ALMS1-IT1: A Key Player in the Novel Disulfidptosis-Related LncRNA Prognostic Signature for Head and Neck Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection and Processing
2.2. Identification of Disulfidptosis-Related lncRNAs
2.3. Establishment and Verification of a Disulfidptosis-Related lncRNA Prognostic Model
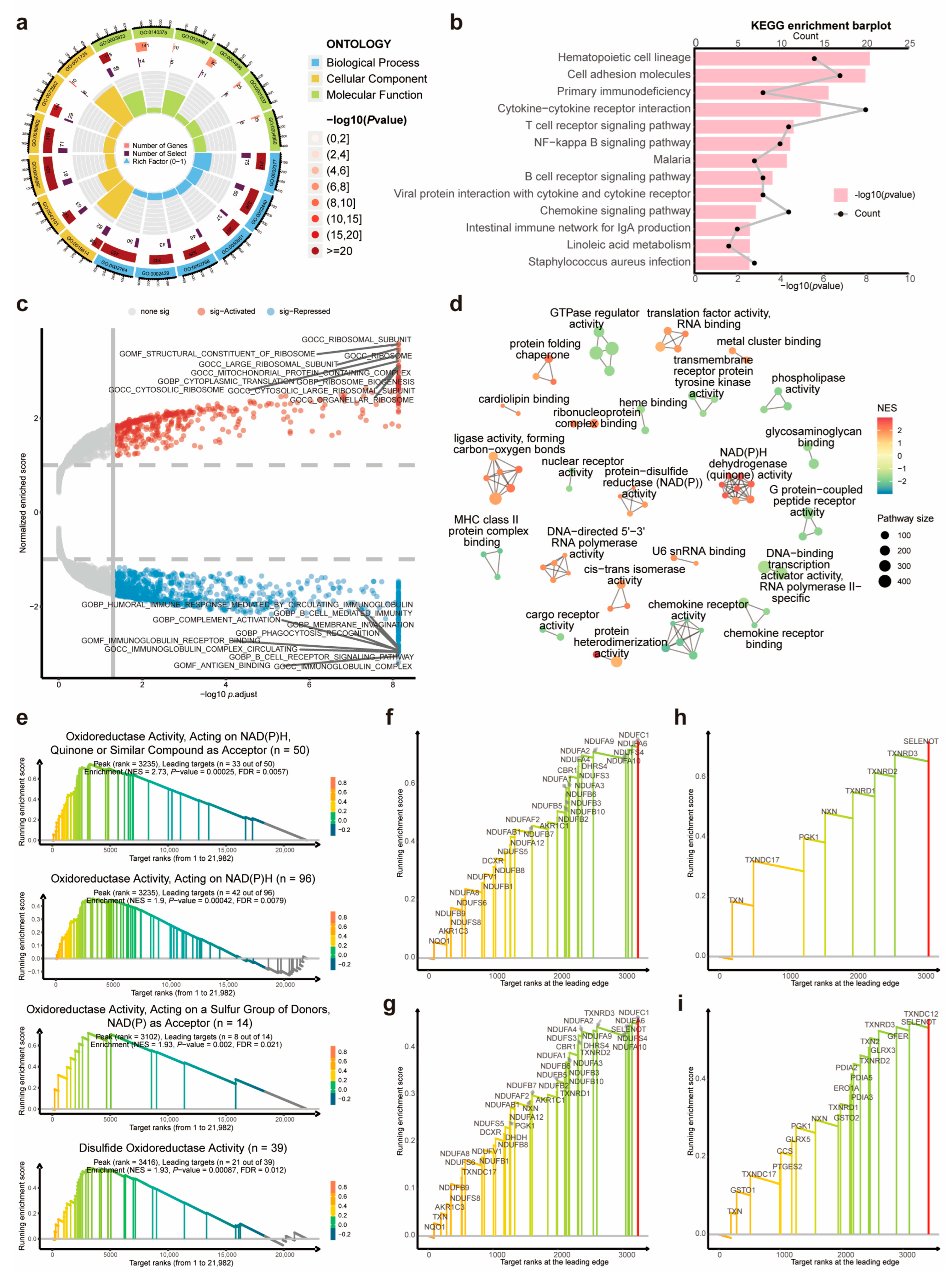
2.4. Functional Enrichment Analysis and Gene Set Enrichment Analysis
2.5. Assessment of Immune Cell Infiltration and Immune Microenvironment
2.6. Cell Lines and Culture
2.7. Small Interfering RNA (siRNA) Transfection
2.8. RNA Extraction and Reverse Transcription–Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
2.9. Cell Death Assays
2.10. Western Blot
2.11. NADP+ and NADPH Measurements
2.12. Immunofluorescence Staining of Actin Filaments
2.13. Statistical Analysis
3. Results
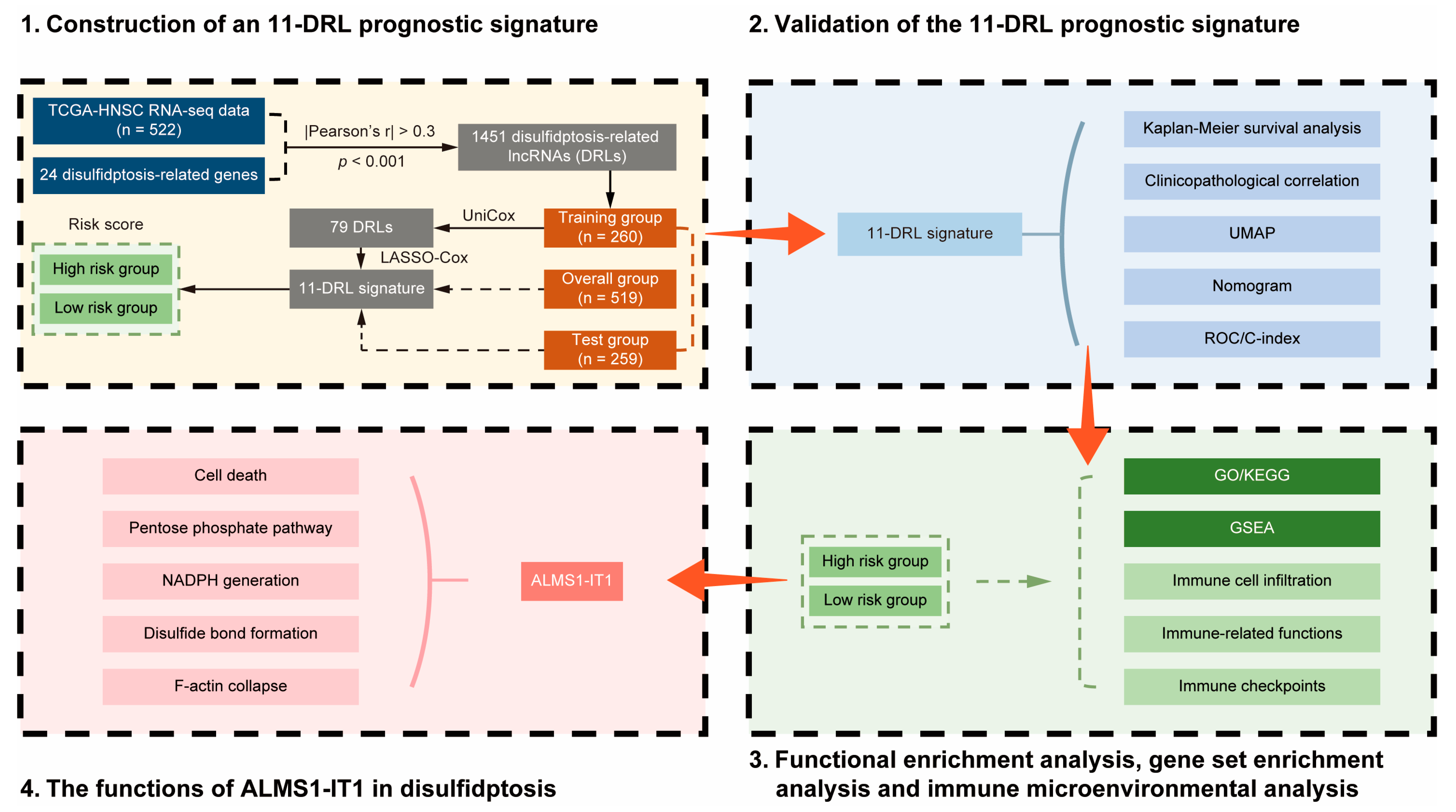
3.1. Publicly Available Data Collection and Identification of Disulfidptosis-Related lncRNAs in HNSCC
3.2. Construction of an 11-DRL Signature as a Novel Prognostic Model for HNSCC
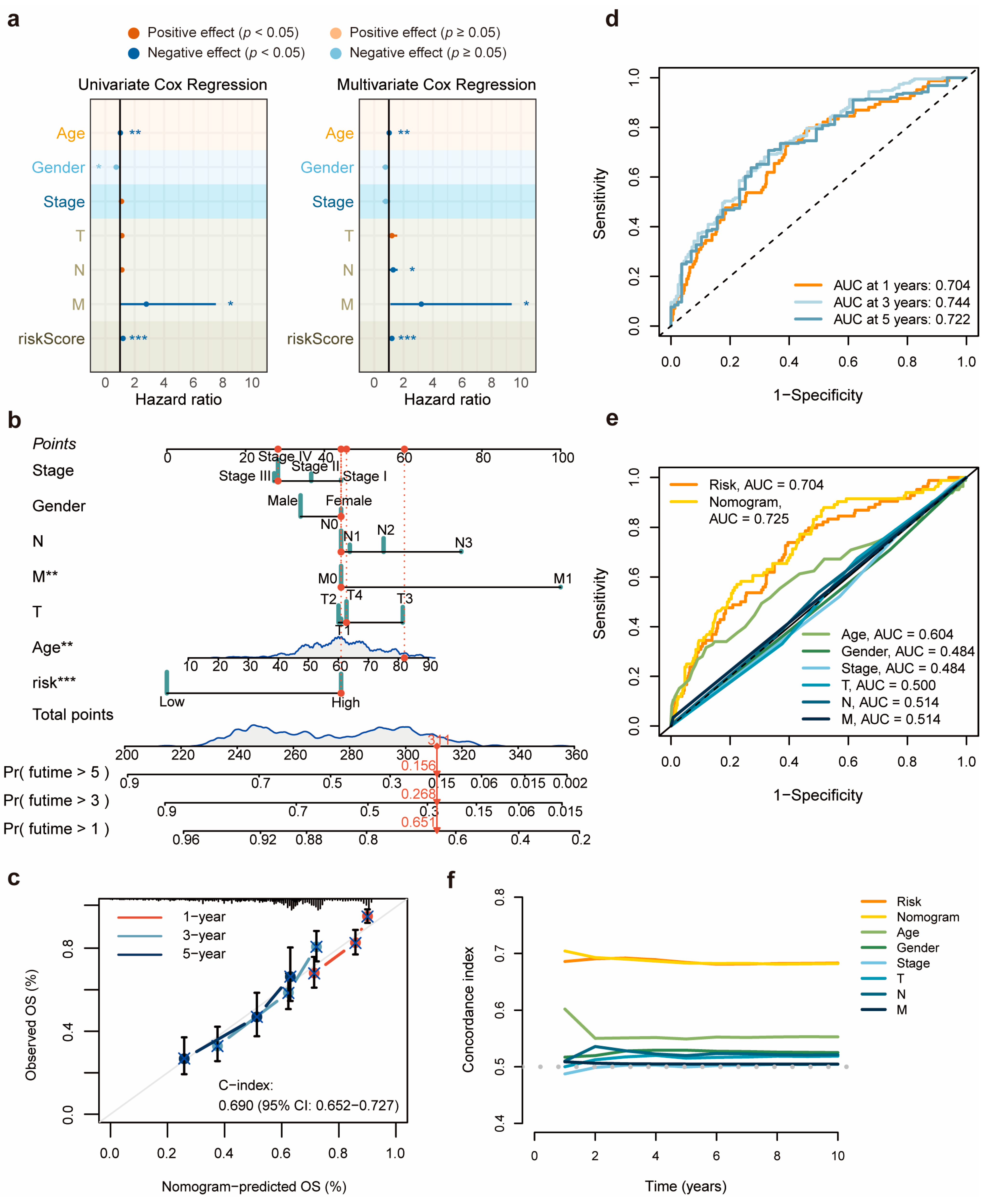
3.3. Validation and Evaluation of the 11-DRL Prognostic Signature in HNSCC
3.4. Dysregulated Immunity-, NADPH-, and Disulfide-Related Pathways between the DRL-Based Risk Clusters
3.5. Immune Landscape in HNSCC and Its Association with the 11-DRL Risk Score
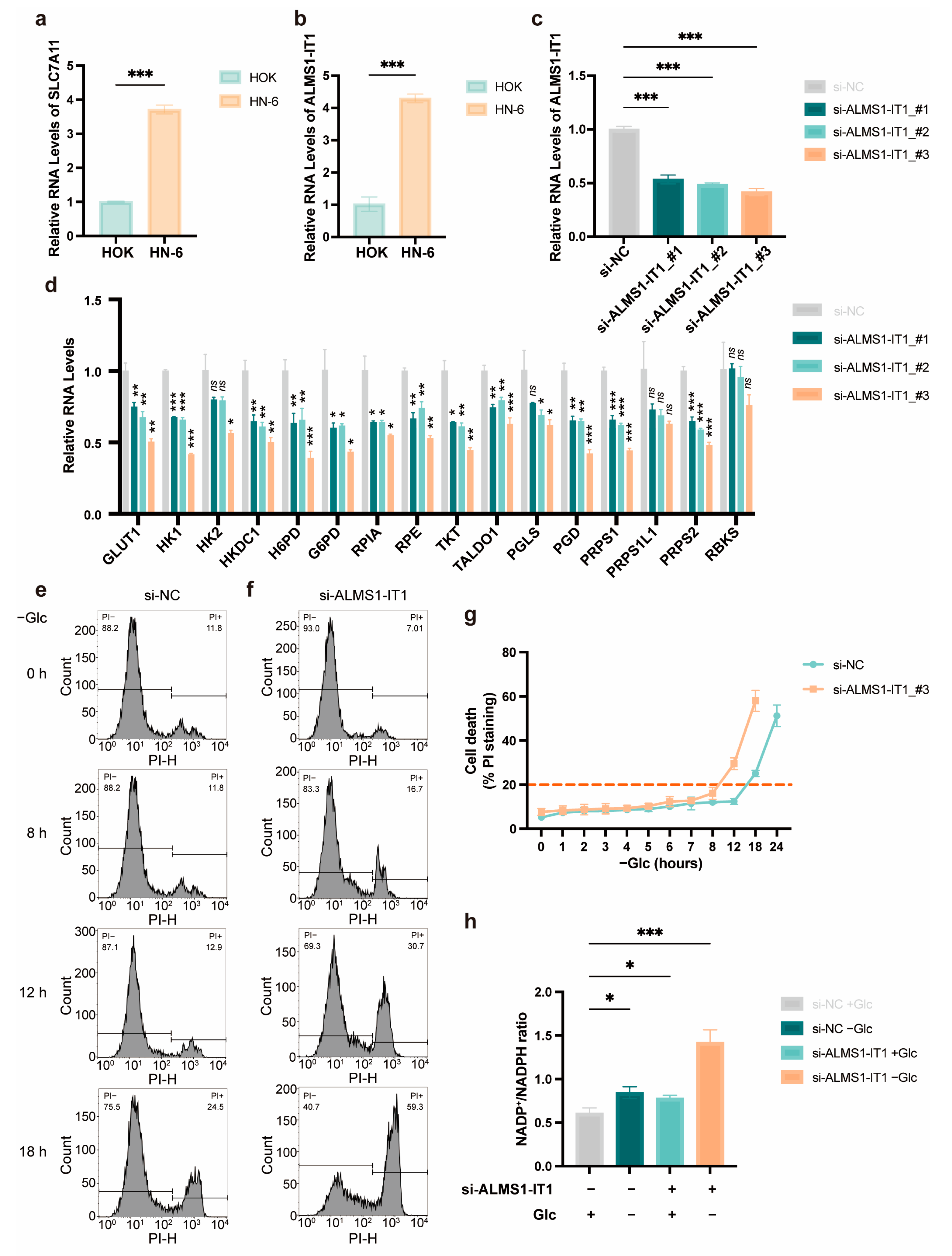
3.6. High Expression of ALMS1-IT1 Is Associated with Poor Prognosis in HNSCC, Modulating Cell Death under Starvation in a PPP-Dependent Manner
3.7. ALMS1-IT1 Regulates Disulfide Bond Formation between Actin Cytoskeleton Proteins during Disulfidptosis
