Assessment of Disordered Linker Predictions in the CAID2 Experiment
Abstract
:1. Introduction
2. Materials and Methods
3. Results
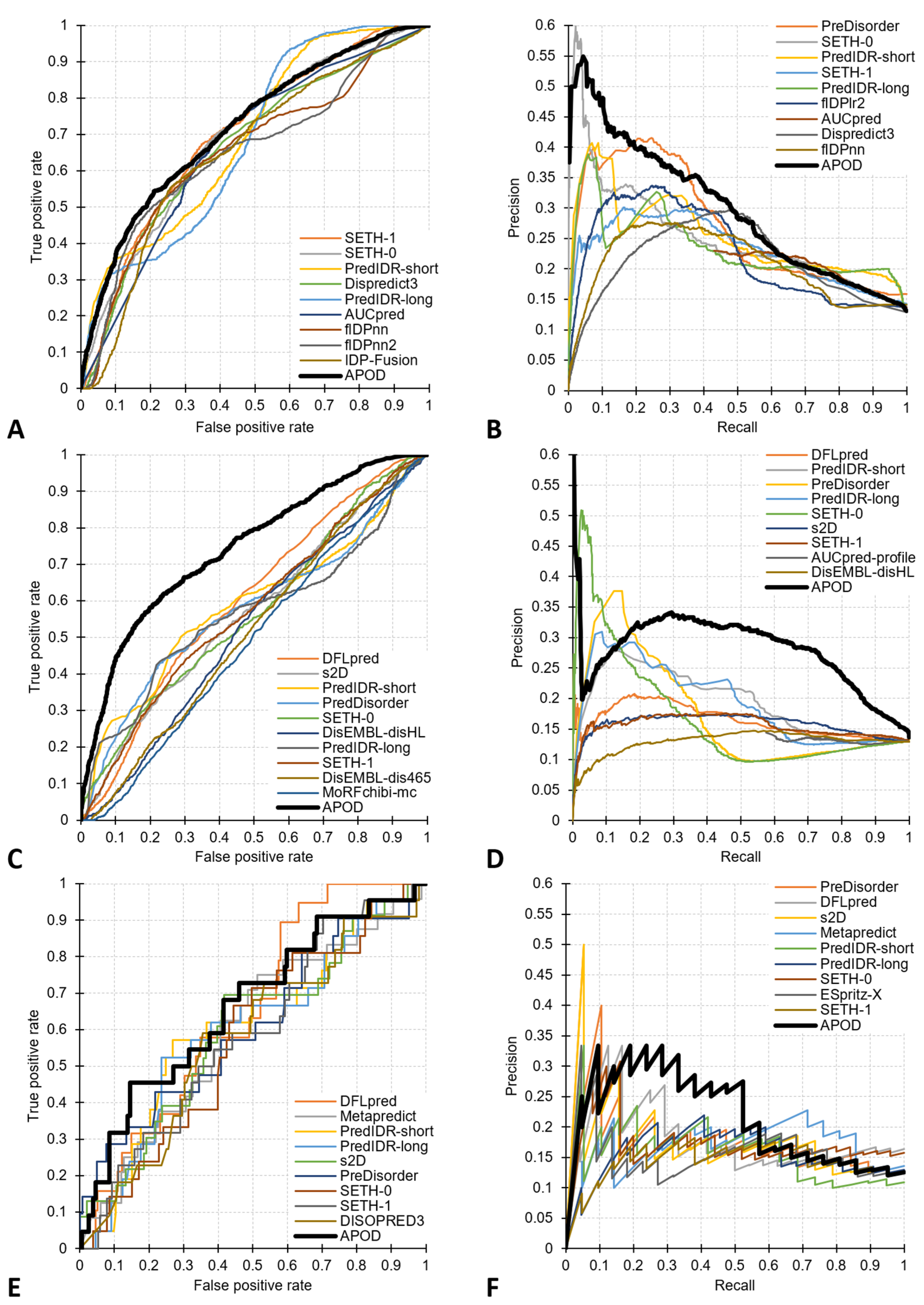
3.1. Prediction of Residues in Disordered Linkers in Protein Sequences
3.2. Prediction of Residues in Disordered Linkers among Disordered Residues
3.3. Prediction of Proteins Harboring Disordered Linkers
4. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Oldfield, C.J.; Uversky, V.N.; Dunker, A.K.; Kurgan, L. Introduction to intrinsically disordered proteins and regions. In Intrinsically Disordered Proteins; Salvi, N., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 1–34. [Google Scholar] [CrossRef]
- Uversky, V.N. Protein intrinsic disorder and structure-function continuum. Prog. Mol. Biol. Transl. Sci. 2019, 166, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Functional unfoldomics: Roles of intrinsic disorder in protein (multi)functionality. Adv. Protein Chem. Struct. Biol. 2024, 138, 179–210. [Google Scholar] [CrossRef] [PubMed]
- Aspromonte, M.C.; Nugnes, M.V.; Quaglia, F.; Bouharoua, A.; DisProt, C.; Tosatto, S.C.E.; Piovesan, D. DisProt in 2024: Improving function annotation of intrinsically disordered proteins. Nucleic Acids Res. 2023, 52, D434–D441. [Google Scholar] [CrossRef]
- Piovesan, D.; Tabaro, F.; Micetic, I.; Necci, M.; Quaglia, F.; Oldfield, C.J.; Aspromonte, M.C.; Davey, N.E.; Davidovic, R.; Dosztanyi, Z.; et al. DisProt 7.0: A major update of the database of disordered proteins. Nucleic Acids Res. 2016, D1, D219–D227. [Google Scholar] [CrossRef] [PubMed]
- Quaglia, F.; Meszaros, B.; Salladini, E.; Hatos, A.; Pancsa, R.; Chemes, L.B.; Pajkos, M.; Lazar, T.; Pena-Diaz, S.; Santos, J.; et al. DisProt in 2022: Improved quality and accessibility of protein intrinsic disorder annotation. Nucleic Acids Res. 2022, 50, D480–D487. [Google Scholar] [CrossRef] [PubMed]
- Hatos, A.; Hajdu-Soltesz, B.; Monzon, A.M.; Palopoli, N.; Alvarez, L.; Aykac-Fas, B.; Bassot, C.; Benitez, G.I.; Bevilacqua, M.; Chasapi, A.; et al. DisProt: Intrinsic protein disorder annotation in 2020. Nucleic Acids Res. 2020, 48, D269–D276. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Brown, C.J.; Lawson, J.D.; Iakoucheva, L.M.; Obradović, Z. Intrinsic Disorder and Protein Function†. Biochemistry 2002, 41, 6573–6582. [Google Scholar] [CrossRef]
- Chen, X.; Zaro, J.L.; Shen, W.C. Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef]
- Meng, F.; Kurgan, L. DFLpred: High-throughput prediction of disordered flexible linker regions in protein sequences. Bioinformatics 2016, 32, i341–i350. [Google Scholar] [CrossRef]
- Sorensen, C.S.; Kjaergaard, M. Effective concentrations enforced by intrinsically disordered linkers are governed by polymer physics. Proc. Natl. Acad. Sci. USA 2019, 116, 23124–23131. [Google Scholar] [CrossRef]
- Shvadchak, V.V.; Subramaniam, V. A Four-Amino Acid Linker between Repeats in the alpha-Synuclein Sequence Is Important for Fibril Formation. Biochemistry 2014, 53, 279–281. [Google Scholar] [CrossRef]
