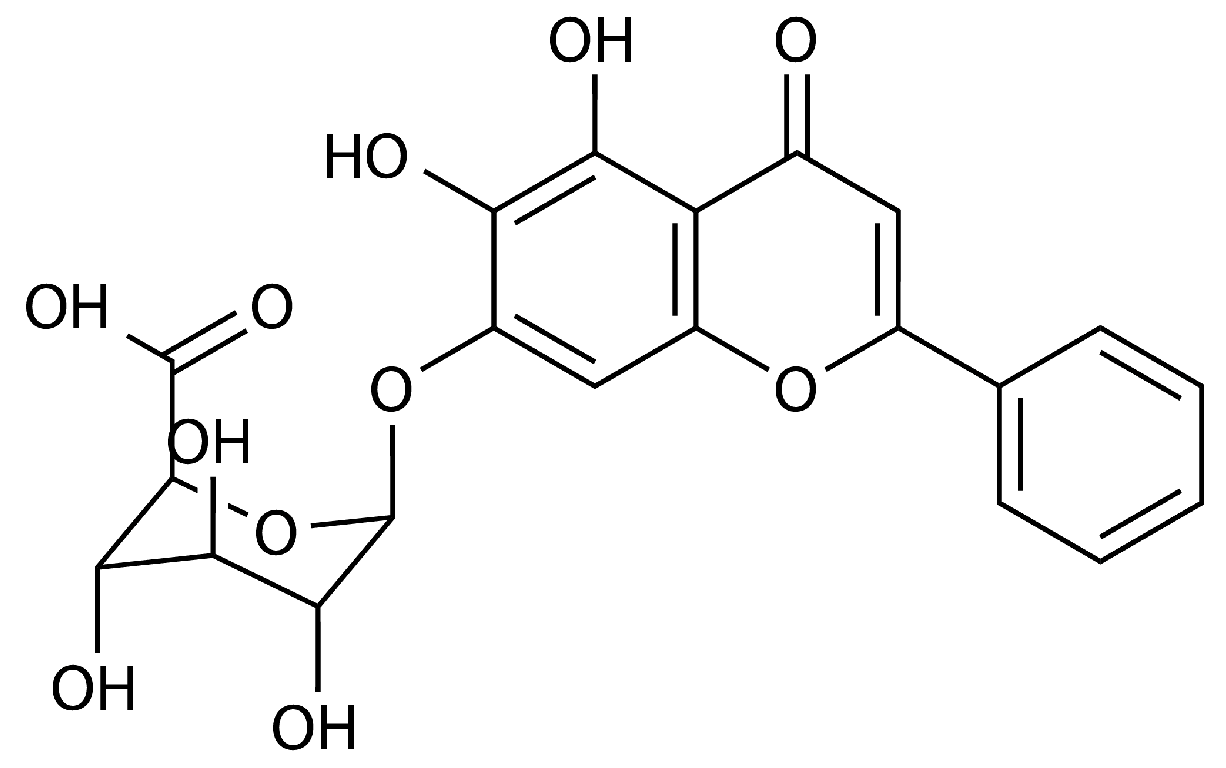
Baicalin Weakens the Virulence of Porcine Extraintestinal Pathogenic Escherichia coli by Inhibiting the LuxS/AI-2 Quorum-Sensing System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Growth Conditions, and Reagents
2.2. Cell Culture
2.3. Construction and Identification of Mutant and Complemented Strain
2.4. Growth Curve Analysis
2.5. Gene Expression Assay
2.6. AI-2 Activity Assay
2.7. Biofilm Formation Assay
2.8. Adherence and Invasion Assay
2.9. Stress Resistance Assay
2.10. Whole-Blood Bactericidal Experiments
2.11. Autoagglutination Assay
2.12. Animal Experiments
2.13. Statistical Methods
3. Results
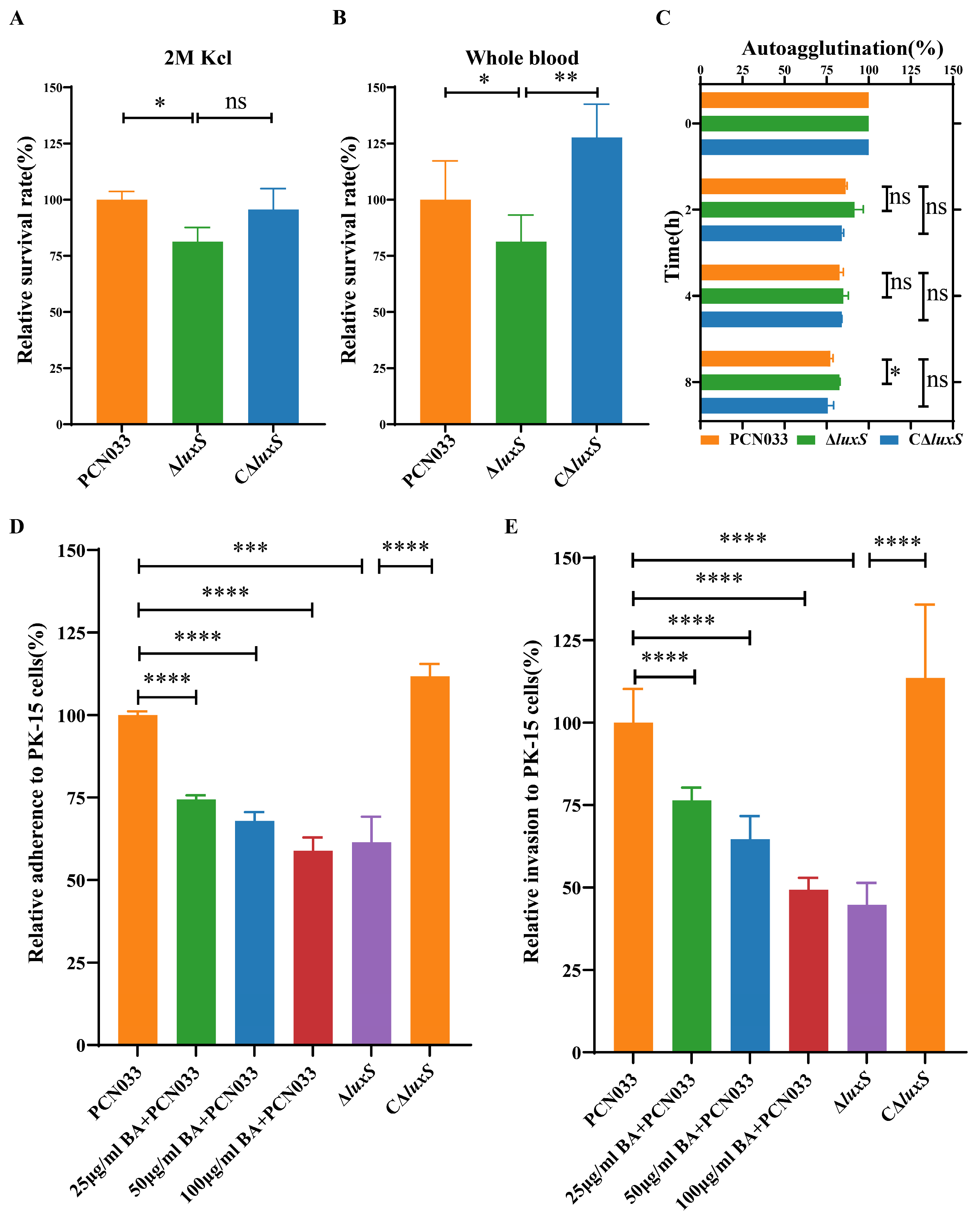
3.1. BA Weakened the Survival Ability of PCN033 under Adverse Environmental Conditions
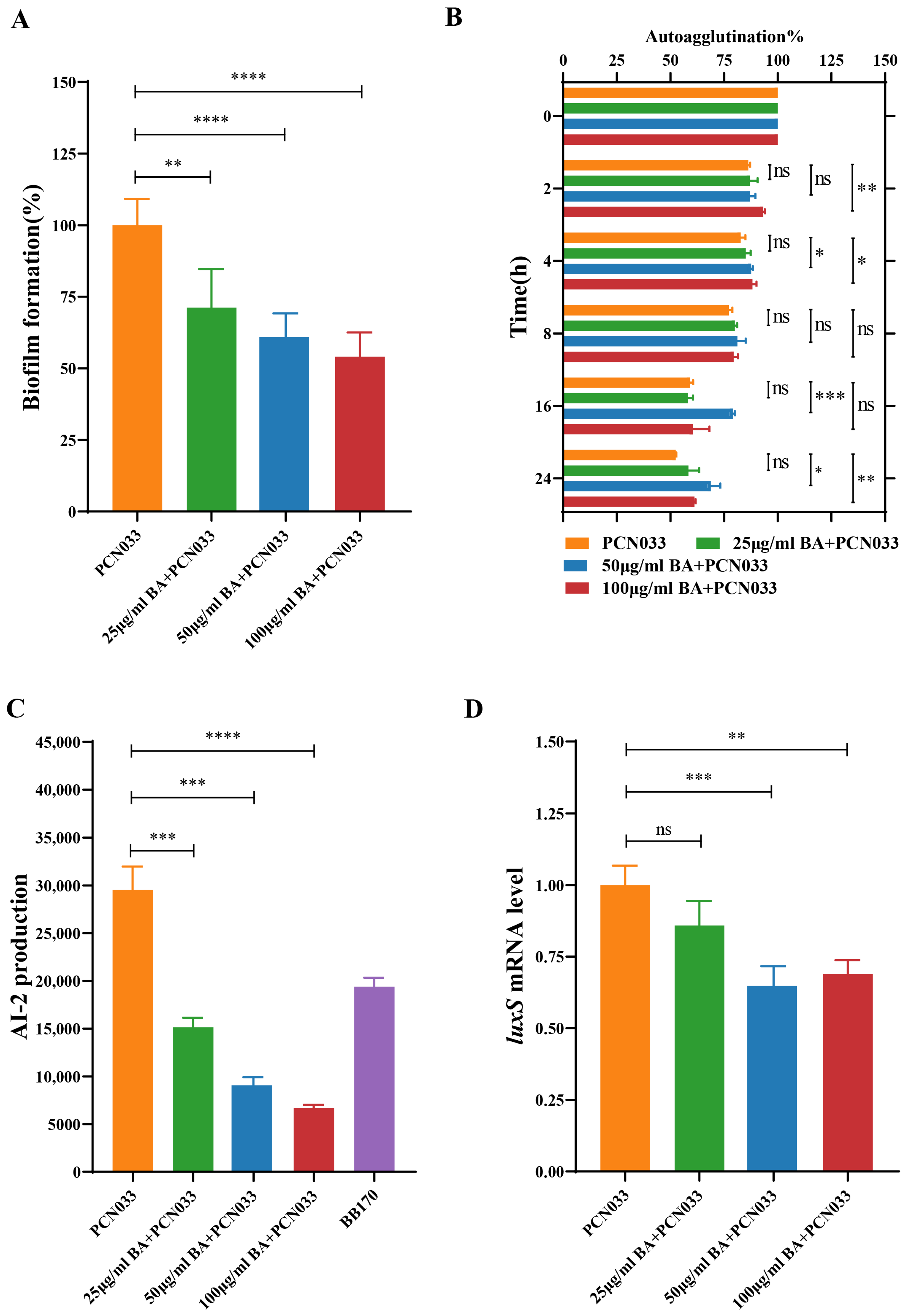
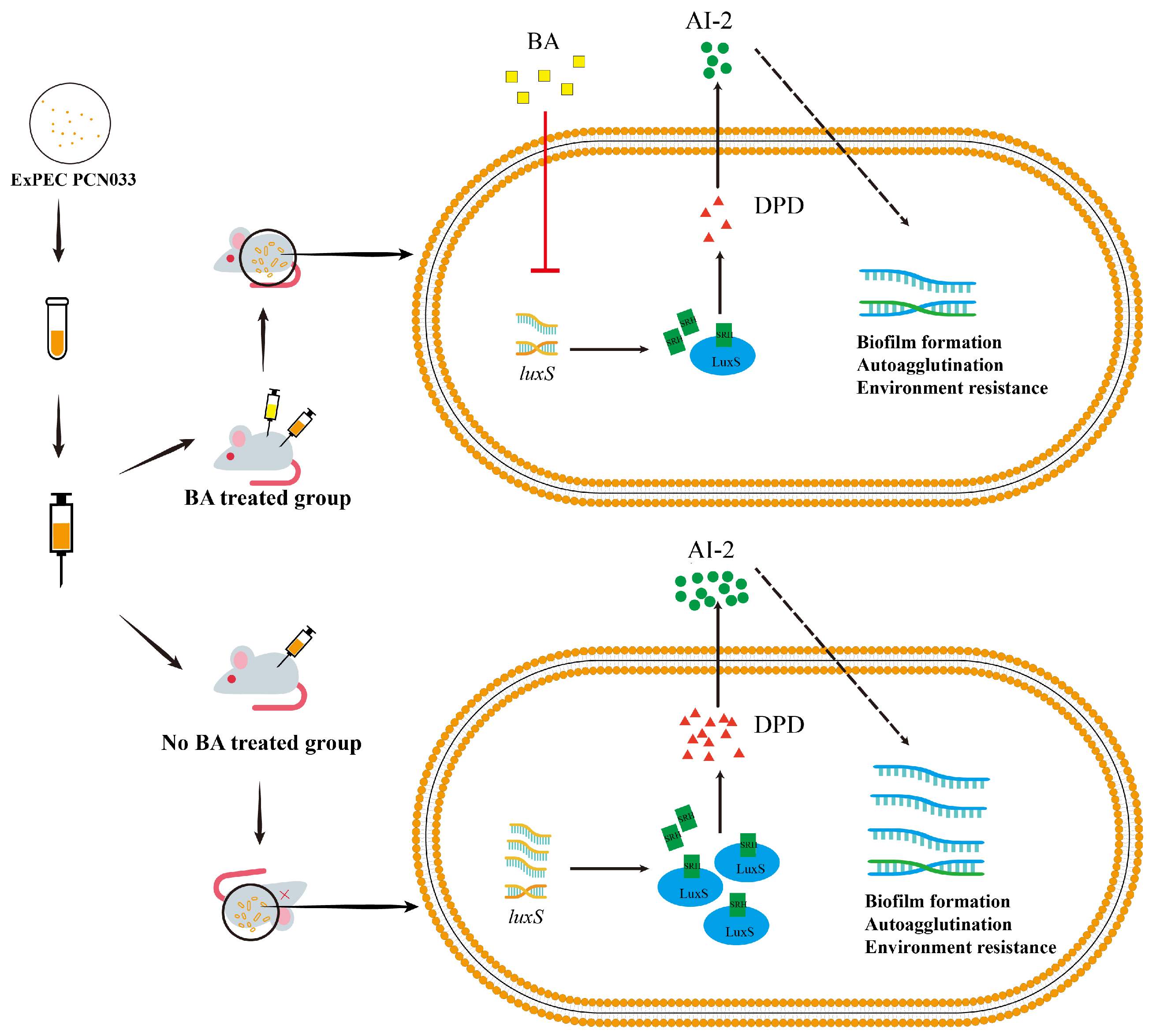
3.2. BA Weakened the Survival and Pathogenesis of PCN033 by Inhibiting the LuxS/AI-2 System
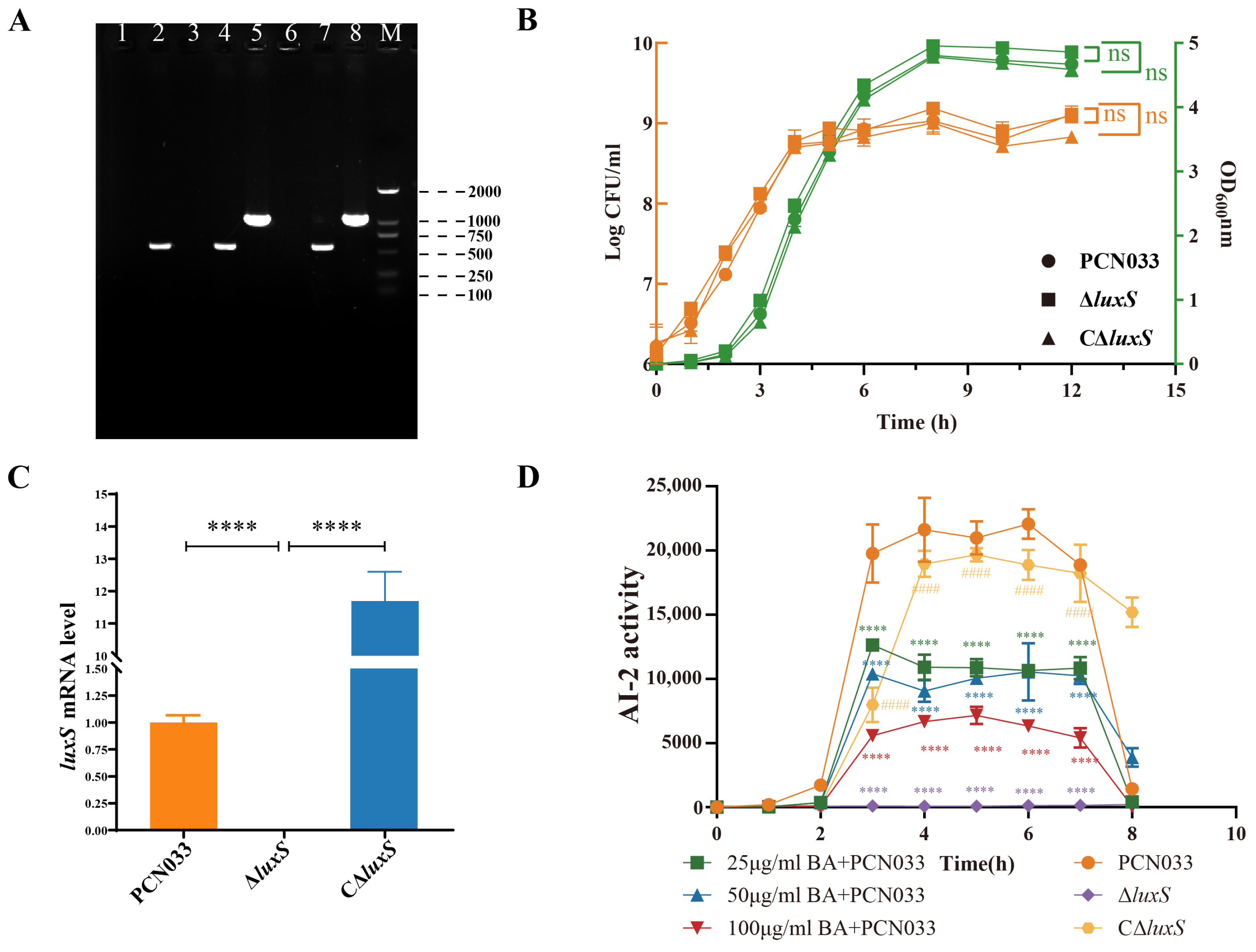
3.3. BA Reduced the Activity of AI-2 by Inhibiting the Expression of the Gene luxS
3.4. Decrease in AI-2 Activity, Caused by BA, Inhibited the Expression Level of luxS and Decreased the Survival Ability of PCN033 In Vitro
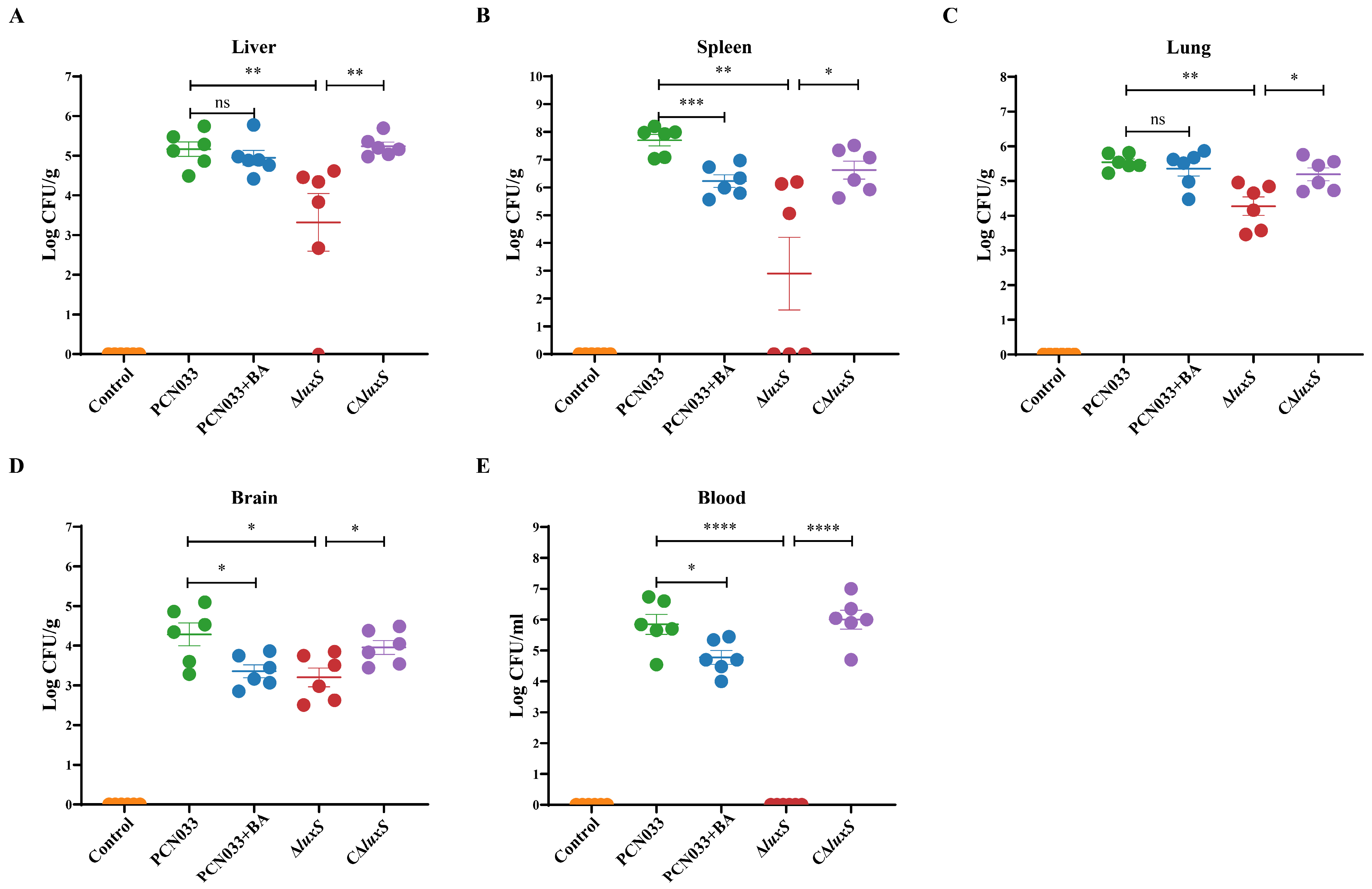
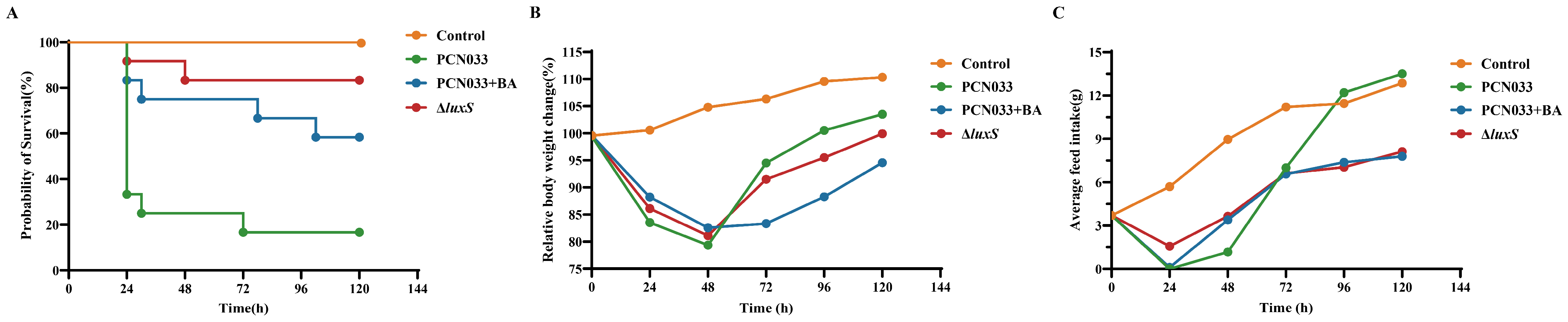
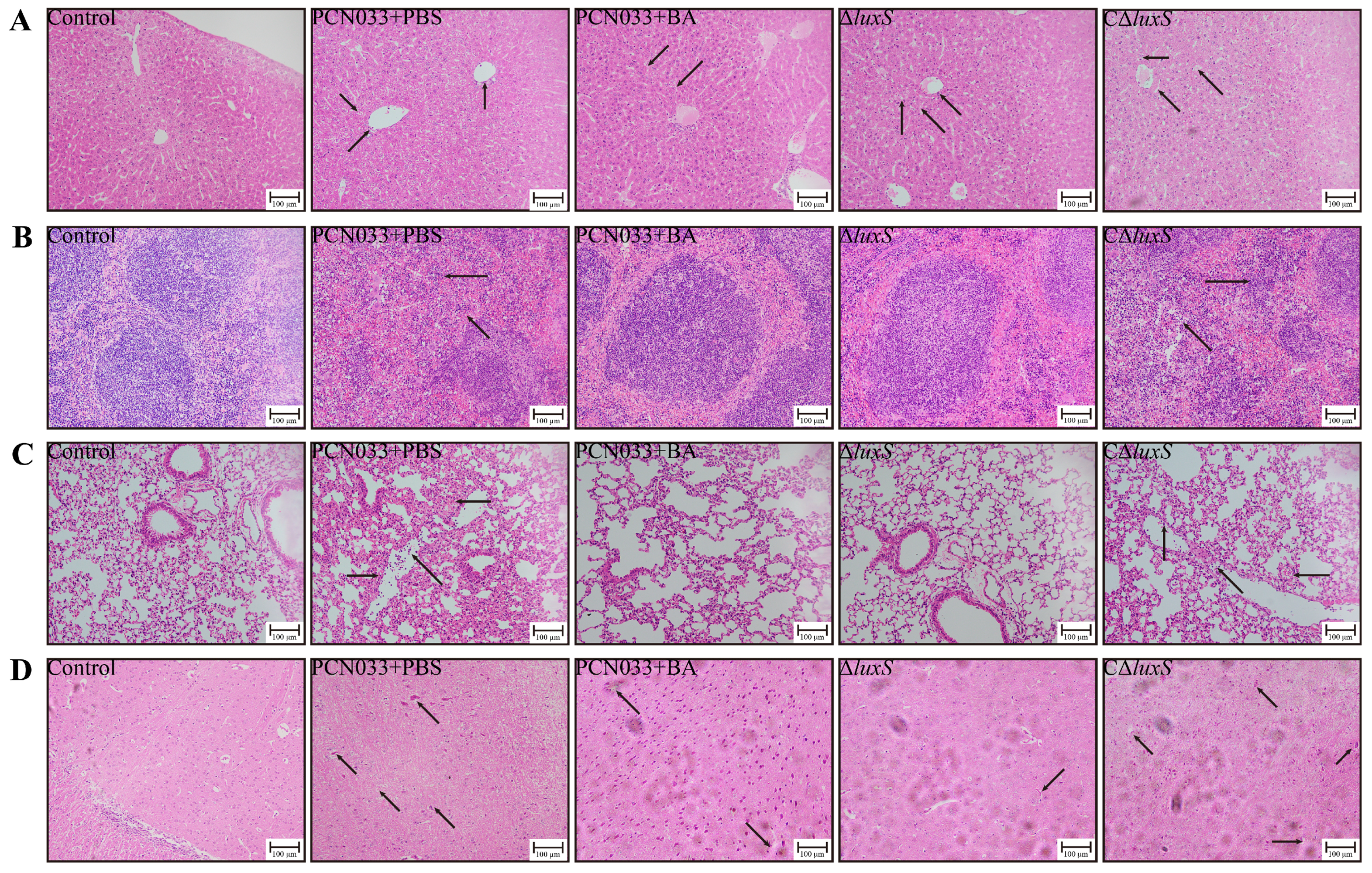
3.5. The Decrease in AI-2 Activity Caused by BA Inhibited the Expression Level of luxS and Decreased the Pathogenesis Ability of PCN033 In Vivo
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, J.L.; Fratamico, P.M.; Gunther, N.W. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 2007, 4, 134–163. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Johnson, J.R. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: Expec. J. Infect. Dis. 2000, 181, 1753–1754. [Google Scholar] [CrossRef] [PubMed]
- Troeger, H.; Richter, J.F.; Beutin, L.; Günzel, D.; Dobrindt, U.; Epple, H.J.; Gitter, A.H.; Zeitz, M.; Fromm, M.; Schulzke, J.D. Escherichia coli alpha-haemolysin induces focal leaks in colonic epithelium: A novel mechanism of bacterial translocation. Cell. Microbiol. 2007, 9, 2530–2540. [Google Scholar] [CrossRef] [PubMed]
