Metabolites from Aerial Parts of Glycyrrhiza foetida as Modulators of Targets Related to Metabolic Syndrome
Abstract
1. Introduction
2. Materials and Methods
2.1. General Experimental Procedure
2.2. Plant Material
2.3. Extraction
2.4. LC-UV-MS2 Analysis
2.5. Purification of Metabolites from the G. foetida Extract
2.5.1. Amorfrtuin N (5)
2.5.2. Exiguaflavone L (21)
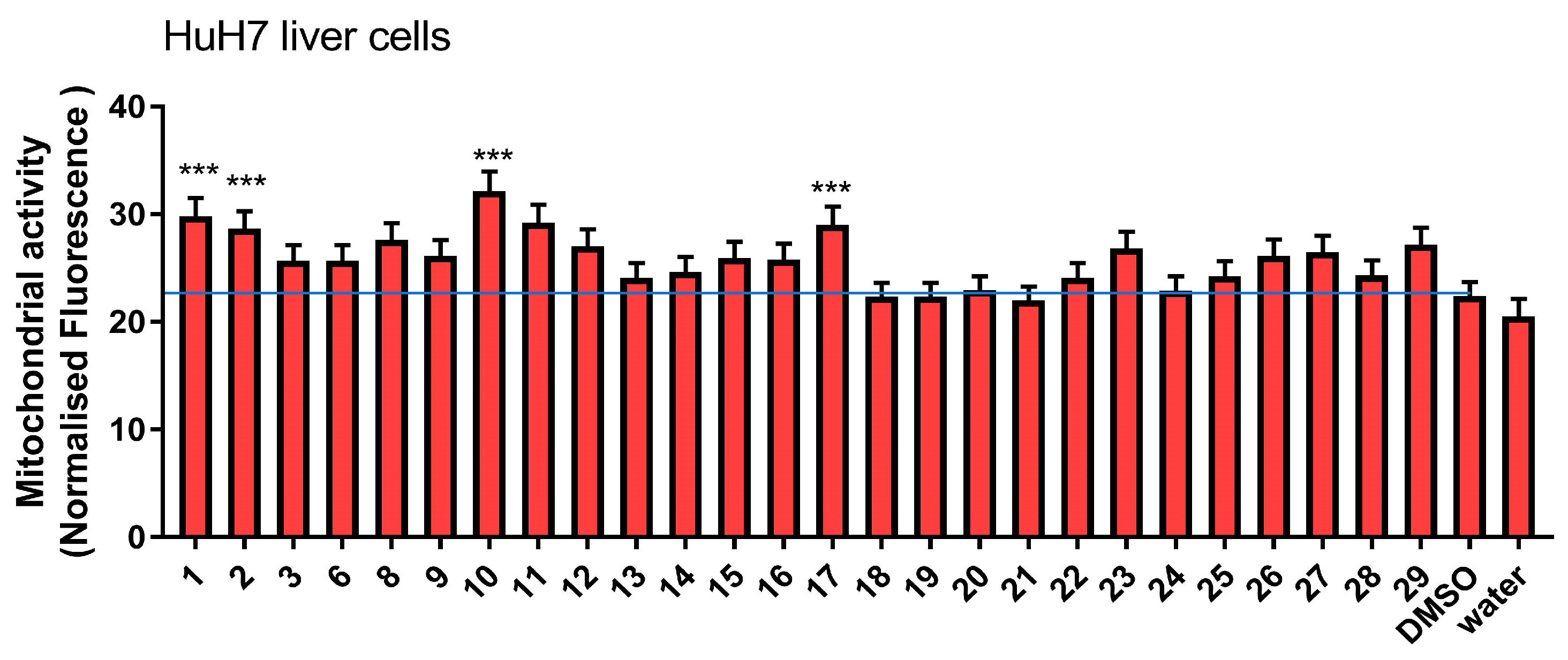
2.6. Mitochondrial Activity
2.7. NBDG Glucose Uptake Assay
2.8. RNA Purification and mRNA Quantification by Real-Time RT-PCR
2.9. GC/MS Analysis
3. Results and Discussion
3.1. Extraction, Isolation, and Structural Elucidation
3.2. Biological Evaluation on Targets Related to Metabolic Syndrome
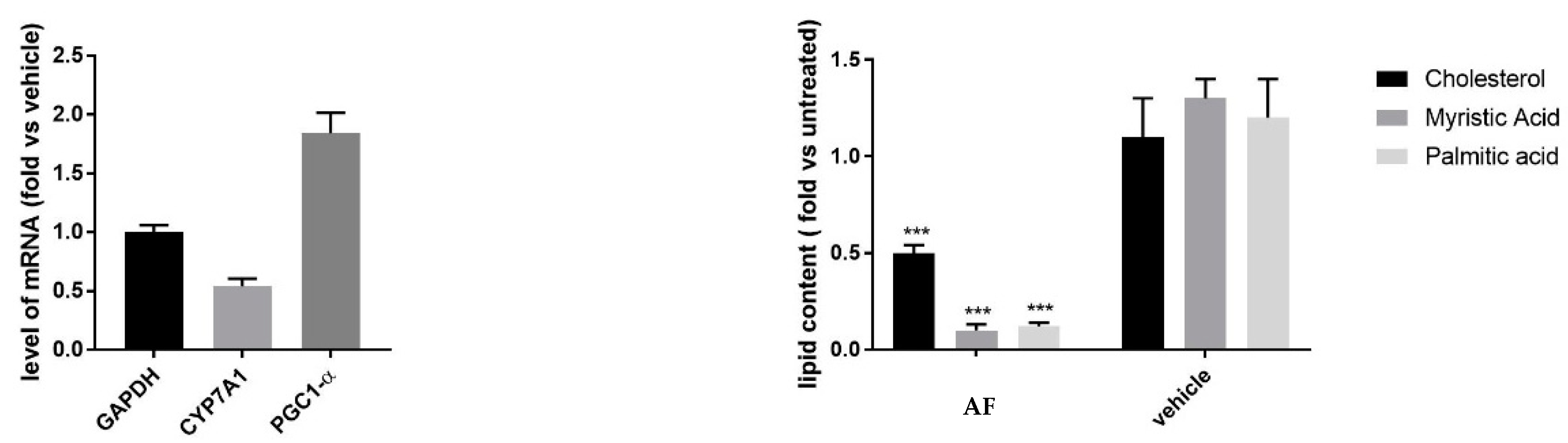
3.2.1. Effect on Mitochondrial Activity