- Guseva, S.; Milles, S.; Jensen, M.R.; Salvi, N.; Kleman, J.P.; Maurin, D.; Ruigrok, R.W.H.; Blackledge, M. Measles virus nucleo- and phosphoproteins form liquid-like phase-separated compartments that promote nucleocapsid assembly. Sci. Adv. 2020, 6, eaaz7095. [Google Scholar] [CrossRef]
- Popovic, A.; Wu, B.; Arrowsmith, C.H.; Edwards, A.M.; Davidson, A.R.; Maxwell, K.L. Structural and Biochemical Characterization of Phage λ FI Protein (gpFI) Reveals a Novel Mechanism of DNA Packaging Chaperone Activity. J. Biol. Chem. 2012, 287, 32085–32095. [Google Scholar] [CrossRef]
- Nsasra, E.; Dahan, I.; Eichler, J.; Yifrach, O. It’s Time for Entropic Clocks: The Roles of Random Chain Protein Sequences in Timing Ion Channel Processes Underlying Action Potential Properties. Entropy 2023, 25, 1351. [Google Scholar] [CrossRef]
- Zandany, N.; Lewin, L.; Nirenberg, V.; Orr, I.; Yifrach, O. Entropic clocks in the service of electrical signaling: ‘Ball and chain’ mechanisms for ion channel inactivation and clustering. FEBS Lett. 2015, 589, 2441–2447. [Google Scholar] [CrossRef] [PubMed]
- Hoshi, T.; Zagotta, W.N.; Aldrich, R.W. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science 1990, 250, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Romero, P.R.; Noutsou, M.; Maurice, M.M.; Rudiger, S.G.; William, A.M., Jr.; Mizianty, M.J.; Kurgan, L.; Uversky, V.N.; Dunker, A.K. Stochastic machines as a colocalization mechanism for scaffold protein function. FEBS Lett. 2013, 587, 1587–1591. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Foutel, N.S.; Glavina, J.; Borcherds, W.M.; Safranchik, M.; Barrera-Vilarmau, S.; Sagar, A.; Estana, A.; Barozet, A.; Garrone, N.A.; Fernandez-Ballester, G.; et al. Conformational buffering underlies functional selection in intrinsically disordered protein regions. Nat. Struct. Mol. Biol. 2022, 29, 781–790. [Google Scholar] [CrossRef]
- Vucetic, S.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Flavors of protein disorder. Proteins 2003, 52, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Dunker, A.K.; Obradovic, Z. The protein trinity--linking function and disorder. Nat. Biotechnol. 2001, 19, 805–806. [Google Scholar] [CrossRef] [PubMed]
- Jakob, U.; Kriwacki, R.; Uversky, V.N. Conditionally and transiently disordered proteins: Awakening cryptic disorder to regulate protein function. Chem. Rev. 2014, 114, 6779–6805. [Google Scholar] [CrossRef] [PubMed]
- Kurgan, L. Resources for computational prediction of intrinsic disorder in proteins. Methods 2022, 204, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Xing, Q.; Kurgan, L. APOD: Accurate sequence-based predictor of disordered flexible linkers. Bioinformatics 2020, 36, i754–i761. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Liu, B. TransDFL: Identification of Disordered Flexible Linkers in Proteins by Transfer Learning. Genom. Proteom. Bioinform. 