- Totsika, M.; Kostakioti, M.; Hannan, T.J.; Upton, M.; Beatson, S.A.; Janetka, J.W.; Hultgren, S.J.; Schembri, M.A. A fimh inhibitor prevents acute bladder infection and treats chronic cystitis caused by multidrug-resistant uropathogenic Escherichia coli st131. J. Infect. Dis. 2013, 208, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Mellata, M. Human and avian extraintestinal pathogenic Escherichia coli: Infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 2013, 10, 916–932. [Google Scholar] [CrossRef] [PubMed]
- Lima-Filho, J.V.; Martins, L.V.; Nascimento, D.C.d.O.; Ventura, R.F.; Batista, J.E.C.; Silva, A.F.B.; Ralph, M.T.; Vaz, R.V.; Rabello, C.B.V.; Da Silva, I.d.M.M.; et al. Zoonotic potential of multidrug-resistant extraintestinal pathogenic Escherichia coli obtained from healthy poultry carcasses in Salvador, Brazil. Braz. J. Infect. Dis. Off. Publ. Braz. Soc. Infect. Dis. 2013, 17, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Cheng, Z.; Bai, Q.; Zhao, K.; Pan, Z.; Yao, H. Screening virulence factors of porcine extraintestinal pathogenic Escherichia coli (an emerging pathotype) required for optimal growth in swine blood. Transbound. Emerg. Dis. 2021, 68, 2005–2016. [Google Scholar] [CrossRef] [PubMed]
- Moulin-Schouleur, M.; Schouler, C.; Tailliez, P.; Kao, M.R.; Brée, A.; Germon, P.; Oswald, E.; Mainil, J.; Blanco, M.; Blanco, J. Common virulence factors and genetic relationships between o18:k1:h7 Escherichia coli isolates of human and avian origin. J. Clin. Microbiol. 2006, 44, 3484–3492. [Google Scholar] [CrossRef]
- Zhu, Y.; Dong, W.; Ma, J.; Yuan, L.; Hejair, H.M.A.; Pan, Z.; Liu, G.; Yao, H. Characterization and virulence clustering analysis of extraintestinal pathogenic Escherichia coli isolated from swine in china. BMC Vet. Res. 2017, 13, 94. [Google Scholar] [CrossRef]
