Peripheral Lymphocytes in Primary Liver Cancers: Elevated NK and CD8+ T Cells and Dysregulated Selenium Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Follow-Up
2.2. Cell Counting of Peripheral Blood Lymphocyte
2.3. Single-Cell Sequencing and iTRAQ
2.4. Bioinformatics Analyses
2.5. Statistics
3. Results
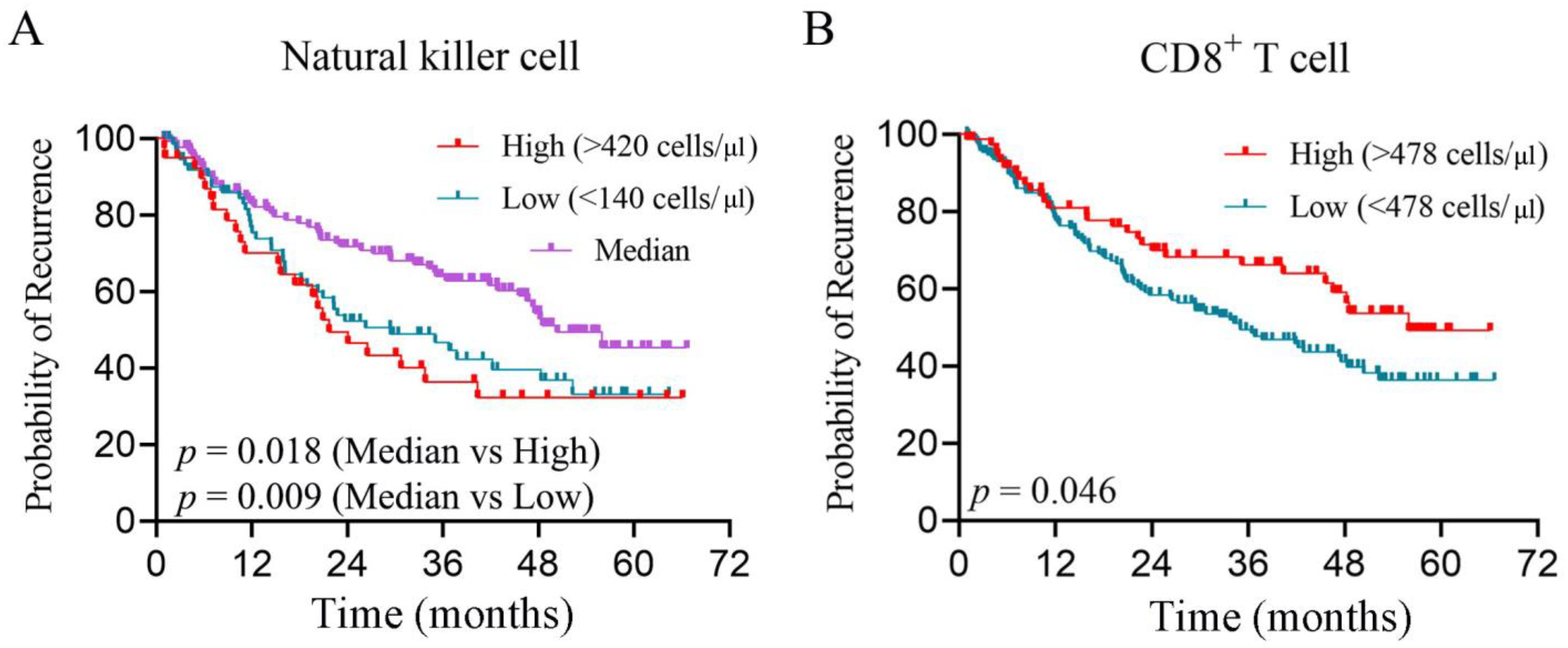
3.1. The Counts of Peripheral NK Cells and CD8+ T Cells Elevated in Primary Liver Cancers
3.2. Peripheral CD8+ T Cell and NK Cells Are Independent Risk Factors for Prognosis
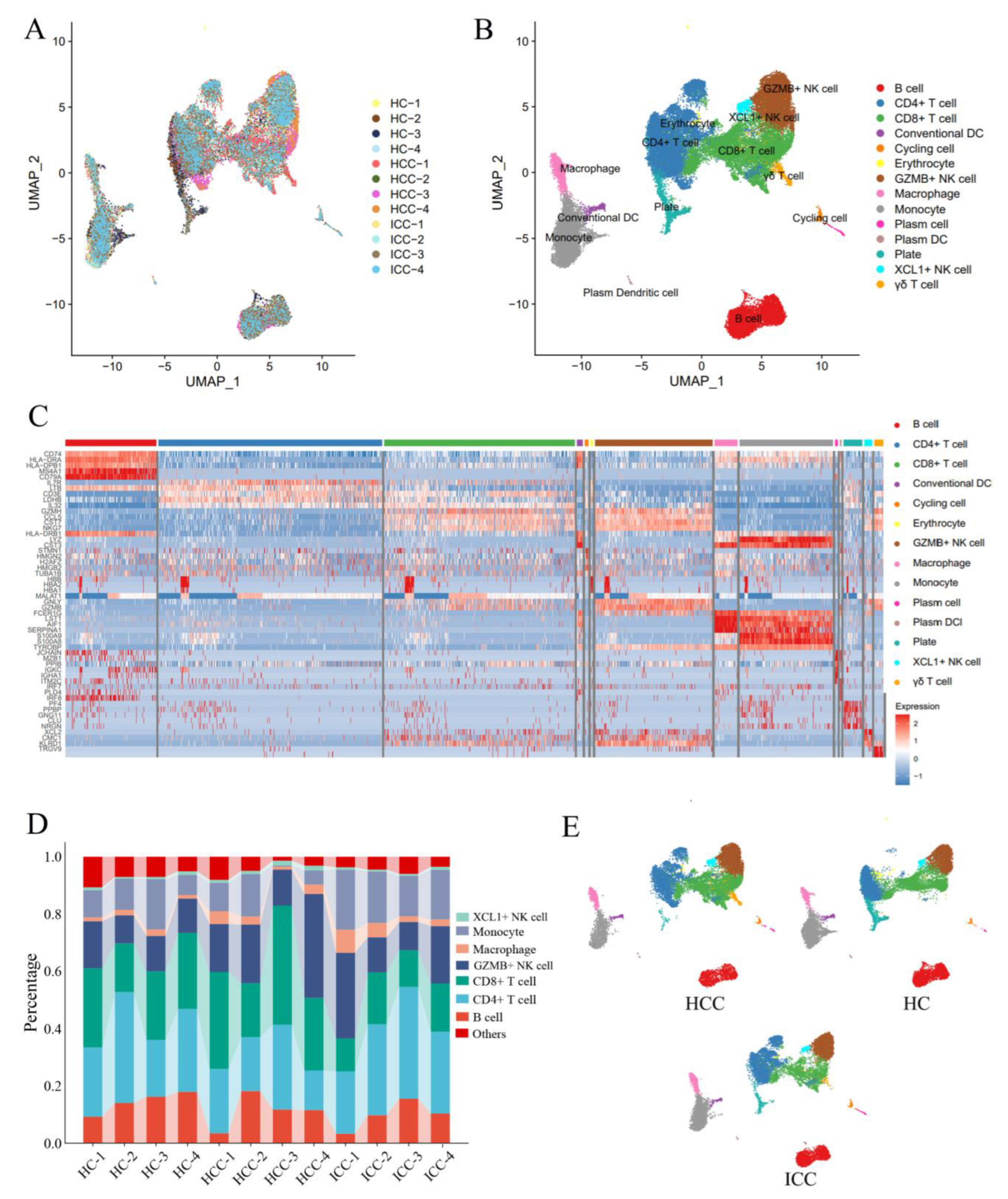
3.3. Single-Cell Sequencing Reveals Transcriptomic Features of Peripheral CD8+ T Cells and NK Cells
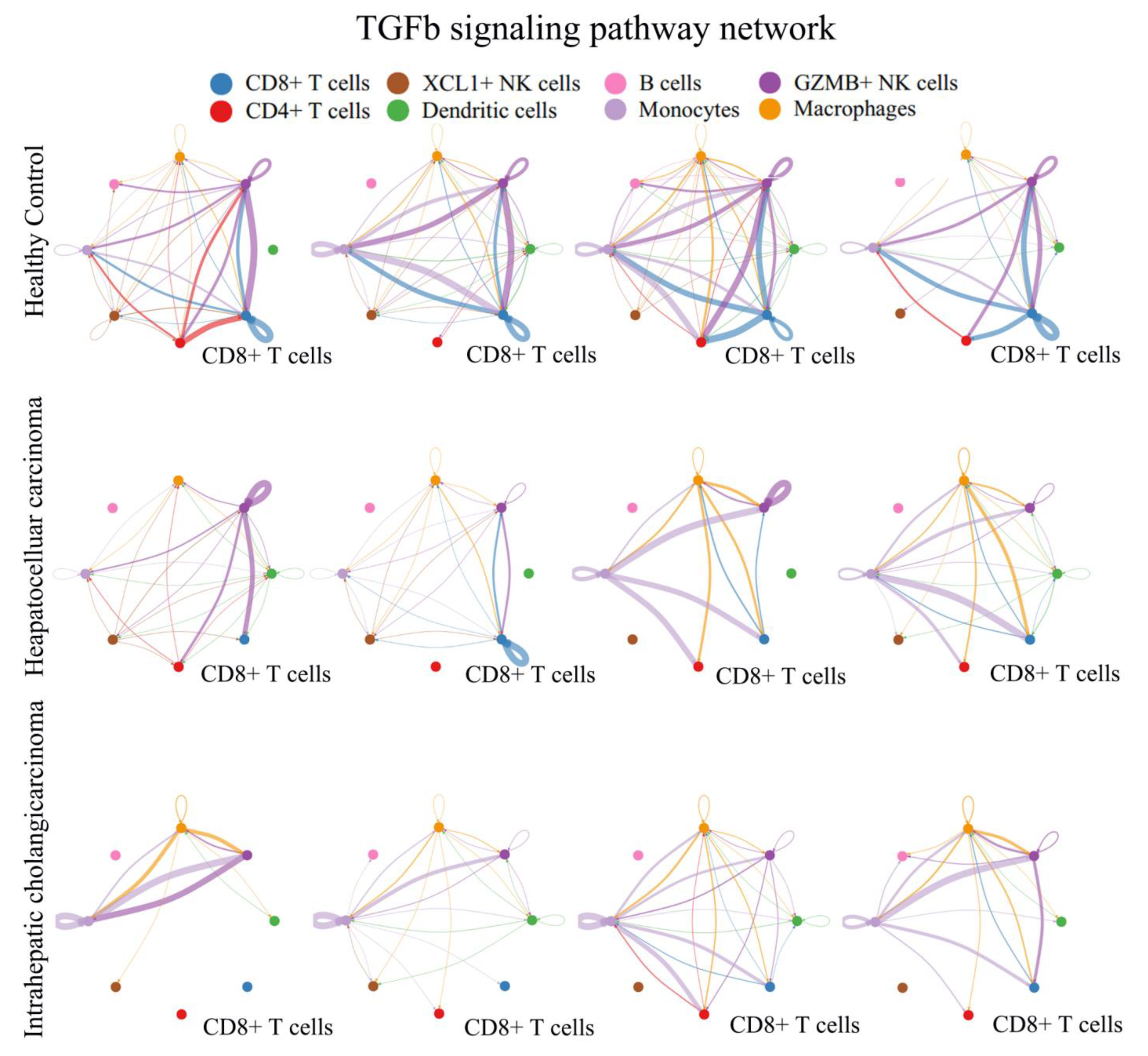
3.4. TGFβ Signal Decreased in Peripheral CD8+ T Cells from PLC
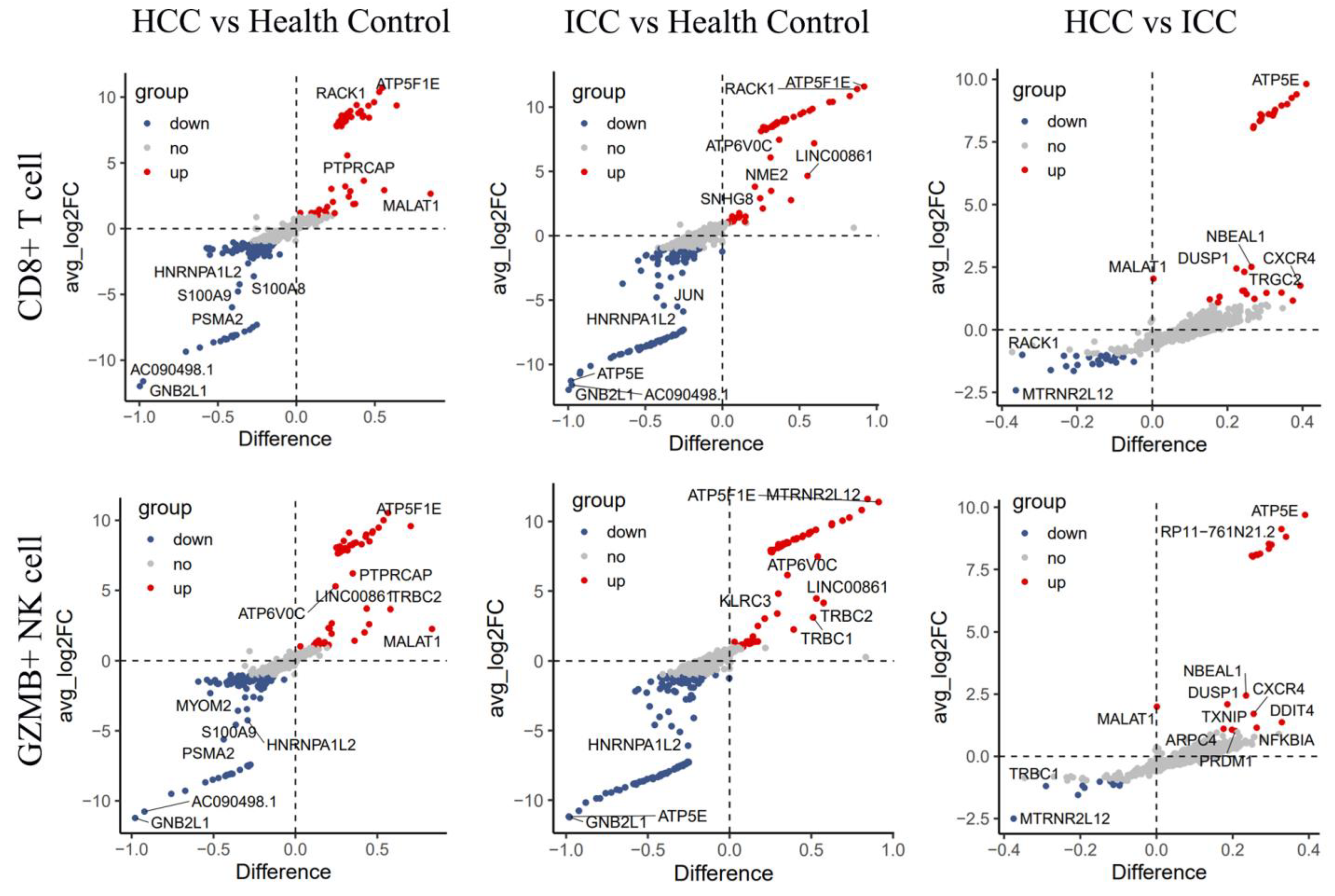
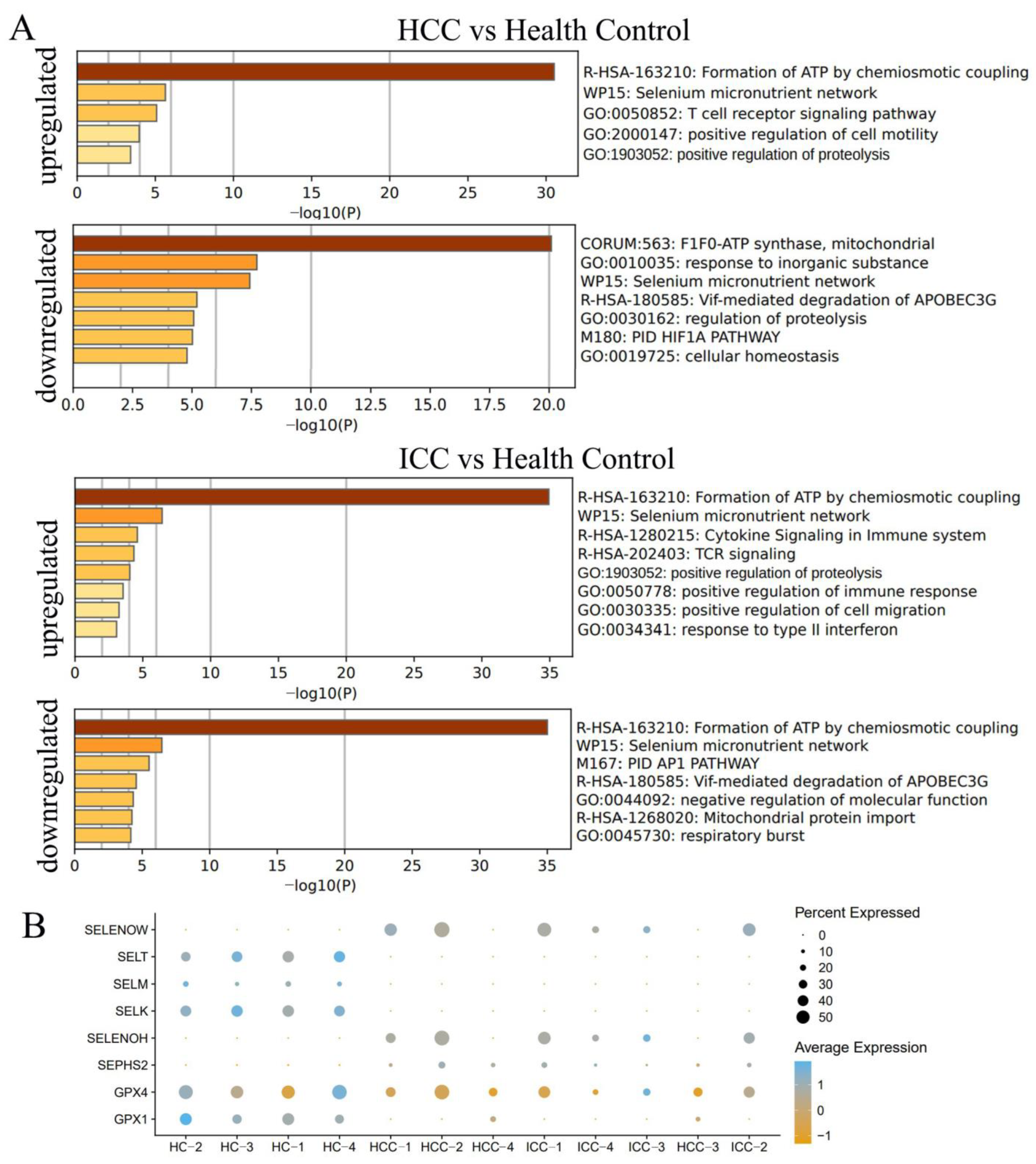
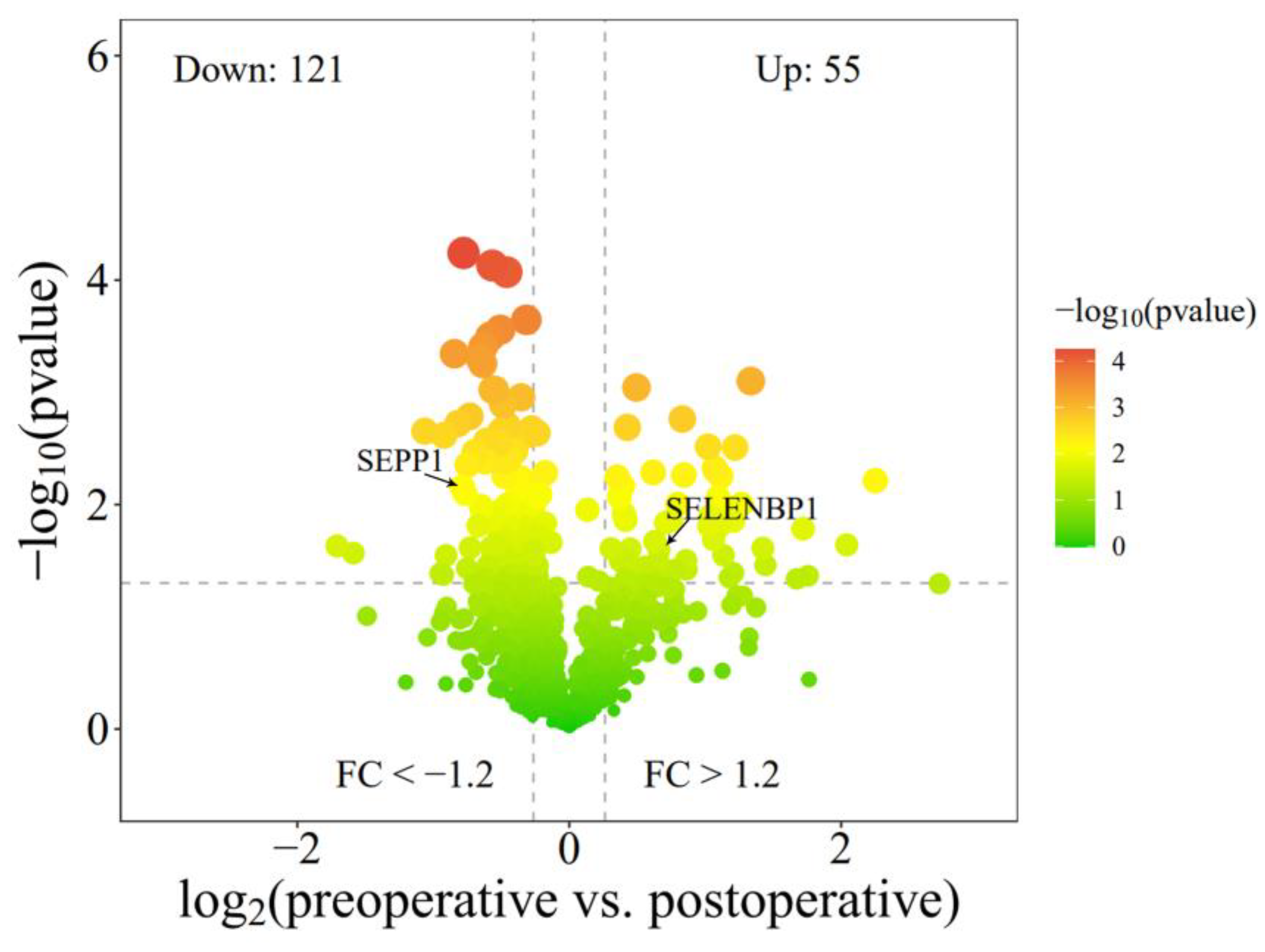
3.5. Differential Genes Enriched in Selenium Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Beaufrère, A.; Calderaro, J.; Paradis, V. Combined hepatocellular-cholangiocarcinoma: An update. J. Hepatol. 