2023, 21, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Kurgan, L. Surveying over 100 predictors of intrinsic disorder in proteins. Expert Rev. Proteom. 2021, 18, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, X.; Liu, B. A comprehensive review and comparison of existing computational methods for intrinsically disordered protein and region prediction. Brief Bioinform. 2019, 20, 330–346. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Kurgan, L. Machine Learning for Intrinsic Disorder Prediction. In Machine Learning in Bioinformatics of Protein Sequences; World Scientific: Singarope, 2023; pp. 205–236. [Google Scholar] [CrossRef]
- Atkins, J.D.; Boateng, S.Y.; Sorensen, T.; McGuffin, L.J. Disorder Prediction Methods, Their Applicability to Different Protein Targets and Their Usefulness for Guiding Experimental Studies. Int. J. Mol. Sci. 2015, 16, 19040–19054. [Google Scholar] [CrossRef]
- Necci, M.; Piovesan, D.; Predictors, C.; DisProt, C.; Tosatto, S.C.E. Critical assessment of protein intrinsic disorder prediction. Nat. Methods 2021, 18, 472–481. [Google Scholar] [CrossRef]
- Lang, B.; Babu, M.M. A community effort to bring structure to disorder. Nat. Methods 2021, 18, 454–455. [Google Scholar] [CrossRef]
- Conte, A.D.; Mehdiabadi, M.; Bouhraoua, A.; Miguel Monzon, A.; Tosatto, S.C.E.; Piovesan, D. Critical assessment of protein intrinsic disorder prediction (CAID)—Results of round 2. Proteins 2023, 91, 1925–1934. [Google Scholar] [CrossRef] [PubMed]
- Hanson, J.; Paliwal, K.K.; Litfin, T.; Zhou, Y. SPOT-Disorder2: Improved Protein Intrinsic Disorder Prediction by Ensembled Deep Learning. Genom. Proteom. Bioinform. 2019, 17, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Ilzhofer, D.; Heinzinger, M.; Rost, B. SETH predicts nuances of residue disorder from protein embeddings. Front. Bioinform. 2022, 2, 1019597. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ma, J.Z.; Xu, J.B. AUCpreD: Proteome-level protein disorder prediction by AUC-maximized deep convolutional neural fields. Bioinformatics 2016, 32, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.J.; Pang, Y.H.; Liu, B. DeepIDP-2L: Protein intrinsically disordered region prediction by combining convolutional attention network and hierarchical attention network. Bioinformatics 2022, 38, 1252–1260. [Google Scholar] [CrossRef]
- Linding, R.; Jensen, L.J.; Diella, F.; Bork, P.; Gibson, T.J.; Russell, R.B. Protein disorder prediction: Implications for structural proteomics. Structure 2003, 11, 1453–1459. [Google Scholar] [CrossRef]
- Orlando, G.; Raimondi, D.; Codice, F.; Tabaro, F.; Vranken, W. Prediction of Disordered Regions in Proteins with Recurrent Neural Networks and Protein Dynamics. J. Mol. Biol. 2022, 434, 167579. [Google Scholar] [CrossRef]
- Jones, D.T.; Cozzetto, D. DISOPRED3: Precise disordered region predictions with annotated protein-binding activity. Bioinformatics 2015, 31, 857–863. [Google Scholar] [CrossRef]