- Tan, C.; Tang, X.; Zhang, X.; Ding, Y.; Zhao, Z.; Wu, B.; Cai, X.; Liu, Z.; He, Q.; Chen, H. Serotypes and virulence genes of extraintestinal pathogenic Escherichia coli isolates from diseased pigs in China. Vet. J. 2012, 192, 483–488. [Google Scholar] [CrossRef]
- Johnson, J.R.; O’Bryan, T.T.; Kuskowski, M.; Maslow, J.N. Ongoing horizontal and vertical transmission of virulence genes and papa alleles among Escherichia coli blood isolates from patients with diverse-source bacteremia. Infect. Immun. 2001, 69, 5363–5374. [Google Scholar] [CrossRef]
- Fan, Q.; Zuo, J.; Wang, H.; Grenier, D.; Yi, L.; Wang, Y. Contribution of quorum sensing to virulence and antibiotic resistance in zoonotic bacteria. Biotechnol. Adv. 2022, 59, 107965. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, J.; Liu, C.; Yang, A.; Qiao, J. Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell. Mol. Life Sci. CMLS 2020, 77, 1319–1343. [Google Scholar] [CrossRef]
- Mayer, C.; Borges, A.; Flament-Simon, S.C.; Simões, M. Quorum sensing architecture network in Escherichia coli virulence and pathogenesis. FEMS Microbiol. Rev. 2023, 47, fuad031. [Google Scholar] [CrossRef]
- Gopishetty, B.; Zhu, J.; Rajan, R.; Sobczak, A.J.; Wnuk, S.F.; Bell, C.E.; Pei, D. Probing the catalytic mechanism of s-ribosylhomocysteinase (luxs) with catalytic intermediates and substrate analogues. J. Am. Chem. Soc. 2009, 131, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, B.; Grenier, D.; Yi, L. Regulatory mechanisms of the luxs/ai-2 system and bacterial resistance. Antimicrob. Agents Chemother. 2019, 63, e01186-19. [Google Scholar] [CrossRef]
- Li, Q.; Mao, S.; Wang, H.; Ye, X. The molecular architecture of pseudomonas aeruginosa quorum-sensing inhibitors. Mar. Drugs 2022, 20, 488. [Google Scholar] [CrossRef]
- Lima, E.M.F.; Winans, S.C.; Pinto, U.M. Quorum sensing interference by phenolic compounds—A matter of bacterial misunderstanding. Heliyon 2023, 9, e17657. [Google Scholar] [CrossRef]
- Huang, T.; Liu, Y.; Zhang, C. Pharmacokinetics and bioavailability enhancement of baicalin: A review. Eur. J. Drug Metab. Pharmacokinet. 2019, 44, 159–168. [Google Scholar] [CrossRef]
