Abstract
Polyamines are polycations derived from amino acids that play an important role in proliferation and growth in almost all living cells. In Streptococcus pneumoniae (the pneumococcus), modulation of polyamine metabolism not only plays an important regulatory role in central metabolism, but also impacts virulence factors such as the capsule and stress responses that affect survival in the host. However, functional annotation of enzymes from the polyamine biosynthesis pathways in the pneumococcus is based predominantly on computational prediction. In this study, we cloned SP_0166, predicted to be a pyridoxal-dependent decarboxylase, from the Orn/Lys/Arg family pathway in S. pneumoniae TIGR4 and expressed and purified the recombinant protein. We performed biochemical characterization of the recombinant SP_0166 and confirmed the substrate specificity. For polyamine analysis, we developed a simultaneous quantitative method using hydrophilic interaction liquid chromatography (HILIC)-based liquid chromatography–tandem mass spectrometry (LC–MS/MS) without derivatization. SP_0166 has apparent Km, kcat, and kcat/Km values of 11.3 mM, 715,053 min−1, and 63,218 min−1 mM−1, respectively, with arginine as a substrate at pH 7.5. We carried out inhibition studies of SP_0166 enzymatic activity with arginine as a substrate using chemical inhibitors DFMO and DFMA. DFMO is an irreversible inhibitor of ornithine decarboxylase activity, while DFMA inhibits arginine decarboxylase activity. Our findings confirm that SP_0166 is inhibited by DFMA and DFMO, impacting agmatine production. The use of arginine as a substrate revealed that the synthesis of putrescine by agmatinase and N-carbamoylputrescine by agmatine deiminase were both affected and inhibited by DFMA. This study provides experimental validation that SP_0166 is an arginine decarboxylase in pneumococci.
1. Introduction
Streptococcus pneumoniae, commonly known as the pneumococcus, is a Gram-positive facultative anaerobe [1]; it is the primary cause of mortality in children under 5 years of age globally and contributes significantly to fatalities across all age groups [2,3]. Based on the unique capsular polysaccharide (CPS) structure, about 100 pneumococcal serotypes are known. Current vaccines target CPS from serotypes most frequently isolated from human infections [4]. Among its arsenal of virulence factors, the capsule stands out as a principal determinant, contributing to immune evasion and survival in the host. In addition to the capsule, S. pneumoniae employs various virulence factors, including adhesins (such as choline-binding protein A, neuraminidase A, and LPXTG proteins), cytotoxins (pneumolysin), and immune-evasive proteins (pneumococcal surface protein A, plasmin-binding, and fibronectin-binding proteins), all of which play crucial roles in adhesion, colonization, cytotoxicity, and evasion of the host immune system [5]. Recently, polyamines have been associated with pneumococcal virulence, and polyamine metabolism in pneumococci serves as an important regulator of central metabolism [6]. Polyamines represent a group of aliphatic polycationic hydrocarbons mostly associated with RNA that play critical roles in diverse biological processes, including cell growth, transcription, and translation [7,8,9]. The primary polyamines encompass putrescine, spermidine, spermine, and cadaverine.
Within the capsular serotypes of S. pneumoniae linked to invasive diseases, the conservation of putative lysine decarboxylase (CadA), spermidine synthase (SpeE), and substrate binding protein of the polyamine transporter protein (PotD) has been confirmed through analysis, exhibiting more than 99% identity across sequenced pneumococcal genomes, and the presence of these genes is shown to be important for in vivo fitness [10]. Initially, our research described the importance of CadA in virulent pneumococcal serotype 4 capsule biosynthesis and its regulatory role in the interplay between stress response and capsule synthesis [11,12]. However, our subsequent studies established that CadA is actually an arginine decarboxylase rather than a lysine decarboxylase [13]. This discrepancy highlights the issues with the current annotation of polyamine metabolism genes in public databases which are primarily based on computational prediction without experimental validation. Current knowledge of polyamine synthesis derives from inferences from E. coli, emphasizing the need for alternative annotations specific to genetically divergent organisms. The primary route for bacterial polyamine synthesis involves the decarboxylation of ornithine to putrescine, a reaction catalyzed by ornithine decarboxylase (ODC), with pyridoxal 5′-phosphate (PLP) serving as a cofactor [14]. Additional routes of putrescine synthesis include a two-step process from arginine, utilizing arginine decarboxylase (ADC) and agmatinase with agmatine as an intermediate, as well as a three-step process involving ADC, agmatine deiminase, and N-carbamoylputrescine amidohydrolase, with agmatine and N-carbamoylputrescine as intermediates [7,14]. Subsequently, spermidine and spermine are produced from putrescine through the sequential actions of spermidine synthase and spermine synthase, respectively. In reversible reactions, spermidine/spermine acetyltransferase can acetylate both spermine and spermidine, while polyamine oxidases convert them back to spermidine and putrescine, respectively. Additionally, a distinct pathway involves the decarboxylation of lysine to cadaverine, catalyzed by lysine decarboxylase.
Ornithine decarboxylation is the rate-limiting step in polyamine biosynthesis [15]. Treatment with the irreversible ODC inhibitor difluoromethylornithine (DFMO) is known to deplete cellular levels of putrescine, spermidine, and agmatine [16,17]. DFMO, acting as a suicide inhibitor, undergoes decarboxylation by ODC akin to ornithine. This process generates a reactive intermediate that forms a stable covalent bond with surrounding groups, resulting in ODC inactivation [18]. DFMO is shown to be an effective chemotherapeutic agent, particularly in hyperproliferative diseases such as cancer [19,20,21]. It is noteworthy that DFMO not only targets ODC but also inhibits ADC activity [16,22]. Difluoromethyl arginine (DFMA) [23], employed as an ADC inhibitor in various microorganisms and plants, leads to reduced agmatine and putrescine levels in diverse organisms [24,25,26]. Genes encoding enzymes for the conversion of ornithine to putrescine and lysine to cadaverine in TIGR4 have not been identified. Current genome annotation of SP_0166 is stated as “pyridoxal-dependent decarboxylase, Orn/Lys/Arg family” with only basic annotation score of 1 out of 5 (https://www.uniprot.org/uniprotkb/A0A0H2UNA7/entry, accessed on 5 March 2024) [27]. Based on this description, SP_0166 has the potential to utilize ornithine, lysine, and arginine. This study is designed to determine the precise catalytic role of SP_0166 in pneumococcal polyamine biosynthesis. Here, we utilized liquid chromatography–tandem mass spectrometry (LC–MS/MS) employing a hydrophilic interaction liquid chromatography (HILIC) to characterize the enzymatic activity of SP_0166. We also measured the kinetic parameters with arginine, lysine, and ornithine substrates and the effect of chemical inhibitors.