3.8. ALMS1-IT1 Modulates F-Actin Collapse during Disulfidptosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACC | Adrenocortical Carcinoma |
| AUC | Area Under Curve |
| BLCA | Bladder Urothelial Carcinoma |
| BRCA | Breast Invasive Carcinoma |
| CESC | Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma |
| CHOL | Cholangiocarcinoma |
| COAD | Colon Adenocarcinoma |
| DEGs | Differentially Expressed Genes |
| DLBC | Lymphoid Neoplasm Diffuse Large B-Cell Lymphoma |
| DRGs | Disulfidptosis-Related Genes |
| DRLs | Disulfidptosis-Related LncRNAs |
| ESCA | Esophageal Carcinoma |
| GBM | Glioblastoma Multiforme |
| GO | Gene Ontology |
| GSEA | Gene Set Enrichment Analysis |
| HNSCC | Head and Neck Squamous Cell Carcinoma |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KICH | Kidney Chromophobe |
| KIRC | Kidney Renal Clear Cell Carcinoma |
| KIRP | Kidney Renal Papillary Cell Carcinoma |
| LAML | Acute Myeloid Leukemia |
| LGG | Brain Lower Grade Glioma |
| LIHC | Liver Hepatocellular Carcinoma |
| LncRNAs | Long Noncoding RNAs |
| LUAD | Lung Adenocarcinoma |
| LUSC | Lung Squamous Cell Carcinoma |
| MESO | Mesothelioma |
| OS | Overall Survival |
| OV | Ovarian Serous Cystadenocarcinoma |
| PAAD | Pancreatic Adenocarcinoma |
| PCPG | Pheochromocytoma and Paraganglioma |
| PEA | Pathway Enrichment Analysis |
| PFS | Progression-Free Survival |
| PRAD | Prostate Adenocarcinoma |
| PPP | Pentose Phosphate Pathway |
| READ | Rectum Adenocarcinoma |
| ROC | Receiver Operating Characteristic Curve |
| SARC | Sarcoma |
| SKCM | Skin Cutaneous Melanoma |
| STAD | Stomach Adenocarcinoma |
| TGCT | Testicular Germ Cell Tumors |
| THCA | Thyroid Carcinoma |
| THYM | Thymoma |
| TME | Tumor Microenvironment |
| UCEC | Uterine Corpus Endometrial Carcinoma |