2021, 74, 1212–1224. [Google Scholar] [CrossRef]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Qing, X.; Xu, W.; Zong, J.; Du, X.; Peng, H.; Zhang, Y. Emerging treatment modalities for systemic therapy in hepatocellular carcinoma. Biomark. Res. 2021, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- Sperandio, R.C.; Pestana, R.C.; Miyamura, B.V.; Kaseb, A.O. Hepatocellular Carcinoma Immunotherapy. Annu. Rev. Med. 2022, 73, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Kallies, A.; Zehn, D.; Utzschneider, D.T. Precursor exhausted T cells: Key to successful immunotherapy? Nat. Rev. Immunol. 2020, 20, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Xia, A.; Zhang, Y.; Xu, J.; Yin, T.; Lu, X.J. T Cell Dysfunction in Cancer Immunity and Immunotherapy. Front. Immunol. 2019, 10, 1719. [Google Scholar] [CrossRef] [PubMed]
- Pauken, K.E.; Sammons, M.A.; Odorizzi, P.M.; Manne, S.; Godec, J.; Khan, O.; Drake, A.M.; Chen, Z.; Sen, D.R.; Kurachi, M.; et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 2016, 354, 1160–1165. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Zheng, L.; Yoo, J.K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q.; et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017, 169, 1342–1356.e16. [Google Scholar] [CrossRef]