- Tan, Y.Q.; Lin, F.; Ding, Y.K.; Dai, S.; Liang, Y.X.; Zhang, Y.S.; Li, J.; Chen, H.W. Pharmacological properties of total flavonoids in Scutellaria baicalensis for the treatment of cardiovascular diseases. Phytomed. Int. J. Phytother. Phytopharm. 2022, 107, 154458. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.Y.; Yuan, F.W.; Liang, T.; Liang, X.C.; Luo, Y.R.; Jiang, M.; Qing, S.Z.; Zhang, W.M. Baicalin inhibits Escherichia coli isolates in bovine mastitic milk and reduces antimicrobial resistance. J. Dairy Sci. 2018, 101, 2415–2422. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Dong, B.; Wang, K.; Cai, S.; Liu, T.; Cheng, X.; Lei, D.; Chen, Y.; Li, Y.; Kong, J.; et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. PLoS ONE 2017, 12, e0176883. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.Y.; Yuan, M.; Wu, Z.M.; Song, K.; Zhang, C.L.; An, Q.; Xia, F.; Yu, J.L.; Yi, P.F.; Fu, B.D.; et al. Anti-bacterial activity of baicalin against apec through inhibition of quorum sensing and inflammatory responses. Sci. Rep. 2019, 9, 4063. [Google Scholar] [CrossRef] [PubMed]
- Zong, B.; Zhang, Y.; Wang, X.; Liu, M.; Zhang, T.; Zhu, Y.; Zheng, Y.; Hu, L.; Li, P.; Chen, H.; et al. Characterization of multiple type-vi secretion system (t6ss) vgrg proteins in the pathogenicity and antibacterial activity of porcine extra-intestinal pathogenic Escherichia coli. Virulence 2019, 10, 118–132. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fan, Q.; Jin, M.; Mao, C.; Zhang, H.; Zhang, X.; Sun, L.; Grenier, D.; Yi, L.; Hou, X.; et al. Paeoniflorin reduce luxs/ai-2 system-controlled biofilm formation and virulence in Streptococcus suis. Virulence 2021, 12, 3062–3073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Ku, X.; Zhang, X.; Zhang, Y.; Chen, G.; Chen, F.; Zeng, W.; Li, J.; Zhu, L.; He, Q. The ai-2/luxs quorum sensing system affects the growth characteristics, biofilm formation, and virulence of Haemophilus parasuis. Front. Cell. Infect. Microbiol. 2019, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.R.; Sun, J.; Du, Y.; Pan, A.; Zeng, L.; Maboudian, R.; Burne, R.A.; Qian, P.Y.; Zhang, W. Mutanofactin promotes adhesion and biofilm formation of cariogenic Streptococcus mutans. Nat. Chem. Biol. 2021, 17, 576–584. [Google Scholar] [CrossRef]