2. Materials and Methods
2.1. Cloning, Expression, and Purification of SP_0166
The SP_0166 gene locus in TIGR4 encodes a protein of 368 amino acids with a predicted molecular weight of ~43 kDa. SP_0166 was amplified from chromosomal DNA of TIGR4 using primers with BamHI and XhoI restriction sites (Table 1). The PCR product was cloned into the pET-28a (+) vector (MilliporeSigma, Burlington, MA, USA) with a 6x -His tag at the C-terminus. The resulting recombinant expression vector was transformed into the E. coli strain BL21 (DE3), which was grown in Terrific broth (Thermo Fisher Scientific, Waltham, MA, USA) containing 30 µg/mL kanamycin and 3% ethanol at 37 °C to an optical density of 1.1 at 600 nm and induced with 1.0 mM 1-thio-β-D-galactopyranoside (IPTG). Five hours post-induction, cells were harvested by centrifugation at 5000× g for 10 min at 4 °C and the cell pellet was stored at −20 °C until further use. The frozen pellet was thawed on ice and resuspended in B-PER Reagent buffer (Thermo Fisher Scientific, Waltham, MA, USA) at 4 mL/g of pellet, with 2 µL of benzonase (Sigma-Aldrich, St. Louis, MO, USA) and 10 µL/mL protease inhibitor (Thermo Fisher Scientific, Waltham, MA, USA), and incubated for 15 min at room temperature.

Table 1.
Primers used for PCR amplification of SP_0166 from S. pneumoniae.
The cell debris was removed by centrifugation at 15,000× g for 5 min and the lysate was loaded onto a HisPur Cobalt Spin Column (Thermo Fisher Scientific, Waltham, MA, USA). After washing with equilibration/wash buffer containing 100 mM imidazole, bound proteins were eluted with 500 mM imidazole in elution buffer. The purified protein was desalted using a Sephadex G-25 PD-10 column (GE Healthcare, Chicago, IL, USA) equilibrated with PBS. Purified protein was evaluated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and staining with Coomassie Brilliant Blue R-250 (Bio-Rad, Hercules, CA, USA). Protein estimation was performed using the BCA method with Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Waltham, MA, USA).
2.2. Sample Preparation to Determine Substrate Specificity
To evaluate the substrate specificity of recombinant SP_0166, arginine, lysine, and ornithine were used as substrates and the reaction products agmatine, cadaverine, and putrescine, respectively, were quantified by LC–MS/MS (Figure 1) [16].
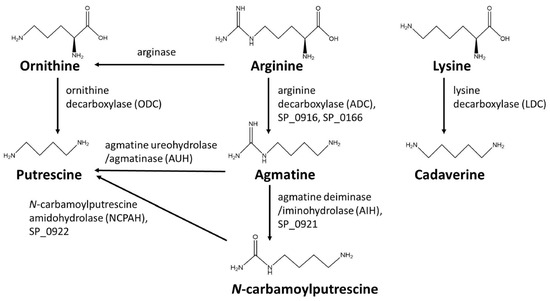
Figure 1.
Polyamine biosynthesis pathway substrates and products.
The reaction mixture consisted of 1.163 µmol/L protein, 10 mM substrate, 0.6 mM pyridoxal-phosphate (PLP), 2.5 mM MgSO4, and 50 mM Tris-HCl (pH 7.5) in a total volume of 500 µL, and was incubated for 30 min at 37 °C in the dark and stopped by addition of 12.5 µL of 70% (w/v) perchloric acid. After incubation on ice for 15 min, the mixture was neutralized by the addition of 25 µL of 10 N KOH. The sample was mixed with 1 mL of 1-butanol containing spermidine-d8 as an internal standard, and the butanol layers were separated by centrifugation for 5 min at 16,100× g. The extracted organic layer was dried under nitrogen and reconstituted with 100 µL of an acetonitrile: methanol: water (40:40:20) solution. To optimize the enzymatic activity of SP_0166, activity measurements were conducted across a pH range of 5.5 to 8.5 using 50 mM Tris-HCl under the same reaction conditions. The enzyme was incubated with each substrate at the indicated pH for 30 min at 37 °C and the reaction products were detected by LC–MS/MS, as described above. Time course activity assays were performed with arginine and lysine substrates for 120 min at 37 °C and the reaction was terminated at different time intervals. The data are shown as the percentage of maximum activity of enzyme with each substrate relative to the highest amount of polyamine product determined by LC–MS/MS.
2.3. LC–MS/MS Analysis of SP_0166 Enzymatic Activity
The polyamines were analyzed on a TSQ Quantum Access triple-quadrupole tandem mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) equipped with the Acquity UPLC system (Waters, Milford, MA, USA). Chromatographic separation was carried out using an Acquity UPLC BEH Amide column (2.1 mm × 150 mm, 1.7 μm) coupled with an Acquity UPLC BEH Amide VanGuard Precolumn (2.1 mm × 5 mm, 1.7 μm) at 40 °C using a column oven, and 10 µL of samples were injected. The mobile phases consisted of water with 0.1% v/v formic acid (A) and acetonitrile with 0.1% v/v formic acid (B). The gradient condition was 0 min (5% A, 95% B), 1 min (5% A, 95% B), 4 min (30% A, 70% B), 6 min (95% A, 5% B), 6.5 min (95% A, 5% B), 7 min (5% A, 95% B), and 10 min (5% A, 95% B). The total run time was 10 min and the flow rate was 0.3 mL/min. The column eluate was directed into the mass spectrometer using an electrospray ionization interface in positive mode. The MS conditions were set as follows: spray voltage = 3500 V, vaporizer temperature = 350 °C, sheath gas pressure = 25 psi, auxiliary gas pressure = 10 psi, and capillary temperature = 350 °C. Samples were run in selected reaction monitoring (SRM) mode and with precursor-to-product ion transitions of m/z 175.1 → m/z 70.4 for arginine, m/z 147.1 → m/z 84.4 for lysine, m/z 133.1 → m/z 116.1 for ornithine, m/z 131.1 → m/z 72.4 for agmatine, m/z 103.2 → m/z 86.4 for cadaverine, m/z 89.2 → m/z 72.4 for putrescine, and m/z 132.1 → m/z 115.2 for N-carbamoylputrescine. Internal standard included spemidine-d8 (m/z 154.2 → m/z 80.4), which was used to normalize the quantified amounts of substrates and products. Scan time was 0.2 s per SRM and the scan width was m/z 0.01. Optimum collision energy and S-lenses conditions were determined for each compound by using Auto-Tune software (Thermo Xcalibur 2.2 SP1.48) for each analyte by post-column infusion of the individual compounds into a 50% A/50% B blend of the mobile phase being pumped at a flow rate of 0.3 mL/min. Xcalibur software (version 2.2 SP1.48) was utilized for data acquisition and processing.