- Iqbal, S.; Hoque, M.T. Estimation of Position Specific Energy as a Feature of Protein Residues from Sequence Alone for Structural Classification. PLoS ONE 2016, 11, e0161452. [Google Scholar] [CrossRef] [PubMed]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Tosatto, S.C. ESpritz: Accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Katuwawala, A.; Wang, K.; Wu, Z.; Ghadermarzi, S.; Gao, J.; Kurgan, L. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 2021, 12, 4438. [Google Scholar] [CrossRef]
- Galzitskaya, O.V.; Garbuzynskiy, S.O.; Lobanov, M.Y. FoldUnfold: Web server for the prediction of disordered regions in protein chain. Bioinformatics 2006, 22, 2948–2949. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, M.Y.; Galzitskaya, O.V. The Ising model for prediction of disordered residues from protein sequence alone. Phys. Biol. 2011, 8, 035004. [Google Scholar] [CrossRef] [PubMed]
- Erdos, G.; Pajkos, M.; Dosztanyi, Z. IUPred3: Prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 2021, 49, W297–W303. [Google Scholar] [CrossRef]
- Emenecker, R.J.; Griffith, D.; Holehouse, A.S. Metapredict: A fast, accurate, and easy-to-use predictor of consensus disorder and structure. Biophys. J. 2021, 120, 4312–4319. [Google Scholar] [CrossRef]
- Necci, M.; Piovesan, D.; Dosztanyi, Z.; Tosatto, S.C.E. MobiDB-lite: Fast and highly specific consensus prediction of intrinsic disorder in proteins. Bioinformatics 2017, 33, 1402–1404. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Eickholt, J.; Cheng, J. PreDisorder: Ab initio sequence-based prediction of protein disordered regions. BMC Bioinform. 2009, 10, 436. [Google Scholar] [CrossRef]
- Bitard-Feildel, T.; Callebaut, I. HCAtk and pyHCA: A Toolkit and Python API for the Hydrophobic Cluster Analysis of Protein Sequences. bioRxiv 2018. [Google Scholar] [CrossRef]
- Mirabello, C.; Wallner, B. rawMSA: End-to-end Deep Learning using raw Multiple Sequence Alignments. PLoS ONE 2019, 14, e0220182. [Google Scholar] [CrossRef]
- Yang, Z.R.; Thomson, R.; McNeil, P.; Esnouf, R.M. RONN: The bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 2005, 21, 3369–3376. [Google Scholar] [CrossRef]
- Hanson, J.; Yang, Y.D.; Paliwal, K.; Zhou, Y.Q. Improving protein disorder prediction by deep bidirectional long short-term memory recurrent neural networks. Bioinformatics 2017, 33, 685–692. [Google Scholar] [CrossRef]
- Hanson, J.; Paliwal, K.; Zhou, Y. Accurate Single-Sequence Prediction of Protein Intrinsic Disorder by an Ensemble of Deep Recurrent and Convolutional Architectures. J. Chem. Inf. Model. 2018, 58, 2369–2376. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Monastyrskyy, B.; Kryshtafovych, A.; Moult, J.; Tramontano, A.; Fidelis, K. Assessment of protein disorder region predictions in CASP10. Proteins 2014, 82 (Suppl. S2), 127–137. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.L.; Kurgan, L. Comprehensive comparative assessment of in-silico predictors of disordered regions. Curr. Protein Pept. Sci. 2012, 13, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Kurgan, L. Deep learning in prediction of intrinsic disorder in proteins. Comput. Struct. Biotechnol. J. 2022, 20, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Katuwawala, A.; Kurgan, L. Comparative Assessment of Intrinsic Disorder Predictions with a Focus on Protein and Nucleic Acid-Binding Proteins. Biomolecules 2020, 10, 1636. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Ghadermarzi, S.; Kurgan, L. Comparative evaluation of AlphaFold2 and disorder predictors for prediction of intrinsic disorder, disorder content and fully disordered proteins. Comput. Struct. Biotechnol. J. 2023, 21, 3248–3258. [Google Scholar] [CrossRef]
- Katuwawala, A.; Peng, Z.; Yang, J.; Kurgan, L. Computational Prediction of MoRFs, Short Disorder-to-order Transitioning Protein Binding Regions. Comput. Struct. Biotechnol. J. 2019, 17, 454–462. [Google Scholar] [CrossRef]
- Kurgan, L.; Hu, G.; Wang, K.; Ghadermarzi, S.; Zhao, B.; Malhis, N.; Erdos, G.; Gsponer, J.; Uversky, V.N.; Dosztanyi, Z. Tutorial: A guide for the selection of fast and accurate computational tools for the prediction of intrinsic disorder in proteins. Nat. Protoc. 2023, 18, 3157–3172. [Google Scholar] [CrossRef]
- Wilson, C.J.; Choy, W.Y.; Karttunen, M. AlphaFold2: A Role for Disordered Protein/Region Prediction? Int. J. Mol. Sci. 2022, 23, 4591. [Google Scholar] [CrossRef]
- Piovesan, D.; Monzon, A.M.; Tosatto, S.C.E. Intrinsic protein disorder and conditional folding in AlphaFoldDB. Protein Sci. 2022, 31, e4466. [Google Scholar] [CrossRef]
- Basu, S.; Hegedus, T.; Kurgan, L. CoMemMoRFPred: Sequence-based Prediction of MemMoRFs by Combining Predictors of Intrinsic Disorder, MoRFs and Disordered Lipid-binding Regions. J. Mol. Biol. 2023, 435, 168272. [Google Scholar] [CrossRef]
- Hong, Y.; Song, J.; Ko, J.; Lee, J.; Shin, W.H. S-Pred: Protein structural property prediction using MSA transformer. Sci. Rep. 2022, 12, 13891. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Li, Z.; Meng, Q.; Zhao, B.; Kurgan, L. CLIP: Accurate prediction of disordered linear interacting peptides from protein sequences using co-evolutionary information. Brief Bioinform. 2023, 24, bbac502. [Google Scholar] [CrossRef]
- Dobson, L.; Tusnady, G.E. MemDis: Predicting Disordered Regions in Transmembrane Proteins. Int. J. Mol. Sci. 2021, 22, 12270. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.H.; Dong, R.; Lee, L.; Rengarajan, S.; Riggi, N.; Boulay, G.; Rivera, M.N. DisP-seq reveals the genome-wide functional organization of DNA-associated disordered proteins. Nat. Biotechnol. 2024, 42, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Del Conte, A.; Bouhraoua, A.; Mehdiabadi, M.; Clementel, D.; Monzon, A.M.; Predictors, C.; Tosatto, S.C.E.; Piovesan, D. CAID prediction portal: A comprehensive service for predicting intrinsic disorder and binding regions in proteins. Nucleic Acids Res. 2023, 51, W62–W69. [Google Scholar] [CrossRef]