- Zhang, X.; Nakaura, Y.; Zhu, J.; Zhang, Z.; Yamamoto, K. Effect of hyperosmotic salt concentration and temperature on viability of Escherichia coli during cold storage. Biocontrol Sci. 2020, 25, 55–62. [Google Scholar] [CrossRef]
- Glaubman, J.; Hofmann, J.; Bonney, M.E.; Park, S.; Thomas, J.M.; Kokona, B.; Ramos Falcón, L.I.; Chung, Y.K.; Fairman, R.; Okeke, I.N. Self-association motifs in the enteroaggregative Escherichia coli heat-resistant agglutinin 1. Microbiology 2016, 162, 1091–1102. [Google Scholar] [CrossRef]
- Misawa, N.; Blaser, M.J. Detection and characterization of autoagglutination activity by Campylobacter jejuni. Infect. Immun. 2000, 68, 6168–6175. [Google Scholar] [CrossRef] [PubMed]
- Zong, B.; Xiao, Y.; Ren, M.; Wang, P.; Fu, S.; Qiu, Y. Baicalin weakens the porcine expec-induced inflammatory response in 3d4/21 cells by inhibiting the expression of nf-κb/mapk signaling pathways and reducing nlrp3 inflammasome activation. Microorganisms 2023, 11, 2126. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Yin, R.; Zuo, S.; Liu, J.; Zhang, Y.; Guo, L.; Qiu, Y.; Ye, C.; Liu, Y.; Wu, Z.; et al. The effects of baicalin on piglets challenged with Glaesserella parasuis. Vet. Res. 2020, 51, 102. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Li, R.; Xu, L.; Qiu, Y.; Fu, S.; Liu, Y.; Wu, Z.; Hou, Y.; Hu, C.A.A. Effects of baicalin on piglet monocytes involving pkc-mapk signaling pathways induced by Haemophilus parasuis. BMC Vet. Res. 2019, 15, 98. [Google Scholar] [CrossRef] [PubMed]