| UCS | Uterine Carcinosarcoma |
| UVM | Uveal Melanoma |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Beck, T.N.; Golemis, E.A. Genomic Insights into Head and Neck Cancer. Cancers Head Neck 2016, 1, 1. [Google Scholar] [CrossRef]
- Liu, X.; Nie, L.; Zhang, Y.; Yan, Y.; Wang, C.; Colic, M.; Olszewski, K.; Horbath, A.; Chen, X.; Lei, G.; et al. Actin Cytoskeleton Vulnerability to Disulfide Stress Mediates Disulfidptosis. Nat. Cell Biol. 2023, 25, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Olszewski, K.; Zhang, Y.; Lim, E.W.; Shi, J.; Zhang, X.; Zhang, J.; Lee, H.; Koppula, P.; Lei, G.; et al. Cystine Transporter Regulation of Pentose Phosphate Pathway Dependency and Disulfide Stress Exposes a Targetable Metabolic Vulnerability in Cancer. Nat. Cell Biol. 2020, 22, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Liu, Q.; Xing, F.; Zeng, C.; Wang, W. Disulfidptosis: A New Form of Programmed Cell Death. J. Exp. Clin. Cancer Res. 2023, 42, 137. [Google Scholar] [CrossRef]
- Koppula, P.; Zhuang, L.; Gan, B. Cystine Transporter SLC7A11/xCT in Cancer: Ferroptosis, Nutrient Dependency, and Cancer Therapy. Protein Cell 2021, 12, 599–620. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, N.; Wu, C.; Prime, S.S. Heterogeneity of Cancer Stem Cells in Tumorigenesis, Metastasis, and Resistance to Antineoplastic Treatment of Head and Neck Tumours. Cells 2021, 10, 3068. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Yang, L. Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol. 2018, 28, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, T.; Zhou, W.; Li, J.; Li, X.; Wang, Q.; Jin, X.; Yin, J.; Chen, L.; Zhang, Y.; et al. Pan-Cancer Characterization of Immune-Related lncRNAs Identifies Potential Oncogenic Biomarkers. Nat. Commun. 2020, 11, 1000. [Google Scholar] [CrossRef]