- Xu, S.; Onoda, A. Accurate and Fast Prediction of Intrinsically Disordered Protein by Multiple Protein Language Models and Ensemble Learning. J. Chem. Inf. Model. 2023. [Google Scholar] [CrossRef]
- Yu, K.; Liu, Z.; Cheng, H.; Li, S.; Zhang, Q.; Liu, J.; Ju, H.Q.; Zuo, Z.; Zhao, Q.; Kang, S.; et al. dSCOPE: A software to detect sequences critical for liquid-liquid phase separation. Brief Bioinform. 2023, 24, bbac550. [Google Scholar] [CrossRef]
- Chaurasiya, D.; Mondal, R.; Lahiri, T.; Tripathi, A.; Ghinmine, T. IDPpred: A new sequence-based predictor for identification of intrinsically disordered protein with enhanced accuracy. J. Biomol. Struct. Dyn. 2023, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Poboinev, V.V.; Khrustalev, V.V.; Khrustaleva, T.A.; Kasko, T.E.; Popkov, V.D. The PentUnFOLD algorithm as a tool to distinguish the dark and the light sides of the structural instability of proteins. Amino Acids 2022, 54, 1155–1171. [Google Scholar] [CrossRef] [PubMed]


| Predictors | AUC | lowAUCratio | AUPRC | F1max | MCCmax |
|---|---|---|---|---|---|
| APOD | 0.723 | 3.82 | 0.292 | 0.381 | 0.281 |
| SETH-1 | 0.712 = | 2.70 + | 0.241 + | 0.349 + | 0.241 + |
| SETH-0 | 0.708 = | 2.96 + | 0.257 + | 0.340 + | 0.230 + |
| PredIDR-short | 0.694 + | 3.15 + | 0.246 + | 0.341 + | 0.244 + |
| PredIDR-long | 0.683 + | 2.90 + | 0.233 + | 0.337 + | 0.246 + |
| Dispredict3 | 0.682 = | 1.61 + | 0.205 + | 0.346 + | 0.234 + |
| AUCpreD | 0.675 + | 1.94 + | 0.210 + | 0.328 + | 0.207 + |
| flDPnn | 0.661 + | 1.81 + | 0.204 + | 0.340 + | 0.225 + |
| flDPnn2 | 0.653 + | 1.91 + | 0.200 + | 0.330 + | 0.214 + |
| IDP-Fusion | 0.652 + | 1.51 + | 0.193 + | 0.328 + | 0.204 + |
| s2D | 0.648 + | 1.72 + | 0.189 + | 0.302 + | 0.171 + |
| flDPlr2 | 0.646 + | 2.48 + | 0.215 + | 0.338 + | 0.221 + |
| DeepIDP-2L | 0.642 + | 1.56 + | 0.197 + | 0.340 + | 0.222 + |
| PreDisorder | 0.641 + | 3.00 + | 0.262 = | 0.350 + | 0.219 + |
| RONN | 0.640 + | 1.51 + | 0.185 + | 0.301 + | 0.171 + |
| flDPtr | 0.635 + | 1.58 + | 0.183 + | 0.301 + | 0.171 + |
| IsUnstruct | 0.631 + | 1.31 + | 0.178 + | 0.303 + | 0.178 + |
| Metapredict | 0.629 + | 0.52 + | 0.163 + | 0.296 + | 0.171 + |
| DisoPred | 0.621 + | 1.84 + | 0.192 + | 0.307 + | 0.179 + |
| SPOT-Disorder-Single | 0.620 + | 1.46 + | 0.176 + | 0.290 + | 0.151 + |
| SPOT-Disorder | 0.617 + | 0.91 + | 0.169 + | 0.301 + | 0.173 + |
| DisEMBL-disHL | 0.616 + | 1.75 + | 0.179 + | 0.278 + | 0.137 + |
| DisoMine | 0.615 + | 0.63 + | 0.161 + | 0.290 + | 0.159 + |
| ESpritz-N | 0.615 + | 1.63 + | 0.179 + | 0.280 + | 0.141 + |
| MobiDB-lite | 0.613 + | 1.74 + | 0.176 + | 0.282 + | 0.142 + |
| DisEMBL-dis465 | 0.610 + | 1.76 + | 0.179 + | 0.279 + | 0.137 + |
| DISOPRED3 | 0.609 + | 0.79 + | 0.163 + | 0.299 + | 0.164 + |
| rawMSA | 0.606 + | 1.70 + | 0.187 + | 0.323 + | 0.197 + |
| VSL2 | 0.605 + | 0.67 + | 0.155 + | 0.290 + | 0.157 + |
| IUPred3 | 0.602 + | 1.21 + | 0.165 + | 0.281 + | 0.143 + |
| ESpritz-X | 0.602 + | 1.25 + | 0.168 + | 0.276 + | 0.132 + |
| AIUPred | 0.595 + | 0.84 + | 0.160 + | 0.283 + | 0.145 + |