- Farhadi, F.; Khameneh, B.; Iranshahi, M.; Iranshahy, M. Antibacterial activity of flavonoids and their structure-activity relationship: An update review. Phytother. Res. 2019, 33, 13–40. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.C.; Chu, X.L.; Su, J.Q.; Cui, Z.Q.; Zhang, L.Y.; Yu, Z.J.; Wu, Z.M.; Cai, M.L.; Li, H.X.; Zhang, Z.J. Baicalin protects mice against Salmonella typhimurium infection via the modulation of both bacterial virulence and host response. Phytomed. Int. J. Phytother. Phytopharm. 2018, 48, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hu, B.; Xu, J.; Ren, Q.; Wang, Z.; Wang, S.; Dong, Y.; Yang, G. Baicalin suppress growth and virulence-related factors of methicillin-resistant Staphylococcus aureus in vitro and vivo. Microb. Pathog. 2020, 139, 103899. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yin, F.; Liu, T.; Li, S.; Tan, C.; Li, L.; Zhou, R.; Huang, Q. The tat system and its dependent cell division proteins are critical for virulence of extra-intestinal pathogenic Escherichia coli. Virulence 2020, 11, 1279–1292. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Hu, Y.; Bu, Z.; Liu, Y.; Zhang, H.; Hu, Y.; Xue, Y.; Li, S.; Tan, C.; Chen, X.; et al. Genome-wide identification of genes critical for in vivo fitness of multi-drug resistant porcine extraintestinal pathogenic Escherichia coli by transposondirected insertion site sequencing using a mouse infection model. Virulence 2023, 14, 2158708. [Google Scholar] [CrossRef]
- Zhu, M.; Dai, X. Bacterial stress defense: The crucial role of ribosome speed. Cell. Mol. Life Sci. CMLS 2020, 77, 853–858. [Google Scholar] [CrossRef]
- Mihaljevic, R.R.; Sikic, M.; Klancnik, A.; Brumini, G.; Mozina, S.S.; Abram, M. Environmental stress factors affecting survival and virulence of Campylobacter jejuni. Microb. Pathog. 2007, 43, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Guerra, M.E.S.; Destro, G.; Vieira, B.; Lima, A.S.; Ferraz, L.F.C.; Hakansson, A.P.; Darrieux, M.; Converso, T.R. Klebsiella pneumoniae biofilms and their role in disease pathogenesis. Front. Cell. Infect. Microbiol. 2022, 12, 877995. [Google Scholar] [CrossRef]