- Guo, Y.; Qu, Z.; Li, D.; Bai, F.; Xing, J.; Ding, Q.; Zhou, J.; Yao, L.; Xu, Q. Identification of a Prognostic Ferroptosis-Related lncRNA Signature in the Tumor Microenvironment of Lung Adenocarcinoma. Cell Death Discov. 2021, 7, 190. [Google Scholar] [CrossRef]
- Lei, G.; Zhang, Y.; Hong, T.; Zhang, X.; Liu, X.; Mao, C.; Yan, Y.; Koppula, P.; Cheng, W.; Sood, A.K.; et al. Ferroptosis as a Mechanism to Mediate P53 Function in Tumor Radiosensitivity. Oncogene 2021, 40, 3533–3547. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cheng, W.; Li, H.; Song, Y. Identification and Validation of Cuproptosis-Related LncRNA Signatures as a Novel Prognostic Model for Head and Neck Squamous Cell Cancer. Cancer Cell Int. 2022, 22, 345. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Huan, Y.; Li, Y.; Gao, X.; Sun, Q.; Zhang, F.; Jiang, T. Transcriptional and Genetic Alterations of Cuproptosis-Related Genes Correlated to Malignancy and Immune-Infiltrate of Esophageal Carcinoma. Cell Death Discov. 2022, 8, 370. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Piao, H.; Zhuang, L.; Sarbassov, D.D.; Ma, L.; Gan, B. FoxO Transcription Factors Promote AKT Ser473 Phosphorylation and Renal Tumor Growth in Response to Pharmacologic Inhibition of the PI3K–AKT Pathway. Cancer Res. 2014, 74, 1682–1693. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.S.; Liu, X.; Jing, J.; Lee, H.; Yadav, R.K.; Liu, J.; Zhou, Y.; Gan, B. STIM2 Interacts with AMPK and Regulates Calcium-induced AMPK Activation. FASEB J. 2019, 33, 2957–2970. [Google Scholar] [CrossRef] [PubMed]