- Cheng, S.; Li, Z.; Gao, R.; Xing, B.; Gao, Y.; Yang, Y.; Qin, S.; Zhang, L.; Ouyang, H.; Du, P.; et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 2021, 184, 792–809.e23. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Seow, J.J.W.; Dutertre, C.A.; Pai, R.; Blériot, C.; Mishra, A.; Wong, R.M.M.; Singh, G.S.N.; Sudhagar, S.; Khalilnezhad, S.; et al. Onco-fetal Reprogramming of Endothelial Cells Drives Immunosuppressive Macrophages in Hepatocellular Carcinoma. Cell 2020, 183, 377–394.e21. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wu, L.; Zhong, Y.; Zhou, K.; Hou, Y.; Wang, Z.; Zhang, Z.; Xie, J.; Wang, C.; Chen, D.; et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell 2021, 184, 404–421.e16. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Wang, D.; Qiu, X.; Luo, G.; Wu, T.; Yang, S.; Li, Z.; Zhu, Y.; Wang, S.; Wu, R.; et al. Trajectory and Functional Analysis of PD-1high CD4+CD8+ T Cells in Hepatocellular Carcinoma by Single-Cell Cytometry and Transcriptome Sequencing. Adv. Sci. 2020, 7, 2000224. [Google Scholar] [CrossRef] [PubMed]
- Germain, P.L.; Lun, A.; Garcia Meixide, C.; Macnair, W.; Robinson, M.D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Research 2021, 10, 979. [Google Scholar] [CrossRef] [PubMed]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.I.