- Laird, W.; Cavanaugh, D. Correlation of autoagglutination and virulence of yersiniae. J. Clin. Microbiol. 1980, 11, 430–432. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, Y.; An, Q.; Sa, R.; Zhang, D.; Xu, R. Research on the role of luxs/ai-2 quorum sensing in biofilm of Leuconostoc citreum 37 based on complete genome sequencing. 3 Biotech 2021, 11, 189. [Google Scholar] [CrossRef]
- Sztajer, H.; Lemme, A.; Vilchez, R.; Schulz, S.; Geffers, R.; Yip, C.Y.Y.; Levesque, C.M.; Cvitkovitch, D.G.; Wagner-Döbler, I. Autoinducer-2-regulated genes in Streptococcus mutans ua159 and global metabolic effect of the luxs mutation. J. Bacteriol. 2008, 190, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Sun, L.; Grenier, D.; Yi, L. The luxs/ai-2 system of Streptococcus suis. Appl. Microbiol. Biotechnol. 2018, 102, 7231–7238. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Hao, L.; Ke, H.; Liang, Z.; Ma, J.; Liu, Z.; Li, Y. Luxs/ai-2 in Streptococcus agalactiae reveals a key role in acid tolerance and virulence. Res. Vet. Sci. 2017, 115, 501–507. [Google Scholar] [CrossRef]
- Stroeher, U.H.; Paton, A.W.; Ogunniyi, A.D.; Paton, J.C. Mutation of luxs of Streptococcus pneumoniae affects virulence in a mouse model. Infect. Immun. 2003, 71, 3206–3212. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Y.; Luo, Q.; Huang, J.; Chen, J.; Zhang, R.; Wang, X. Luxs/ai-2 quorum sensing system in Edwardsiella piscicida promotes biofilm formation and pathogenicity. Infect. Immun. 2020, 88, e00907-19. [Google Scholar] [CrossRef] [PubMed]

| Strain or Plasmid | Description | Source or Reference |
|---|---|---|
| Strain | ||
| PCN033 | Wild-type (WT), porcine origin, O11, D CmS | [10] |
| ΔluxS | luxS gene mutant strain of PCN033 | This study |
| CΔluxS | The complemented strain of ΔluxS of PCN033, CmR | This Study |
| DH5α | F−, φ80dlacZΔM15, Δ(lacZYAargF)U169, deoR, recA1, endA1, hsdR17 (rk−, mk+), phoA, supE44, λ−, thi-1, gyrA96, rel1 | Takara Bio |
| χ7213 | Thi-1 thr-1 leuB6 fhuA21 lacY1 glnV44ΔasdA4 recA1 RP4 2-Tc::Mu[λpir] KmR | Dr. Roy Curtiss, USA |
| plasmid | ||
| pRE112 | riT oriV Δasd CmR SacB, suicide vector | Dr. Roy Curtiss, USA |