2.4. Enzyme Kinetics Analysis
Kinetic parameters of SP_0166 were analyzed by detecting the amount of the product generated over a range of substrate concentrations from 0.01 mM to 30 mM at pH 7.5 for 1 h. For quantification, calibration standards were prepared for agmatine, cadaverine, and putrescine. The nonlinear regression method of the Michaelis–Menten equation with Sigma Plot v.12 was used to estimate the kinetics parameters (kcat, Km, and kcat/Km). All experiments were performed with three independent replicates and the values represent the mean ± the standard deviation of three separate measurements.
2.5. Enzyme Inhibition Assays
To investigate the effect of decarboxylase inhibitors on SP_0166 activity, DFMO was added to the reaction mixture in a range between 0.1 to100 mM and DFMA between 0.003 to 3 mM. The inhibitors were stored as 2 M stock solutions in water at −20 °C. The stocks were diluted with reaction buffer on the day of the experiment. The inhibitors were prepared by diluting each concentration in water and adding it to the reaction mixture after 5 min preincubation at 37 °C in the dark. A parallel control reaction was incubated without inhibitors. The enzyme and inhibitor were incubated at 37 °C for 1 h and polyamine reaction products were extracted as described earlier. All experiments were performed in triplicate. The concentration of inhibitor that inhibited 50% of the control activity (IC50 value) was determined by varying the concentration of the inhibitor DFMA or DFMO and measuring the decarboxylase activity. Sigma Plot v.12 was used to fit the curve through the points and IC50 values were interpolated from the fitted curve.
3. Results
3.1. Expression and Purification of SP_0166
Annotation of SP_0166 gene in S. pneumoniae serotype 4 in bioinformatic databases such as in UniProt, KEGG, and BioCyc [27,28,29] indicates that it is potentially a pyridoxal-dependent decarboxylase with a broad substrate specificity, which includes ornithine, lysine, and arginine. The 368 amino acid protein has a predicted molecular weight of 42,757 Da. Purified recombinant SP_0166 is ~43 kDa, as expected (Figure 2 and Figure S1). The yield of recombinant SP_0166 (4.09 mg protein/100 mL culture) is comparable with previously characterized recombinant SP_0916/arginine decarboxylase (3.42 mg protein/100 mL culture) [13].
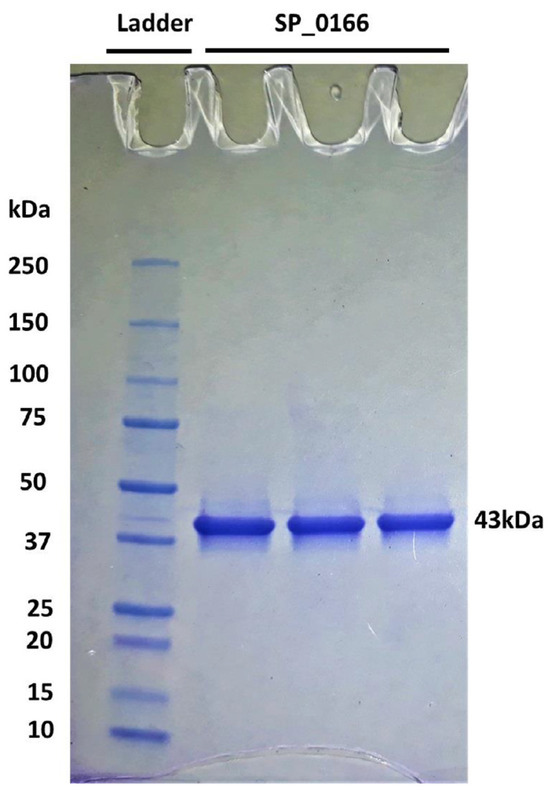
Figure 2.
Expression and purification of recombinant SP_0166. The overexpressed and purified recombinant 43 kDa SP_0166 from S. pneumoniae TIGR4 was resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
3.2. Substrate Specificity of SP_0166
To determine the substrate specificity of SP_0166, the recombinant enzyme was incubated with different substrates and the reaction end products were measured. With arginine, lysine, and ornithine as substrates, the corresponding products would be agmatine, cadaverine, and putrescine, respectively, and these were measured by LC–MS/MS. HILIC columns in LC–MS/MS streamline the sample preparation process by eliminating the need for derivatization of the target polyamines. The analysis achieved simultaneous quantification of polyamines within a 10 min run time, and the chromatogram peaks corresponding to the polyamines were detected (Figure 3).
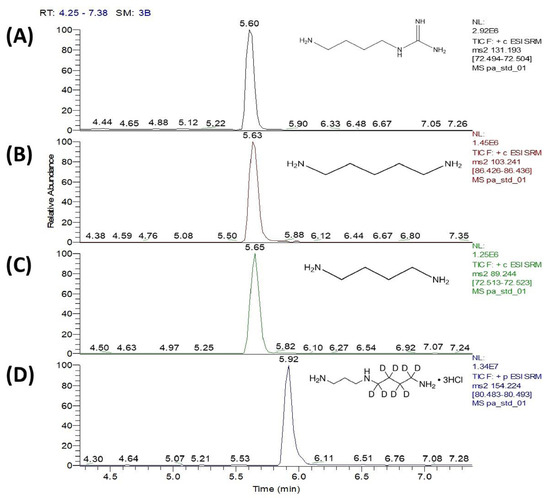
Figure 3.