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Zhou, C.; Sun, B.Y.; Zhou, P.Y.; Yang, Z.F.; Wang, Z.T.; Liu, G.; Gan, W.; Wang, Z.; Zhou, J.; Fan, J.; et al. MAIT cells confer resistance to Lenvatinib plus anti-PD1 antibodies in hepatocellular carcinoma through TNF-TNFRSF1B pathway. Clin. Immunol. 2023, 256, 109770. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Xu, R.H.; Wei, W.; Krawczyk, M.; Wang, W.; Luo, H.; Flagg, K.; Yi, S.; Shi, W.; Quan, Q.; Li, K.; et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017, 16, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.F.; Wu, L.; Liu, S.P.; Jiang, M.M.; Hu, B.; Zhou, K.Q.; Guo, W.; Xu, Y.; Zhong, Y.; Zhou, X.R.; et al. Dissecting spatial heterogeneity and the immune-evasion mechanism of CTCs by single-cell RNA-seq in hepatocellular carcinoma. Nat. Commun. 2021, 12, 4091. [Google Scholar] [CrossRef]
- Heinrich, B.; Ruf, B.; Subramanyam, V.; Myojin, Y.; Lai, C.W.; Craig, A.J.; Craig, A.J.; Fu, J.; Xie, C.; Kroemer, A.; et al. Checkpoint Inhibitors Modulate Plasticity of Innate Lymphoid Cells in Peripheral Blood of Patients with Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 849958. [Google Scholar] [CrossRef]
- Hu, R.; Zhang, W.; Han, Z.; Ma, M.; Huang, Q.; Lv, M.; Ma, W.; Sun, X.; Feng, W.; Li, J.; et al. Identification of immune-related target and prognostic biomarkers in PBMC of hepatocellular carcinoma. BMC Gastroenterol. 2023, 23, 234. [Google Scholar] [CrossRef]