- Kerseviciute, I.; Gordevicius, J. aPEAR: An R Package for Autonomous Visualisation of Pathway Enrichment Networks. bio-Rxiv 2023. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust Enumeration of Cell Subsets from Tissue Expression Profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Sutton, G.J.; Poppe, D.; Simmons, R.K.; Walsh, K.; Nawaz, U.; Lister, R.; Gagnon-Bartsch, J.A.; Voineagu, I. Comprehensive Evaluation of Deconvolution Methods for Human Brain Gene Expression. Nat. Commun. 2022, 13, 1358. [Google Scholar] [CrossRef]
- Qie, Y.; Yuan, H.; Von Roemeling, C.A.; Chen, Y.; Liu, X.; Shih, K.D.; Knight, J.A.; Tun, H.W.; Wharen, R.E.; Jiang, W.; et al. Surface Modification of Nanoparticles Enables Selective Evasion of Phagocytic Clearance by Distinct Macrophage Phenotypes. Sci. Rep. 2016, 6, 26269. [Google Scholar] [CrossRef] [PubMed]
- Yuen, G.J.; Demissie, E.; Pillai, S. B Lymphocytes and Cancer: A Love–Hate Relationship. Trends Cancer 2016, 2, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Grimbaldeston, M.; Tsai, M. Immunomodulatory Mast Cells: Negative, as Well as Positive, Regulators of Immunity. Nat. Rev. Immunol. 2008, 8, 478–486. [Google Scholar] [CrossRef]
- Gutiérrez-Melo, N.; Baumjohann, D. T Follicular Helper Cells in Cancer. Trends Cancer 2023, 9, 309–325. [Google Scholar] [CrossRef]
- Ohkura, N.; Sakaguchi, S. Transcriptional and Epigenetic Basis of Treg Cell Development and Function: Its Genetic Anomalies or Variations in Autoimmune Diseases. Cell Res. 2020, 30, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Donini, C.; D’Ambrosio, L.; Grignani, G.; Aglietta, M.; Sangiolo, D. Next Generation Immune-Checkpoints for Cancer Therapy. J. Thorac. Dis. 2018, 10, S1581–S1601. [Google Scholar] [CrossRef]