UHPLC–MSMS chromatograms of (A) agmatine, (B) cadaverine, and (C) putrescine, which are the decarboxylation products of arginine, lysine, and ornithine, respectively, and (D) spermidine-d8 (internal standard). Four separate single monitoring (SRM) transitions are shown for synthesized polyamine standards within 10 min running time.
Similar to SP_0916, LC–MS/MS analysis of SP_0166 reaction products showed that arginine is the preferred substrate, as the relative activity of the enzyme to produce agmatine, the end product of arginine decarboxylation, was 12-fold higher than for cadaverine, the end product of lysine decarboxylation (Figure 4) [16]. The SP_0166 enzyme in TIGR4 is annotated broadly as a pyridoxal-dependent decarboxylase within the Orn/Lys/Arg family. Our results clearly establish that SP_0166 preferentially decarboxylates arginine and has lower affinity for lysine and ornithine.
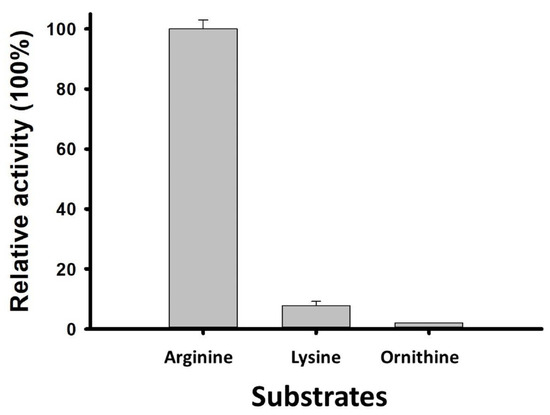
Figure 4.
SP_0166 is an arginine decarboxylase. SP_0166 functions as an arginine decarboxylase, demonstrating the highest relative activity with arginine as a substrate. It exhibits relatively lower activity with lysine and negligible activity with ornithine.
3.3. Optimization of pH and Incubation Time to Enhance the ADC Activity
ADCs in E. coli, a model organism for bacterial polyamine metabolic pathways, are annotated to be either constitutive (optimal catalytic activity at pH 5.2) or inducible (optimal pH is 8.23) [30,31,32]. To characterize the enzyme activity of SP_0166, optimal pH was determined. The maximum activity of SP_0166 was found at pH 7.5 and the effect of pH on each substrate is shown in Figure 5A. The enzyme activity increases steadily from pH 6.5 to pH 7.5 and rapidly decreases as pH increases, except when ornithine is the substrate. Generally, the effect of pH on SP_0166 activity with arginine and lysine is similar between pH 6.5–8.0.
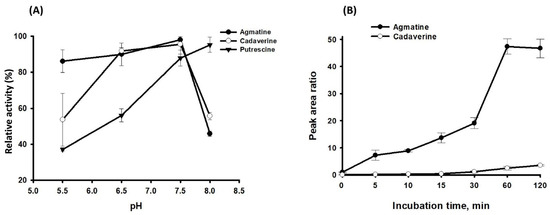
Figure 5.
Optimization of reaction conditions for polyamine analysis by LC–MS/MS. Panel (A) depicts the impact of pH on the activity of recombinant SP_0166, while panel (B) presents the time course of agmatine and cadaverine production from arginine and lysine substrates, respectively.
We measured the arginine and lysine decarboxylase activity of SP_0166 at pH 7.5 at different time points up to 120 min. As shown in Figure 5B, both agmatine and cadaverine levels gradually increased in 60 min. Agmatine synthesis from arginine increased between 30 min and 60 min and decreased slightly after 60 min.
3.4. Enzyme Kinetics Analysis
The enzyme kinetics of the SP_0166 were measured at various concentrations of arginine, lysine, or ornithine that ranged from 0.03 mM to 30 mM. The SP_0166 catalyzed decarboxylation of substrates, and the concentration of each substrate was expressed in the form of the concentration of the converted product, as shown in Figure 6A. The kinetic parameters determined using the Michaelis–Menten equation are summarized in Figure 6B, and goodness of fit (R2) was 0.9962, 0.9998, and 0.9201 for arginine, lysine, and ornithine, respectively. The Km for catalysis of the substrate–enzyme pairs was in this order: lysine > ornithine > arginine, demonstrating that SP_0166 is most efficient at decarboxylating arginine. Also, the catalytic efficiency (kcat/Km) of SP_0166 for the conversion of arginine to agmatine is about 64-fold and 368-fold higher than for the conversion of lysine to cadaverine and ornithine to putrescine, respectively (Figure 6B). These findings unequivocally establish SP_0166 as primarily functioning as an arginine decarboxylase. While SP_0166 exhibits the capacity to decarboxylate lysine and ornithine substrates, it does so with a markedly lower catalytic efficiency.
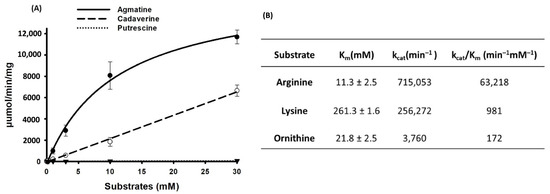
Figure 6.
LC–MS/MS analysis of SP_0166 enzyme kinetics. (A) Comparison of the enzyme kinetics for the conversion of arginine to agmatine (●), lysine to cadaverine (○), and ornithine to putrescine (▼) at different substrate concentrations. The lines represent curve-fitting for each point. (B) Comparison of kinetic parameters. The results shown are the means of triplicate experiments, and data represent mean ± standard deviation.
3.5. Inhibition of Decarboxylase Activity by DFMA and DFMO
To study the effects of DFMO and DFMA, the recombinant SP_0166 protein was preincubated with arginine, lysine, and ornithine for 5 min and followed by the addition of each inhibitor at different concentrations and incubation for an additional 1 h. The samples were extracted and analyzed by LC–MS/MS for the polyamine quantitation. Enzyme activity without inhibitor was used for control and relative activity values were calculated for each concentration. Corrected activity was plotted against nominal inhibitor concentration and fitted to determine the concentration of the inhibitor required for half-maximal inhibition of substrate generation (IC50) [33,34]. Figure 7A,B and Table 2 show DFMA and DFMO with IC50 values in the µM and mM range, respectively, for the inhibition pattern of the polyamines produced when arginine, lysine, and ornithine were used as substrates. The reported information on DFMA and DFMO suggests that these compounds primarily bind to and inhibit ADC and ODC, respectively, and our experimental results align with this observation. When SP_0166 was incubated with arginine and DFMA, the amount of agmatine produced was significantly reduced compared with cadaverine and putrescine. This result indicates that SP_0166 ADC activity is more sensitive to DFMA inhibition than LDC or ODC. In the case of DFMO, agmatine, cadaverine, and putrescine all showed similar inhibition results, showing that SP_0166 enzymatic activity is inhibited by DFMO.