- Shen, P.; Wang, A.; He, M.; Wang, Q.; Zheng, S. Increased circulating Lin(−/low) CD33(+) HLA-DR(−) myeloid-derived suppressor cells in hepatocellular carcinoma patients. Hepatol. Res. 2014, 44, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Couvert, P.; Carrié, A.; Tezenas du Montcel, S.; Vaysse, J.; Sutton, A.; Barget, N.; Trinchet, J.C.; Beaugrand, M.; Ganne, N.; Giral, P.; et al. Insulin-like growth factor 2 gene methylation in peripheral blood mononuclear cells of patients with hepatitis C related cirrhosis or hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2012, 36, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Lu, J.; Li, K.; Zhang, H.; Zhao, X.; Li, G.; Li, N. Diagnostic and prognostic value of STAP1 and AHNAK methylation in peripheral blood immune cells for HBV-related hepatopathy. Front. Immunol. 2022, 13, 1091103. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Yu, M.; Ma, X.; Sun, J.; Liu, C.; Wang, C.; Wu, S.; Fu, P.; Yang, Z.; He, Y.; et al. IFNα Potentiates Anti-PD-1 Efficacy by Remodeling Glucose Metabolism in the Hepatocellular Carcinoma Microenvironment. Cancer Discov. 2022, 12, 1718–1741. