nucleosome Retention during Transcription Elongation. Science 2022, 376, 1313–1316. [Google Scholar] [CrossRef]
- Squazzo, S.L. The Paf1 Complex Physically and Functionally Associates with Transcription Elongation Factors in Vivo. EMBO J. 2002, 21, 1764–1774. [Google Scholar] [CrossRef]
- Krogan, N.J.; Kim, M.; Ahn, S.H.; Zhong, G.; Kobor, M.S.; Cagney, G.; Emili, A.; Shilatifard, A.; Buratowski, S.; Greenblatt, J.F. RNA Polymerase II Elongation Factors of Saccharomyces Cerevisiae: A Targeted Proteomics Approach. Mol. Cell. Biol. 2002, 22, 6979–6992. [Google Scholar] [CrossRef]
- Lee, Y.; Park, D.; Iyer, V.R. The ATP-Dependent Chromatin Remodeler Chd1 Is Recruited by Transcription Elongation Factors and Maintains H3K4me3/H3K36me3 Domains at Actively Transcribed and Spliced Genes. Nucleic Acids Res. 2017, 45, 7180–7190. [Google Scholar] [CrossRef]
- Warner, M.H.; Roinick, K.L.; Arndt, K.M. Rtf1 Is a Multifunctional Component of the Paf1 Complex That Regulates Gene Expression by Directing Cotranscriptional Histone Modification. Mol. Cell. Biol. 2007, 27, 6103–6115. [Google Scholar] [CrossRef]
- Laribee, R.N.; Krogan, N.J.; Xiao, T.; Shibata, Y.; Hughes, T.R.; Greenblatt, J.F.; Strahl, B.D. BUR Kinase Selectively Regulates H3 K4 Trimethylation and H2B Ubiquitylation through Recruitment of the PAF Elongation Complex. Curr. Biol. 2005, 15, 1487–1493. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Kao, C.-F.; Krogan, N.J.; Sun, Z.-W.; Greenblatt, J.F.; Osley, M.A.; Strahl, B.D. Histone H2B Ubiquitylation Is Associated with Elongating RNA Polymerase II. Mol. Cell. Biol. 2005, 25, 637–651. [Google Scholar] [CrossRef]
- Fetian, T.; McShane, B.M.; Horan, N.L.; Shodja, D.N.; True, J.D.; Mosley, A.L.; Arndt, K.M. Paf1 Complex Subunit Rtf1 Stimulates H2B Ubiquitylation by Interacting with the Highly Conserved N-Terminal Helix of Rad6. Proc. Natl. Acad. Sci. USA 2023, 120, e2220041120. [Google Scholar] [CrossRef]
- Heidemann, M.; Hintermair, C.; Voß, K.; Eick, D. Dynamic Phosphorylation Patterns of RNA Polymerase II CTD during Transcription. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2013, 1829, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Yoh, S.M.; Cho, H.; Pickle, L.; Evans, R.M.; Jones, K.A. The Spt6 SH2 Domain Binds Ser2-P RNAPII to Direct Iws1-Dependent mRNA Splicing and Export. Genes Dev. 2007, 21, 160–174. [Google Scholar] [CrossRef]
- David, C.J.; Boyne, A.R.; Millhouse, S.R.; Manley, J.L. The RNA Polymerase II C-Terminal Domain Promotes Splicing Activation through Recruitment of a U2AF65–Prp19 Complex. Genes Dev. 2011, 25, 972–983. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.P.; Greenleaf, A.L. The Splicing Factor, Prp40, Binds the Phosphorylated Carboxyl-Terminal Domain of RNA Polymerase II. J. Biol. Chem. 2000, 275, 39935–39943. [Google Scholar] [CrossRef] [PubMed]
- MacKellar, A.L.; Greenleaf, A.L. Cotranscriptional Association of mRNA Export Factor Yra1 with C-Terminal Domain of RNA Polymerase II. J. Biol. Chem. 2011, 286, 36385–36395. [Google Scholar] [CrossRef]