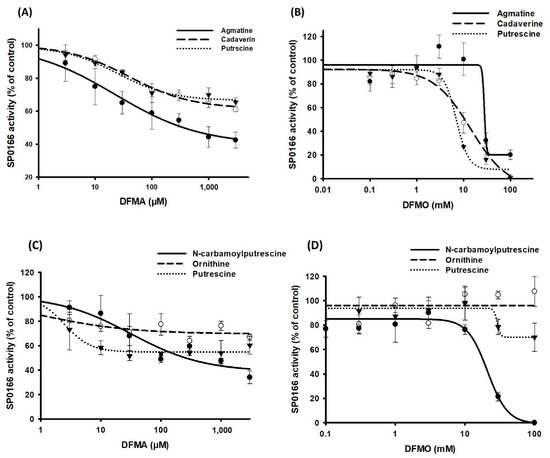
Figure 7.
Inhibition of recombinant SP_0166 decarboxylase activity by DFMA and DFMO. Inhibition potency of different concentrations of (A) DFMA and (B) DFMO on polyamine synthesis from arginine (●), lysine (○), and ornithine (▼) substrates. Inhibition potency of different concentrations of (C) DFMA and (D) DFMO on synthesis of N-carbamoyl putrescine (●), ornithine (○), and putrescine (▼) from arginine. The results shown are the means of triplicate experiments, and data represent mean ± standard deviation.

Table 2.
Inhibition potency of DFMO and DFMA against enzymatic activity of SP_0166 with different substrates.
Two distinct enzymes are known to catalyze the conversion of agmatine to putrescine or N-carbamoyl putrescine in prokaryotes (Figure 1). Agmatinase and agmatine deiminase convert agmatine to putrescine and N-carbamoylputrescine, respectively [6,12,35]. When SP_0166 was incubated with arginine along with DFMA and DFMO, we confirmed that there was a decrease in N-carbamoylputrescine levels in a pattern like that of agmatine (Figure 7C,D). Therefore, it could be confirmed that agmatine deiminase was inhibited by DFMA and DFMO. Under the same conditions, the amount of putrescine produced decreased as the concentration of DFMA increased, but there was negligible impact by DFMO. Putrescine is produced from agmatine directly by agmatinase or indirectly via N-carbamoylputrescine by N-carbamoylputrescine amidohydrolase. Based on our results, N-carbamoylputrescine amidohydrolase and agmatine deiminase are affected by DFMA but not by DFMO. Arginase is an enzyme that hydrolyzes arginine to ornithine and urea [36,37]. However, there was no inhibition of ornithine synthesis from arginine, indicating that DFMA had no effect on arginase activity. This result also supports that arginase has not yet been annotated in the TIGR4 genome. Based on these results, it can be expected that agmatine deiminase is affected by both DFMA and DFMO, like ADC, but agmatinase and N-carbamoylputrescine amidohydrolase are inhibited only by DFMA.
4. Discussion
In numerous human bacterial pathogens, polyamines play a pivotal role in shaping host–pathogen interactions. For instance, pretreatment of eukaryotic cells with cadaverine renders them less susceptible to the impact of Shigella enterotoxins [37]. Additionally, cadaverine functions to impede the release of Shigella flexneri from the phagocytic vacuole [38]. Notably, polyamines exert significant influence on biofilm synthesis and maturation in various pathogens, including Yersinia pestis [39], Vibrio cholerae [40], and Pseudomonas aeruginosa [41]. Furthermore, polyamines are integral to the survival and colonization strategies employed by Helicobacter pylori within the gastric mucosa. Their role extends to facilitating the adaptation of the bacterium to the acidic conditions prevailing in the stomach [42]. Additionally, these versatile molecules contribute significantly to the adaptive processes undertaken by Salmonella in the host environment. Polyamines are actively involved in modulating the expression of genes associated with Salmonella invasion, intracellular survival, and systemic infection [43].
We and others have shown that polyamine synthesis and transport mechanisms are important for various bacterial responses under different physiological conditions. Specifically, the deletion of the spermidine synthesis has been linked to a delayed onset of autolysis in S. pneumoniae [44], while deletion of agmatine biosynthesis genes has been associated with prolonged lag phases during its growth [11,13]. Moreover, the disruption of the polyamine transport, as observed in the transport operon deletion strain ΔpotABCD, results in reduced putrescine and spermidine levels, and attenuation in vivo [13,45]. Similarly, deletion of the spermidine biosynthesis impedes the growth of Campylobacter jejuni [46], and simultaneous deletion of genes responsible for agmatine and putrescine production leads to growth impairment in Pseudomonas aeruginosa [35]. Furthermore, mutations in spermidine biosynthesis genes render P. aeruginosa more susceptible to antibiotics and oxidative stress [47]. Most importantly, deletion of either the polyamine transport operon or the gene involved in agmatine production in S. pneumoniae results in the loss of capsule [11,12,13], the predominant virulence factor in pneumococci, which explains the reported in vivo attenuation.
To understand the contribution of individual and combined effects of genes of the polyamine biosynthesis pathway on pneumococcal metabolism and virulence, it is critical to accurately annotate the function of genes from this pathway. The current annotation of the polyamine biosynthesis pathways in the pneumococci is limited and could be inaccurate (as shown with the annotation of cadA in our previous work [11,13]). In the genome annotation of S. pneumoniae TIGR4, the open reading frame SP_0166 is annotated to be a pyridoxal-dependent decarboxylase within the Orn-Lys-Arg family. This study establishes that SP_0166 is an arginine decarboxylase (ADC) that catalyzes agmatine synthesis and shows the inhibition of polyamine synthesis by DFMA and DFMO using a relatively sensitive LC–MS/MS approach.