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.J.; Mao, X.T.; Li, Y.Y.; Liu, D.D.; Fan, K.Q.; Liu, R.B.; Wu, T.T.; Wang, H.L.; Zhang, Y.; Yang, B.; et al. Multiomics analyses reveal a critical role of selenium in controlling T cell differentiation in Crohn’s disease. Immunity 2021, 54, 1728–1744.e7. [Google Scholar] [CrossRef] [PubMed]
- Carlson, B.A.; Yoo, M.H.; Shrimali, R.K.; Irons, R.; Gladyshev, V.N.; Hatfield, D.L.; Park, J.M. Role of selenium-containing proteins in T-cell and macrophage function. Proc. Nutr. Soc. 2010, 69, 300–310. [Google Scholar] [CrossRef]
- Lai, H.; Zeng, D.; Liu, C.; Zhang, Q.; Wang, X.; Chen, T. Selenium-containing ruthenium complex synergizes with natural killer cells to enhance immunotherapy against prostate cancer via activating TRAIL/FasL signaling. Biomaterials 2019, 219, 119377. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Liu, T.; Chen, S.; Luo, Y.; Ma, M.; Xue, F.; Zhang, L.; Bao, W.; Chen, H. Targeted Therapeutic-Immunomodulatory Nanoplatform Based on Noncrystalline Selenium. ACS Appl. Mater. Interfaces 2019, 11, 45404–45415. [Google Scholar] [CrossRef] [PubMed]


| Univariate Analysis | Multivariate Analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| HR | Confidence Interval | p-Value | HR | Confidence Interval | p-Value | |||
| Lower | Upper | Lower | Upper | |||||