- Mayer, A.; Heidemann, M.; Lidschreiber, M.; Schreieck, A.; Sun, M.; Hintermair, C.; Kremmer, E.; Eick, D.; Cramer, P. CTD Tyrosine Phosphorylation Impairs Termination Factor Recruitment to RNA Polymerase II. Science 2012, 336, 1723–1725. [Google Scholar] [CrossRef]
- Schreieck, A.; Easter, A.D.; Etzold, S.; Wiederhold, K.; Lidschreiber, M.; Cramer, P.; Passmore, L.A. RNA Polymerase II Termination Involves C-Terminal-Domain Tyrosine Dephosphorylation by CPF Subunit Glc7. Nat. Struct. Mol. Biol. 2014, 21, 175–179. [Google Scholar] [CrossRef]
- Shah, N.; Maqbool, M.A.; Yahia, Y.; El Aabidine, A.Z.; Esnault, C.; Forné, I.; Decker, T.-M.; Martin, D.; Schüller, R.; Krebs, S.; et al. Tyrosine-1 of RNA Polymerase II CTD Controls Global Termination of Gene Transcription in Mammals. Mol. Cell 2018, 69, 48–61.e6. [Google Scholar] [CrossRef]
- Hintermair, C.; Heidemann, M.; Koch, F.; Descostes, N.; Gut, M.; Gut, I.; Fenouil, R.; Ferrier, P.; Flatley, A.; Kremmer, E.; et al. Threonine-4 of Mammalian RNA Polymerase II CTD Is Targeted by Polo-like Kinase 3 and Required for Transcriptional Elongation: CTD Thr4 Is Required for Transcription Elongation. EMBO J. 2012, 31, 2784–2797. [Google Scholar] [CrossRef] [PubMed]
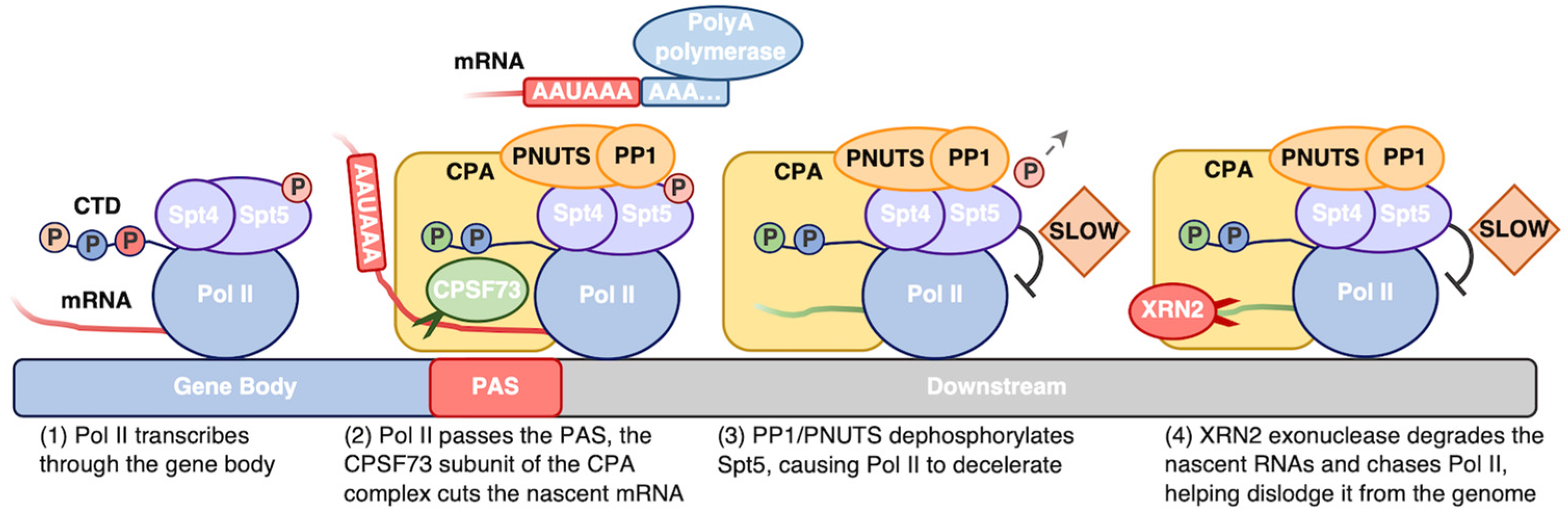
- Rodríguez-Molina, J.B.; West, S.; Passmore, L.A. Knowing When to Stop: Transcription Termination on Protein-Coding Genes by Eukaryotic RNAPII. Mol. Cell 2023, 83, 404–415. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Libri, D.; Porrua, O. Mechanisms of Eukaryotic Transcription Termination at a Glance. J. Cell Sci. 2023, 136, jcs259873. [Google Scholar] [CrossRef] [PubMed]
- Eaton, J.D.; West, S. Termination of Transcription by RNA Polymerase II: BOOM! Trends Genet. 2020, 36, 664–675. [Google Scholar] [CrossRef]
- Proudfoot, N.J. Transcriptional Termination in Mammals: Stopping the RNA Polymerase II Juggernaut. Science 2016, 352, aad9926. [Google Scholar] [CrossRef] [PubMed]
- Cortazar, M.A.; Sheridan, R.M.; Erickson, B.; Fong, N.; Glover-Cutter, K.; Brannan, K.; Bentley, D.L. Control of RNA Pol II Speed by PNUTS-PP1 and Spt5 Dephosphorylation Facilitates Termination by a “Sitting Duck Torpedo” Mechanism. Mol. Cell 2019, 76, 896–908.e4. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Brewer, G. The Regulation of mRNA Stability in Mammalian Cells: 2.0. Gene 2012, 500, 10–21. [Google Scholar] [CrossRef]
- Neve, J.; Patel, R.; Wang, Z.; Louey, A.; Furger, A.M. Cleavage and Polyadenylation: Ending the Message Expands Gene Regulation. RNA Biol. 2017, 14, 865–890. [Google Scholar] [CrossRef]
- Nordick, K.; Hoffman, M.G.; Betz, J.L.; Jaehning, J.A. Direct Interactions between the Paf1 Complex and a Cleavage and Polyadenylation Factor Are Revealed by Dissociation of Paf1 from RNA Polymerase II. Eukaryot. Cell 2008, 7, 1158–1167. [Google Scholar] [CrossRef]
- Penheiter, K.L.; Washburn, T.M.; Porter, S.E.; Hoffman, M.G.; Jaehning, J.A. A Posttranscriptional Role for the Yeast Paf1-RNA Polymerase II Complex Is Revealed by Identification of Primary Targets. Mol. Cell 2005, 20, 213–223. [Google Scholar] [CrossRef]
- Nagaike, T.; Logan, C.; Hotta, I.; Rozenblatt-Rosen, O.; Meyerson, M.; Manley, J.L. Transcriptional Activators Enhance Polyadenylation of mRNA Precursors. Mol. Cell 2011, 41, 409–418. [Google Scholar] [CrossRef]