- Lee, Y.; Martin-Orozco, N.; Zheng, P.; Li, J.; Zhang, P.; Tan, H.; Park, H.J.; Jeong, M.; Chang, S.H.; Kim, B.-S.; et al. Inhibition of the B7-H3 Immune Checkpoint Limits Tumor Growth by Enhancing Cytotoxic Lymphocyte Function. Cell Res. 2017, 27, 1034–1045. [Google Scholar] [CrossRef] [PubMed]
- Picarda, E.; Ohaegbulam, K.C.; Zang, X. Molecular Pathways: Targeting B7-H3 (CD276) for Human Cancer Immunotherapy. Clin. Cancer Res. 2016, 22, 3425–3431. [Google Scholar] [CrossRef] [PubMed]
- Luan, T.; Zhang, T.; Lv, Z.; Guan, B.; Xu, J.; Li, J.; Li, M.; Hu, S. The lncRNA ALMS1-IT1 May Promote Malignant Progression of Lung Adenocarcinoma via AVL9-mediated Activation of the Cyclin-dependent Kinase Pathway. FEBS Open Bio. 2021, 11, 1504–1515. [Google Scholar] [CrossRef]
- Patra, K.C.; Hay, N. The Pentose Phosphate Pathway and Cancer. Trends Biochem. Sci. 2014, 39, 347–354. [Google Scholar] [CrossRef]
- Reckzeh, E.S.; Karageorgis, G.; Schwalfenberg, M.; Ceballos, J.; Nowacki, J.; Stroet, M.C.M.; Binici, A.; Knauer, L.; Brand, S.; Choidas, A.; et al. Inhibition of Glucose Transporters and Glutaminase Synergistically Impairs Tumor Cell Growth. Cell Chem. Biol. 2019, 26, 1214–1228.e25. [Google Scholar] [CrossRef]
- Stine, Z.E.; Schug, Z.T.; Salvino, J.M.; Dang, C.V. Targeting Cancer Metabolism in the Era of Precision Oncology. Nat. Rev. Drug Discov. 2022, 21, 141–162. [Google Scholar] [CrossRef]
- Temre, M.K.; Yadav, S.; Goel, Y.; Pandey, S.K.; Kumar, A.; Singh, S.M. Glutor, a Glucose Transporter Inhibitor, Exerts Antineoplastic Action on Tumor Cells of Thymic Origin: Implication of Modulated Metabolism, Survival, Oxidative Stress, Mitochondrial Membrane Potential, pH Homeostasis, and Chemosensitivity. Front. Oncol. 2022, 12, 925666. [Google Scholar] [CrossRef]