Compared to the PLP-dependent inducible and constitutive arginine decarboxylases (ADC) in E. coli, which require optimum pH values of 5.2 and 8.4, respectively, for catalytic activity and Paramecium bursaria chlorella virus-1 with an optimum pH of 8.2 [48], the optimum pH for SP_0166 is 7.5. The Km values of ADCs from E. coli and chlorella virus-1 range between 0.03 to 0.65 mM. Despite a higher Km value for arginine (11.3 mM), catalytic efficiency (~1.05 × 106 s−1 M−1) of SP_0166 (adapted from Figure 6) is comparable to that of E. coli ADC (1.1 × 106 s−1 M−1), and is higher than that of Paramecium bursaria chlorella virus-1 (3.3 × 104 s−1 M−1). Interestingly, while optimum pH of Shewanella algae ADC is 7.5 and its Km is 14.55 mM [49], like SP_0166, its catalytic efficiency of 8.67 × 102 s−1 M−1 is significantly lower compared to SP_0166 (1.05 × 106 s−1 M−1).
In the previous study, we demonstrated SP_0916 (~54 kDa) in pneumococcal serotype 4 to be an ADC [13]. The discovery here, that SP_0166 is also an ADC, is intriguing but not entirely surprising, considering that recent reports on convergent evolution have revealed the emergence of ADCs from at least four different protein folds. Two forms of ADC are pyridoxal 5′-phosphate (PLP)-dependent enzymes, while the other two utilize pyruvoyl cofactors [50]. However, this means that the gene cadA, which encodes an enzyme for the biosynthesis of cadaverine, is still lacking in the pneumococcal genomes. Moreover, the current annotation of polyamine metabolism in the pneumococcal genome shows other notable deficiencies. ODC, a major biosynthetic enzyme that catalyzes the conversion of ornithine to putrescine, is not annotated in pneumococcal genomes in bioinformatic databases. Interestingly, ODC in Ca. Pelagibacter ubique and Ca. Fonsibacter ubiquis have been shown recently to evolve into ADC [51,52], which could additionally explain our discovery of two different ADCs in pneumococcus. Information on polyamine acetyltransferases that regulate motility and biofilm formation [53], and deacetylases that utilize acetylated polyamine substrates, is also poorly described [6]. Enzymes of the pyridoxal-dependent decarboxylase family, which catalyze the synthesis of polyamines, are expected to potentially utilize a variety of substrates amino acids. Characterizing SP_0166 as an ADC within the pyridoxal-dependent decarboxylase family here has significantly enhanced the annotation of the polyamine synthesis pathway in Spn TIGR4.
Agmatine produced by ADC is an intermediate in the synthesis of putrescine and spermidine and plays an important role in regulating CPS in pneumococci [13]. We already reported that DFMO inhibits CPS in multiple pneumococcal serotypes, and our results demonstrate that DFMO could inhibit ADC activity and play a prominent role in elucidating mechanisms related to altered polyamine synthesis and CPS inhibition [16]. Building upon our previous findings with SP_0916/ADC and SP_0166 in this work, future studies will focus on characterizing the role of SP_0166 in pneumococcal growth in vitro, its impact on the expression of other genes and proteins, its influence on the response to stressors, and potential impact on CPS, and, ultimately, its contribution to virulence in vivo. These studies will facilitate direct comparisons of the significance and roles of the two established ADCs in S. pneumoniae, which are beyond the scope of this study.
In conclusion, polyamine biosynthesis and transport genes are highly conserved across various pneumococcal serotypes. Therefore, understanding the polyamine–CPS nexus could help identify new serotype-independent vaccine antigens. Thus, a comprehensive understanding of the polyamine metabolic pathway can provide new approaches for the prevention and treatment of pneumococcal infection.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biom14040463/s1, Figure S1: Expression and purification of recombinant SP_0166 resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Author Contributions
Conceptualization, B.N.; methodology, J.H.L., M.B.A. and L.A.S.; software, J.H.L.; validation, J.H.L., M.B.A. and L.A.S.; formal analysis, J.H.L.; investigation, J.H.L., M.B.A., L.A.S., E.S. and B.N.; resources, E.S. and B.N.; data curation, J.H.L., M.B.A. and L.A.S.; writing—original draft preparation, J.H.L.; writing—review and editing, all authors; visualization, J.H.L. and M.B.A.; supervision, E.S. and B.N.; project administration, B.N.; funding acquisition, B.N. All authors have read and agreed to the published version of the manuscript.
Funding
This analysis was supported by the Omics and Bioinformatics core of the Center for Biomedical Research Excellence in Pathogen Host Interactions (grant # P20GM103646) funded by the National Institute for General Medical Sciences (NIGMS). Core B is also supported by the Mississippi INBRE, funded by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under grant number P20GM103476. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the NIH.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
We thank Patrick M. Woster (Medical University of South Carolina) for DFMO.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- André, A.C.; Debande, L.; Marteyn, B.S. The selective advantage of facultative anaerobes relies on their unique ability to cope with changing oxygen levels during infection. Cell. Microbiol. 2021, 23, e13338. [Google Scholar] [CrossRef] [PubMed]
- Troeger, C.; Blacker, B.; Khalil, I.A.; Rao, P.C.; Cao, J.; Zimsen, S.R.; Albertson, S.B.; Deshpande, A.; Farag, T.; Abebe, Z. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1191–1210. [Google Scholar] [CrossRef] [PubMed]