| pHSG396 | ori lacZ CmR | Takara Bio (Beijing, China) |
| Primer | Sequence | Remark |
|---|---|---|
| luxS-u-F | AATTCCCGGGAGAGCTCATACCTTTGAACCGGGTATG(SACI) | Upstream flanking of luxS |
| luxS-u-R | AAATTACCGGAGGTGGCTAATCAGTAAACTATCTTCACAATT | |
| luxS-d-F | AATTGTGAAGATAGTTTACTGATTAGCCACCTCCGGTAATTT | Downstream flanking of luxS |
| luxS-d-R | TCCCAAGCTTCTTCTAGAGTAAAGATCTGTTCCGCGAT(XBAI) | |
| Out-ΔluxS-F | TATAGTCAACTGGAAGGGCTTG | External source primers for ΔluxS |
| Out-ΔluxS-R | GCGCGAAGAGGATTTTGTAG | |
| In-ΔluxS-F | ATGCCGTTGTTAGATAGCTT | Internal source primers forΔluxS |
| In-ΔluxS-R | CTGCAACTTCTCTTTCGGCA | |
| luxSRF | CTTCCATTGCCGCTTTCCAG | Primer of luxS for q RT-PCR |
| luxSRR | TACCCTGGAGCACCTGTTTG | |
| CluxS-F | CGAGGGGTCGACTCTAGAATGCCGTTGTTAGATAGCTTCAC | The sequence of luxS with the fragment of plasmid pHSG396 |
| CluxS-R | ATTCGAGCTATCGGTACCCTAGATGTGCAGTTCCTGCA | |
| VF | ATGACCATGATTACGCCAAG | Validation primer for C∆luxS |
| VR | CTACAGCGTGAGCATTGAGAA | |
| 16SRNAF | GAATGCCACGGTGAATAC | Primer of 16SRNA for q RT-PCR |
| 16SRNAR | GGTTACCTTGTTACGACTTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zong, B.; Xiao, Y.; Wang, P.; Liu, W.; Ren, M.; Li, C.; Fu, S.; Zhang, Y.; Qiu, Y. Baicalin Weakens the Virulence of Porcine Extraintestinal Pathogenic Escherichia coli by Inhibiting the LuxS/AI-2 Quorum-Sensing System. Biomolecules 2024, 14, 452. https://doi.org/10.3390/biom14040452
Zong B, Xiao Y, Wang P, Liu W, Ren M, Li C, Fu S, Zhang Y, Qiu Y. Baicalin Weakens the Virulence of Porcine Extraintestinal Pathogenic Escherichia coli by Inhibiting the LuxS/AI-2 Quorum-Sensing System. Biomolecules. 2024; 14(4):452. https://doi.org/10.3390/biom14040452
Chicago/Turabian StyleZong, Bingbing, Yong Xiao, Peiyi Wang, Wei Liu, Mingxing Ren, Changyan Li, Shulin Fu, Yanyan Zhang, and Yinsheng Qiu. 2024. "Baicalin Weakens the Virulence of Porcine Extraintestinal Pathogenic Escherichia coli by Inhibiting the LuxS/AI-2 Quorum-Sensing System" Biomolecules 14, no. 4: 452. https://doi.org/10.3390/biom14040452