- Dieguez-Acuña, F.J.; Woods, J.S. Inhibition of NF-κB–DNA Binding by Mercuric Ion: Utility of the Non-Thiol Reductant, Tris(2-Carboxyethyl)Phosphine Hydrochloride (TCEP), on Detection of Impaired NF-κB–DNA Binding by Thiol-Directed Agents. Toxicol. Vitr. 2000, 14, 7–16. [Google Scholar] [CrossRef]
- Chen, H.; Yang, W.; Li, Y.; Ma, L.; Ji, Z. Leveraging a Disulfidptosis-Based Signature to Improve the Survival and Drug Sensitivity of Bladder Cancer Patients. Front. Immunol. 2023, 14, 1198878. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Zhang, Y.; Yan, Z.; Wang, Y.; Li, Y.; Qiu, Q.; Du, Y.; Chen, Z.; Liu, X. Identification of Disulfidptosis Related Subtypes, Characterization of Tumor Microenvironment Infiltration, and Development of DRG Prognostic Prediction Model in RCC, in Which MSH3 Is a Key Gene during Disulfidptosis. Front. Immunol. 2023, 14, 1205250. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Liao, P.; Liu, X.; Yang, Z.; Wang, Y.; Zhong, W.; Wang, B. Construction and Validation of a Reliable Disulfidptosis-Related LncRNAs Signature of the Subtype, Prognostic, and Immune Landscape in Colon Cancer. IJMS 2023, 24, 12915. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Qiu, K.; Dong, B.; Guo, D.; Fu, J.; Zhu, C.; Niu, Z. Disulfidptosis-Associated Long Non-Coding RNA Signature Predicts the Prognosis, Tumor Microenvironment, and Immunotherapy and Chemotherapy Options in Colon Adenocarcinoma. Cancer Cell Int. 2023, 23, 218. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Huang, J.; Yan, Y.; Jiang, C.; Zhang, S.; Chen, H.; Jiang, L.; Zhang, J.; Zhang, Q.; Yang, G.; et al. Unraveling the Role of Disulfidptosis-Related LncRNAs in Colon Cancer: A Prognostic Indicator for Immunotherapy Response, Chemotherapy Sensitivity, and Insights into Cell Death Mechanisms. Front. Mol. Biosci. 2023, 10, 1254232. [Google Scholar] [CrossRef]