- McAllister, D.A.; Liu, L.; Shi, T.; Chu, Y.; Reed, C.; Burrows, J.; Adeloye, D.; Rudan, I.; Black, R.E.; Campbell, H.; et al. Global, regional, and national estimates of pneumonia morbidity and mortality in children younger than 5 years between 2000 and 2015: A systematic analysis. Lancet Glob. Health 2019, 7, e47–e57. [Google Scholar] [CrossRef] [PubMed]
- Ganaie, F.; Saad, J.S.; McGee, L.; van Tonder, A.J.; Bentley, S.D.; Lo, S.W.; Gladstone, R.A.; Turner, P.; Keenan, J.D.; Breiman, R.F.; et al. A New Pneumococcal Capsule Type, 10D, is the 100th Serotype and Has a Large cps Fragment from an Oral Streptococcus. mBio 2020, 11, e00937-20. [Google Scholar] [CrossRef]
- Feldman, C.; Anderson, R. Pneumococcal virulence factors in community-acquired pneumonia. Curr. Opin. Pulm. Med. 2020, 26, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Nanduri, B.; Swiatlo, E. The expansive effects of polyamines on the metabolism and virulence of Streptococcus pneumoniae. Pneumonia 2021, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Kashiwagi, K. Effects of polyamines on protein synthesis and growth of Escherichia coli. J. Biol. Chem. 2018, 293, 18702–18709. [Google Scholar] [CrossRef] [PubMed]
- Gevrekci, A.O. The roles of polyamines in microorganisms. World J. Microbiol. Biotechnol. 2017, 33, 204. [Google Scholar] [CrossRef]
- Shah, P.; Nanduri, B.; Swiatlo, E.; Ma, Y.; Pendarvis, K. Polyamine biosynthesis and transport mechanisms are crucial for fitness and pathogenesis of Streptococcus pneumoniae. Microbiology 2011, 157, 504–515. [Google Scholar] [CrossRef]
- Nakamya, M.F.; Ayoola, M.B.; Park, S.; Shack, L.A.; Swiatlo, E.; Nanduri, B. The Role of Cadaverine Synthesis on Pneumococcal Capsule and Protein Expression. Med. Sci. 2018, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Ayoola, M.B.; Shack, L.A.; Nakamya, M.F.; Thornton, J.A.; Swiatlo, E.; Nanduri, B. Polyamine Synthesis Effects Capsule Expression by Reduction of Precursors in Streptococcus pneumoniae. Front. Microbiol. 2019, 10, 476138. [Google Scholar] [CrossRef] [PubMed]
- Ayoola, M.B.; Nakamya, M.F.; Shack, L.A.; Park, S.; Lim, J.; Lee, J.H.; Ross, M.K.; Eoh, H.; Nanduri, B. SP_0916 Is an Arginine Decarboxylase That Catalyzes the Synthesis of Agmatine, Which Is Critical for Capsule Biosynthesis in Streptococcus pneumoniae. Front. Microbiol. 2020, 11, 578533. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.J. Biosynthesis of polyamines and polyamine-containing molecules. Biochem. J. 2016, 473, 2315–2329. [Google Scholar] [CrossRef] [PubMed]
- Park, M.G.; Kim, S.Y.; Lee, C.J. DMSO-tolerant ornithine decarboxylase (ODC) tandem assay optimised for high-throughput screening. J. Enzym. Inhib. Med. Chem. 2023, 38, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Ayoola, M.B.; Shack, L.A.; Lee, J.H.; Lim, J.; Eoh, H.; Swiatlo, E.; Phanstiel, O.; Nanduri, B. Difluoromethylornithine (DFMO) and AMXT 1501 inhibit capsule biosynthesis in pneumococci. Sci. Rep. 2022, 12, 11804. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.M.; Cirenajwis, H.; Wallace, H.M.; Oredsson, S.; Persson, L. A role for antizyme inhibitor in cell proliferation. Amino Acids 2015, 47, 1341–1352. [Google Scholar] [CrossRef]
- Pegg, A.E.; McGovern, K.A.; Wiest, L. Decarboxylation of alpha-difluoromethylornithine by ornithine decarboxylase. Biochem. J. 1987, 241, 305–307. [Google Scholar] [CrossRef]
- Meyskens, F.L., Jr.; Gerner, E.W. Development of difluoromethylornithine (DFMO) as a chemoprevention agent. Clin. Cancer Res. 1999, 5, 945–951. [Google Scholar]
- Madka, V.; Patlolla, J.M.R.; Venkatachalam, K.; Zhang, Y.; Pathuri, G.; Stratton, N.; Lightfoot, S.; Janakiram, N.B.; Mohammed, A.; Rao, C.V. Chemoprevention of Colon Cancer by DFMO, Sulindac, and NO-Sulindac Administered Individually or in Combinations in F344 Rats. Cancers 2023, 15, 4001. [Google Scholar] [CrossRef]
- Somani, R.R.; Rai, P.R.; Kandpile, P.S. Ornithine Decarboxylase Inhibition: A Strategy to Combat Various Diseases. Mini Rev. Med. Chem. 2018, 18, 1008–1021. [Google Scholar] [CrossRef] [PubMed]
- Tassoni, A.; Awad, N.; Griffiths, G. Effect of ornithine decarboxylase and norspermidine in modulating cell division in the green alga Chlamydomonas reinhardtii. Plant Physiol. Biochem. 2018, 123, 125–131. [Google Scholar] [CrossRef]
- Hyvönen, M.T.; Keinänen, T.A.; Nuraeva, G.K.; Yanvarev, D.V.; Khomutov, M.; Khurs, E.N.; Kochetkov, S.N.; Vepsäläinen, J.; Zhgun, A.A.; Khomutov, A.R. Hydroxylamine Analogue of Agmatine: Magic Bullet for Arginine Decarboxylase. Biomolecules 2020, 10, 406. [Google Scholar] [CrossRef] [PubMed]
- Rollins-Smith, L.A.; Ruzzini, A.C.; Fites, J.S.; Reinert, L.K.; Hall, E.M.; Joosse, B.A.; Ravikumar, V.I.; Huebner, M.I.; Aka, A.; Kehs, M.H.; et al. Metabolites Involved in Immune Evasion by Batrachochytrium dendrobatidis Include the Polyamine Spermidine. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed]
- Tailor, A.; Bhatla, S.C. Polyamine homeostasis modulates plasma membrane- and tonoplast-associated aquaporin expression in etiolated salt-stressed sunflower (Helianthus annuus L.) seedlings. Protoplasma 2021, 258, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Yarlett, N.; Waters, W.R.; Harp, J.A.; Wannemuehler, M.J.; Morada, M.; Bellcastro, J.; Upton, S.J.; Marton, L.J.; Frydman, B.J. Activities of DL-alpha-difluoromethylarginine and polyamine analogues against Cryptosporidium parvum infection in a T-cell receptor alpha-deficient mouse model. Antimicrob. Agents Chemother. 2007, 51, 1234–1239. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Paley, S.; Karp, P.D. The BioCyc metabolic network explorer. BMC Bioinform. 2021, 22, 208. [Google Scholar] [CrossRef]
- Blethen, S.L.; Boeker, E.A.; Snell, E. Arginine Decarboxylase from Escherichia coli: I. Purification and Specificity for Substrates and Coenzyme. J. Biol. Chem. 1968, 243, 1671–1677. [Google Scholar] [CrossRef]