| FoldUnfold | 0.581 + | 1.39 + | 0.154 + | 0.263 + | 0.109 + |
| Dispredict2 | 0.573 + | 1.22 + | 0.160 + | 0.274 + | 0.121 + |
| pyHCA | 0.569 + | 1.54 + | 0.165 + | 0.266 + | 0.136 + |
| DFLpred | 0.526 + | 1.51 + | 0.153 + | 0.235 + | 0.070 + |
| ESpritz-D | 0.512 + | 0.99 + | 0.138 + | 0.253 + | 0.109 + |
| Predictors | AUC | lowAUCratio | AUPRC | F1max | MCCmax |
|---|---|---|---|---|---|
| APOD | 0.724 | 3.00 | 0.269 | 0.367 | 0.264 |
| DFLpred | 0.614 + | 1.63 + | 0.181 + | 0.279 + | 0.136 + |
| s2D | 0.541 + | 1.03 + | 0.142 + | 0.249 + | 0.076 + |
| PredIDR-short | 0.530 + | 2.17 + | 0.173 + | 0.290 + | 0.159 + |
| PreDisorder | 0.517 + | 1.98 + | 0.172 + | 0.270 + | 0.135 + |
| SETH-0 | 0.512 + | 1.88 + | 0.167 + | 0.251 + | 0.108 + |
| DisEMBL-disHL | 0.506 + | 0.76 + | 0.128 + | 0.237 + | 0.043 + |
| PredIDR-long | 0.505 + | 2.11 + | 0.168 + | 0.284 + | 0.160 + |
| SETH-1 | 0.503 + | 1.07 + | 0.136 + | 0.248 + | 0.077 + |
| DisEMBL-dis465 | 0.495 + | 0.68 + | 0.126 + | 0.239 + | 0.046 + |
| RONN | 0.482 + | 0.61 + | 0.124 + | 0.241 + | 0.059 + |
| AUCpreD | 0.477 + | 1.01 + | 0.128 + | 0.236 + | 0.039 + |
| DISOPRED3 | 0.474 + | 0.55 + | 0.120 + | 0.239 + | 0.052 + |
| Dispredict2 | 0.465 + | 0.47 + | 0.114 + | 0.232 + | 0.055 + |
| IsUnstruct | 0.449 + | 0.46 + | 0.113 + | 0.236 + | 0.043 + |
| ESpritz-N | 0.447 + | 0.81 + | 0.119 + | 0.233 + | 0.048 + |
| FoldUnfold | 0.437 + | 0.83 + | 0.119 + | 0.186 + | 0.003 + |
| DeepIDP-2L | 0.427 + | 0.18 + | 0.109 + | 0.237 + | 0.042 + |
| Dispredict3 | 0.427 + | 0.08 + | 0.106 + | 0.227 + | 0.042 + |
| flDPlr2 | 0.421 + | 0.10 + | 0.108 + | 0.236 + | 0.051 + |
| ESpritz-X | 0.420 + | 0.80 + | 0.117 + | 0.234 + | 0.042 + |
| MobiDB-lite | 0.419 + | 0.30 + | 0.113 + | 0.206 + | 0.009 + |
| Metapredict | 0.415 + | 0.46 + | 0.112 + | 0.239 + | 0.063 + |
| SPOT-Disorder-Single | 0.414 + | 0.42 + | 0.109 + | 0.233 + | 0.040 + |
| flDPnn | 0.412 + | 0.01 + | 0.104 + | 0.235 + | 0.045 + |
| VSL2 | 0.409 + | 0.19 + | 0.103 + | 0.234 + | 0.036 + |
| rawMSA | 0.401 + | 0.00 + | 0.103 + | 0.235 + | 0.035 + |
| flDPnn2 | 0.400 + | 0.07 + | 0.102 + | 0.234 + | 0.043 + |
| IUPred3 | 0.397 + | 0.62 + | 0.106 + | 0.233 + | 0.042 + |
| IDP-Fusion | 0.390 + | 0.11 + | 0.102 + | 0.236 + | 0.032 + |
| SPOT-Disorder | 0.383 + | 0.28 + | 0.101 + | 0.230 + | 0.030 + |
| AIUPred | 0.380 + | 0.50 + | 0.105 + | 0.233 + | 0.045 + |
| pyHCA | 0.368 + | 0.60 + | 0.105 + | 0.227 + | 0.028 + |
| flDPtr | 0.367 + | 0.10 + | 0.096 + | 0.231 + | 0.033 + |
| DisoPred | 0.360 + | 0.37 + | 0.101 + | 0.238 + | 0.031 + |
| DisoMine | 0.340 + | 0.04 + | 0.092 + | 0.233 + | 0.046 + |
| ESpritz-D | 0.282 + | 0.16 + | 0.087 + | 0.229 + | 0.033 + |
| Predictors | AUC | lowAUCratio | AUPRC | F1max | MCCmax |
|---|---|---|---|---|---|
| APOD | 0.664 | 2.65 | 0.226 | 0.326 | 0.219 |
| DFLpred | 0.633 = | 1.75 + | 0.191 + | 0.280 + | 0.171 + |
| Metapredict | 0.605 + | 1.99 + | 0.179 + | 0.288 + | 0.173 + |