- Shriwas, P.; Roberts, D.; Li, Y.; Wang, L.; Qian, Y.; Bergmeier, S.; Hines, J.; Adhicary, S.; Nielsen, C.; Chen, X. A Small-Molecule Pan-Class I Glucose Transporter Inhibitor Reduces Cancer Cell Proliferation in Vitro and Tumor Growth in Vivo by Targeting Glucose-Based Metabolism. Cancer Metab. 2021, 9, 14. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.-Y.; Xiao, M.; Fu, M.; Gao, Q.; Li, R.-F.; Wang, J.; Li, S.-L.; Ge, X.-Y. ALMS1-IT1: A Key Player in the Novel Disulfidptosis-Related LncRNA Prognostic Signature for Head and Neck Squamous Cell Carcinoma. Biomolecules 2024, 14, 266. https://doi.org/10.3390/biom14030266
Sun X-Y, Xiao M, Fu M, Gao Q, Li R-F, Wang J, Li S-L, Ge X-Y. ALMS1-IT1: A Key Player in the Novel Disulfidptosis-Related LncRNA Prognostic Signature for Head and Neck Squamous Cell Carcinoma. Biomolecules. 2024; 14(3):266. https://doi.org/10.3390/biom14030266
Chicago/Turabian StyleSun, Xin-Yi, Mian Xiao, Min Fu, Qian Gao, Rui-Feng Li, Jing Wang, Sheng-Lin Li, and Xi-Yuan Ge. 2024. "ALMS1-IT1: A Key Player in the Novel Disulfidptosis-Related LncRNA Prognostic Signature for Head and Neck Squamous Cell Carcinoma" Biomolecules 14, no. 3: 266. https://doi.org/10.3390/biom14030266
APA StyleSun, X.-Y., Xiao, M., Fu, M., Gao, Q., Li, R.-F., Wang, J., Li, S.-L., & Ge, X.-Y. (2024). ALMS1-IT1: A Key Player in the Novel Disulfidptosis-Related LncRNA Prognostic Signature for Head and Neck Squamous Cell Carcinoma. Biomolecules, 14(3), 266. https://doi.org/10.3390/biom14030266