- Hong, E.Y.; Lee, S.-G.; Yun, H.; Kim, B.-G. Improving the Stability and Activity of Arginine Decarboxylase at Alkaline pH for the Production of Agmatine. Front. Catal. 2021, 1, 774512. [Google Scholar] [CrossRef]
- Fried, R.; Carlton, R.M.; Fried, D.A. Starving Cancer Cells: Evidence-Based Strategies to Slow Cancer Progression: A Selection of Readings for Health Services Providers; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Ross, M.K.; Pluta, K.; Bittles, V.; Borazjani, A.; Allen Crow, J. Interaction of the serine hydrolase KIAA1363 with organophosphorus agents: Evaluation of potency and kinetics. Arch. Biochem. Biophys. 2016, 590, 72–81. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Regunathan, S.; Reis, D.J. Characterization of arginine decarboxylase in rat brain and liver: Distinction from ornithine decarboxylase. J. Neurochem. 2000, 74, 2201–2208. [Google Scholar] [CrossRef] [PubMed]
- Nakada, Y.; Itoh, Y. Identification of the putrescine biosynthetic genes in Pseudomonas aeruginosa and characterization of agmatine deiminase and N-carbamoylputrescine amidohydrolase of the arginine decarboxylase pathway. Microbiology 2003, 149, 707–714. [Google Scholar] [CrossRef]
- Rath, M.; Müller, I.; Kropf, P.; Closs, E.I.; Munder, M. Metabolism via arginase or nitric oxide synthase: Two competing arginine pathways in macrophages. Front. Immunol. 2014, 5, 532. [Google Scholar] [CrossRef] [PubMed]
- Maurelli, A.T.; Fernandez, R.E.; Bloch, C.A.; Rode, C.K.; Fasano, A. “Black holes” and bacterial pathogenicity: A large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 1998, 95, 3943–3948. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, I.M.; Silva, M.; Schuch, R.; Walker, W.A.; Siber, A.M.; Maurelli, A.T.; McCormick, B.A. Cadaverine prevents the escape of Shigella flexneri from the phagolysosome: A connection between bacterial dissemination and neutrophil transepithelial signaling. J. Infect. Dis. 2001, 184, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Patel, C.N.; Wortham, B.W.; Lines, J.L.; Fetherston, J.D.; Perry, R.D.; Oliveira, M.A. Polyamines are essential for the formation of plague biofilm. J. Bacteriol. 2006, 188, 2355–2363. [Google Scholar] [CrossRef] [PubMed]
- Karatan, E.; Duncan, T.R.; Watnick, P.I. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 2005, 187, 7434–7443. [Google Scholar] [CrossRef]
- Liu, Z.; Hossain, S.S.; Morales Moreira, Z.; Haney, C.H. Putrescine and Its Metabolic Precursor Arginine Promote Biofilm and c-di-GMP Synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2022, 204, e0029721. [Google Scholar] [CrossRef]
- McNamara, K.M.; Gobert, A.P.; Wilson, K.T. The role of polyamines in gastric cancer. Oncogene 2021, 40, 4399–4412. [Google Scholar] [CrossRef]
- Guerra, P.R.; Liu, G.; Lemire, S.; Nawrocki, A.; Kudirkiene, E.; Møller-Jensen, J.; Olsen, J.E.; Jelsbak, L. Polyamine depletion has global effects on stress and virulence gene expression and affects HilA translation in Salmonella enterica serovar typhimurium. Res. Microbiol. 2020, 171, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Potter, A.J.; Paton, J.C. Spermidine biosynthesis and transport modulate pneumococcal autolysis. J. Bacteriol. 2014, 196, 3556–3561. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.N.; Thornton, J.A.; Stokes, J.; Sunesara, I.; Swiatlo, E.; Nanduri, B. Polyamine transporter in Streptococcus pneumoniae is essential for evading early innate immune responses in pneumococcal pneumonia. Sci. Rep. 2016, 6, 26964. [Google Scholar] [CrossRef] [PubMed]
- Hanfrey, C.C.; Pearson, B.M.; Hazeldine, S.; Lee, J.; Gaskin, D.J.; Woster, P.M.; Phillips, M.A.; Michael, A.J. Alternative spermidine biosynthetic route is critical for growth of Campylobacter jejuni and is the dominant polyamine pathway in human gut microbiota. J. Biol. Chem. 2011, 286, 43301–43312. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.; Mulcahy, H.; Kanevets, U.; Shi, Y.; Lewenza, S. Surface-localized spermidine protects the Pseudomonas aeruginosa outer membrane from antibiotic treatment and oxidative stress. J. Bacteriol. 2012, 194, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Giles, T.N.; Graham, D.E. Crenarchaeal arginine decarboxylase evolved from an S-adenosylmethionine decarboxylase enzyme. J. Biol. Chem. 2008, 283, 25829–25838. [Google Scholar] [CrossRef]
- Pei, X.D.; Lu, L.H.; Yue, S.Y.; Li, Y.; Liu, X.L.; Li, F.; Wu, K.J.; Wang, C.H. Characterization of a Novel Shewanella algae Arginine Decarboxylase Expressed in Escherichia coli. Mol. Biotechnol. 2022, 64, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liang, J.; Hanfrey, C.C.; Phillips, M.A.; Michael, A.J. Discovery of ancestral L-ornithine and L-lysine decarboxylases reveals parallel, pseudoconvergent evolution of polyamine biosynthesis. J. Biol. Chem. 2021, 297, 101219. [Google Scholar] [CrossRef]
- Li, B.; Liang, J.; Baniasadi, H.R.; Phillips, M.A.; Michael, A.J. Functional polyamine metabolic enzymes and pathways encoded by the virosphere. Proc. Natl. Acad. Sci. USA 2023, 120, e2214165120. [Google Scholar] [CrossRef]
- Li, B.; Liang, J.; Phillips, M.A.; Michael, A.J. Neofunctionalization of S-adenosylmethionine decarboxylase into pyruvoyl-dependent L-ornithine and L-arginine decarboxylases is widespread in bacteria and archaea. J. Biol. Chem. 2023, 299, 105005. [Google Scholar] [CrossRef] [PubMed]
- Armalytė, J.; Čepauskas, A.; Šakalytė, G.; Martinkus, J.; Skerniškytė, J.; Martens, C.; Sužiedėlienė, E.; Garcia-Pino, A.; Jurėnas, D. A polyamine acetyltransferase regulates the motility and biofilm formation of Acinetobacter baumannii. Nat. Commun. 2023, 14, 3531. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).