| Sex (male) | 2.46 | 1.24 | 4.86 | 0.010 | 2.33 | 1.14 | 4.75 | 0.02 |
| Age (>60 years) | 0.91 | 0.63 | 1.32 | 0.635 | ||||
| Total Bilirubin (>20.4 μmol/L) | 0.97 | 0.54 | 1.73 | 0.919 | ||||
| Albumin (<35 g/L) | 0.84 | 0.37 | 1.91 | 0.670 | ||||
| Alanine aminotran-sferase (>50 U/L) | 0.93 | 0.62 | 1.39 | 0.724 | ||||
| γ-glutamyltransferas (>60 U/L) | 1.85 | 1.27 | 2.68 | 0.001 | 1.39 | 0.91 | 2.13 | 0.126 |
| Prothrombin time (>14 s) | 0.94 | 0.49 | 1.80 | 0.855 | ||||
| AFP (>400 ng/mL) | 2.04 | 1.38 | 3.02 | <0.001 | 1.69 | 1.09 | 2.60 | 0.018 |
| HBsAg (+) | 1.18 | 0.70 | 1.97 | 0.534 | ||||
| Perihepatic invasion | 3.50 | 1.29 | 9.52 | 0.014 | 4.09 | 1.40 | 11.92 | 0.010 |
| PVTT | 1.69 | 0.93 | 3.08 | 0.085 | 1.23 | 0.64 | 2.37 | 0.528 |
| Tumor number (>2) | 1.56 | 1.08 | 2.26 | 0.018 | 1.45 | 0.98 | 2.14 | 0.064 |
| Maximum tumor diameter (>5 cm) | 2.05 | 1.42 | 2.98 | 0.000 | 1.38 | 0.89 | 2.14 | 0.151 |
| B cell (>166 cells/μL) | 0.66 | 0.46 | 0.95 | 0.026 | 0.72 | 0.49 | 1.06 | 0.097 |
| CD4+ T cell (>333 cells/μL) | 1.61 | 0.75 | 3.46 | 0.221 | ||||
| CD8+ T cell (>478 cells/μL) | 0.65 | 0.42 | 0.99 | 0.046 | 0.62 | 0.40 | 0.96 | 0.031 |
| CD4/CD8(>1.05) | 1.35 | 0.74 | 2.46 | 0.321 | ||||
| NK cell | 0.010 | 0.004 | ||||||