- Katahira, J. Nuclear Export of Messenger RNA. Genes 2015, 6, 163–184. [Google Scholar] [CrossRef]
- Logan, J.; Falck-Pedersen, E.; Darnell, J.E.; Shenk, T. A Poly(A) Addition Site and a Downstream Termination Region Are Required for Efficient Cessation of Transcription by RNA Polymerase II in the Mouse Beta Maj-Globin Gene. Proc. Natl. Acad. Sci. USA 1987, 84, 8306–8310. [Google Scholar] [CrossRef]
- Zhang, H.; Rigo, F.; Martinson, H.G. Poly(A) Signal-Dependent Transcription Termination Occurs through a Conformational Change Mechanism That Does Not Require Cleavage at the Poly(A) Site. Mol. Cell 2015, 59, 437–448. [Google Scholar] [CrossRef][Green Version]
- Eaton, J.D.; Francis, L.; Davidson, L.; West, S. A Unified Allosteric/Torpedo Mechanism for Transcriptional Termination on Human Protein-Coding Genes. Genes Dev. 2020, 34, 132–145. [Google Scholar] [CrossRef]
| Protein | Subunits | Size (kDa) | Main Binding Partners | Function |
|---|---|---|---|---|
| TFIID | TBP, 13 TAFs | ~1300 | promoter, Pol II | Nucleates PIC assembly by binding multiple core promoter elements |
| TFIIA | TFIIAα, TFIIAβ, TFIIAɣ | 35, 19, and 12 | TBP, TFIID | Stabilizes the TFIID-DNA interaction; enhances the effects of transcriptional co-activators |
| TFIIB | TFIIB | 33 | promoter, TBP, Pol II | Helps to define the start site of transcription and orient Pol II in the proper direction |
| TFIIF | RAP30, RAP74 | 30 and 74 | promoter, Pol II, GTFs | Guides Pol II to the PIC and facilitates elongation |
| Pol II | Rpb1–Rpb12 | ~514 | promoter, all GTFs | Catalyzes RNA synthesis; phosphorylation of the CTD tail of Rpb1 serves a regulatory role |
| TFIIE | TFIIEα, TFIIEβ | 56 and 34 | promoter, TFIIH, Pol II, TFIIF | Recruits TFIIH to the PIC; stimulates enzymatic activities of TFIIH; stabilizes the open DNA conformation |
| TFIIH | Core domain: XPD, XBP, p62, p52, p44, p34, p8; CAK domain: CDK7, MAT1, cyclin H | ~500 | downstream DNA, TFIIE, Pol II | CDK7 kinase phosphorylates the CTD; ATP-dependent XPB translocase opens the promoter DNA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Archuleta, S.R.; Goodrich, J.A.; Kugel, J.F. Mechanisms and Functions of the RNA Polymerase II General Transcription Machinery during the Transcription Cycle. Biomolecules 2024, 14, 176. https://doi.org/10.3390/biom14020176
Archuleta SR, Goodrich JA, Kugel JF. Mechanisms and Functions of the RNA Polymerase II General Transcription Machinery during the Transcription Cycle. Biomolecules. 2024; 14(2):176. https://doi.org/10.3390/biom14020176
Chicago/Turabian StyleArchuleta, Stephen R., James A. Goodrich, and Jennifer F. Kugel. 2024. "Mechanisms and Functions of the RNA Polymerase II General Transcription Machinery during the Transcription Cycle" Biomolecules 14, no. 2: 176. https://doi.org/10.3390/biom14020176
APA StyleArchuleta, S. R., Goodrich, J. A., & Kugel, J. F. (2024). Mechanisms and Functions of the RNA Polymerase II General Transcription Machinery during the Transcription Cycle. Biomolecules, 14(2), 176. https://doi.org/10.3390/biom14020176