| PredIDR-short | 0.603 + | 1.09 + | 0.179 + | 0.304 = | 0.186 = |
| PredIDR-long | 0.594 + | 0.99 + | 0.173 + | 0.299 = | 0.179 = |
| s2D | 0.562 + | 1.66 = | 0.188 = | 0.263 + | 0.141 + |
| PreDisorder | 0.559 + | 2.41 = | 0.203 = | 0.246 + | 0.149 + |
| SETH-0 | 0.558 + | 1.22 + | 0.164 + | 0.242 + | 0.112 + |
| SETH-1 | 0.551 + | 0.51 + | 0.144 + | 0.247 + | 0.106 + |
| DISOPRED3 | 0.550 + | 0.83 + | 0.138 + | 0.258 + | 0.125 + |
| ESpritz-X | 0.548 + | 1.25 + | 0.159 + | 0.253 + | 0.124 + |
| FoldUnfold | 0.547 + | 1.08 + | 0.133 + | 0.250 + | 0.136 + |
| IsUnstruct | 0.540 + | 1.31 + | 0.142 + | 0.249 + | 0.106 + |
| DisEMBL-dis465 | 0.534 + | 0.45 + | 0.134 + | 0.251 + | 0.114 + |
| rawMSA | 0.533 + | 0.39 + | 0.134 + | 0.252 + | 0.111 + |
| ESpritz-N | 0.533 + | 0.54 + | 0.140 + | 0.243 + | 0.098 + |
| SPOT-Disorder | 0.524 + | 0.57 + | 0.141 + | 0.235 + | 0.091 + |
| DisEMBL-disHL | 0.518 + | 0.30 + | 0.130 + | 0.241 + | 0.088 + |
| RONN | 0.517 + | 0.40 + | 0.130 + | 0.242 + | 0.097 + |
| AUCpred | 0.514 + | 1.04 + | 0.126 + | 0.224 + | 0.050 + |
| MobiDB-lite | 0.513 + | 1.19 + | 0.137 + | 0.241 + | 0.106 + |
| SPOT-Disorder-Single | 0.503 + | 0.06 + | 0.123 + | 0.236 + | 0.076 + |
| pyHCA | 0.498 + | 1.04 + | 0.136 + | 0.233 + | 0.084 + |
| flDPlr2 | 0.494 + | 0.08 + | 0.124 + | 0.236 + | 0.088 + |
| VSL2 | 0.488 + | 0.09 + | 0.118 + | 0.237 + | 0.087 + |
| AIUPred | 0.481 + | 0.37 + | 0.120 + | 0.232 + | 0.077 + |
| IUPred3 | 0.479 + | 0.10 + | 0.117 + | 0.231 + | 0.073 + |
| DisoPred | 0.470 + | 0.21 + | 0.119 + | 0.225 + | 0.048 + |
| DeepIDP-2L | 0.436 + | 0.00 + | 0.105 + | 0.229 + | 0.068 + |
| DisoMine | 0.435 + | 0.00 + | 0.109 + | 0.223 + | 0.047 + |
| Dispredict3 | 0.430 + | 0.00 + | 0.105 + | 0.219 + | 0.037 + |
| flDPnn2 | 0.424 + | 0.03 + | 0.106 + | 0.219 + | 0.035 + |
| ESpritz-D | 0.421 + | 0.24 + | 0.108 + | 0.214 + | 0.026 + |
| IDP-Fusion | 0.417 + | 0.34 + | 0.113 + | 0.228 + | 0.057 + |
| flDPtr | 0.405 + | 0.00 + | 0.101 + | 0.221 + | 0.041 + |
| flDPnn | 0.389 + | 0.00 + | 0.097 + | 0.219 + | 0.036 + |
| Dispredict2 | 0.383 + | 0.04 + | 0.103 + | 0.221 + | 0.037 + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.; Hu, G.; Wu, Z.; Uversky, V.N.; Kurgan, L. Assessment of Disordered Linker Predictions in the CAID2 Experiment. Biomolecules 2024, 14, 287. https://doi.org/10.3390/biom14030287
Wang K, Hu G, Wu Z, Uversky VN, Kurgan L. Assessment of Disordered Linker Predictions in the CAID2 Experiment. Biomolecules. 2024; 14(3):287. https://doi.org/10.3390/biom14030287
Chicago/Turabian StyleWang, Kui, Gang Hu, Zhonghua Wu, Vladimir N. Uversky, and Lukasz Kurgan. 2024. "Assessment of Disordered Linker Predictions in the CAID2 Experiment" Biomolecules 14, no. 3: 287. https://doi.org/10.3390/biom14030287
APA StyleWang, K., Hu, G., Wu, Z., Uversky, V. N., & Kurgan, L. (2024). Assessment of Disordered Linker Predictions in the CAID2 Experiment. Biomolecules, 14(3), 287. https://doi.org/10.3390/biom14030287