| lower (<120 cells/μL) | 1.92 | 1.18 | 3.14 | 0.009 | 2.23 | 1.34 | 3.70 | 0.002 |
| upper (>420 cells/μL) | 1.64 | 1.09 | 2.48 | 0.018 | 1.57 | 1.03 | 2.41 | 0.038 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, C.; Lu, Z.; Sun, B.; Yi, Y.; Zhang, B.; Wang, Z.; Qiu, S.-J. Peripheral Lymphocytes in Primary Liver Cancers: Elevated NK and CD8+ T Cells and Dysregulated Selenium Metabolism. Biomolecules 2024, 14, 222. https://doi.org/10.3390/biom14020222
Zhou C, Lu Z, Sun B, Yi Y, Zhang B, Wang Z, Qiu S-J. Peripheral Lymphocytes in Primary Liver Cancers: Elevated NK and CD8+ T Cells and Dysregulated Selenium Metabolism. Biomolecules. 2024; 14(2):222. https://doi.org/10.3390/biom14020222
Chicago/Turabian StyleZhou, Cheng, Zhufeng Lu, Baoye Sun, Yong Yi, Boheng Zhang, Zheng Wang, and Shuang-Jian Qiu. 2024. "Peripheral Lymphocytes in Primary Liver Cancers: Elevated NK and CD8+ T Cells and Dysregulated Selenium Metabolism" Biomolecules 14, no. 2: 222. https://doi.org/10.3390/biom14020222
APA StyleZhou, C., Lu, Z., Sun, B., Yi, Y., Zhang, B., Wang, Z., & Qiu, S.-J. (2024). Peripheral Lymphocytes in Primary Liver Cancers: Elevated NK and CD8+ T Cells and Dysregulated Selenium Metabolism. Biomolecules, 14(2), 222. https://doi.org/10.3390/biom14020222