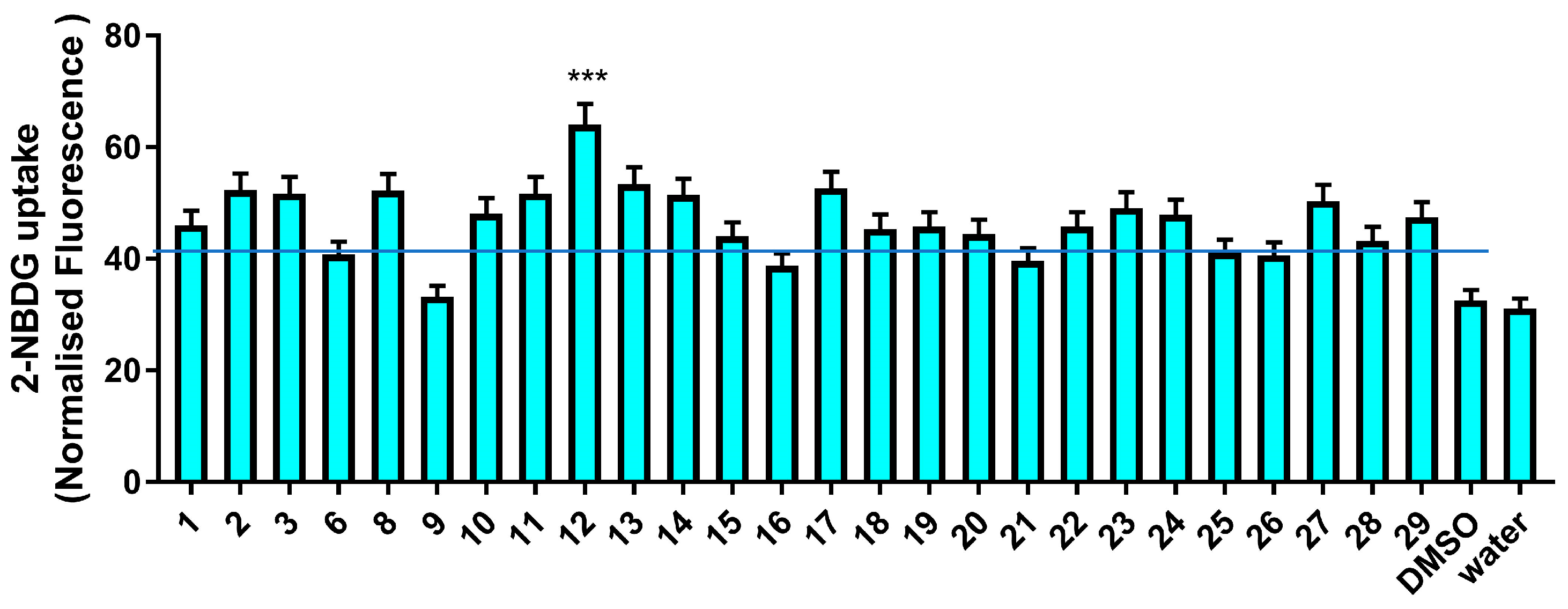
3.2.2. Glucose Uptake Modulatory Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mohamed, S.M.; Shalaby, M.A.; El-Shiekh, R.A.; El-Banna, H.A.; Emam, S.R.; Bakr, A.F. Metabolic syndrome: Risk factors, diagnosis, pathogenesis, and management with natural approaches. Food Chem. Adv. 2023, 3, 100335. [Google Scholar]
- Santa, K.; Kumazawa, Y.; Nagaoka, I. Prevention of metabolic syndrome by phytochemicals and vitamin D. Int. J. Mol. Sci. 2023, 24, 2627. [Google Scholar] [CrossRef]
- Weidner, C.; de Groot, J.C.; Prasad, A.; Freiwald, A.; Quedenau, C.; Kliem, M.; Witzke, A.; Kodelja, V.; Han, C.T.; Giegold, S.; et al. Amorfrutins are potent antidiabetic dietary natural products. Proc. Natl. Acad. Sci. USA 2012, 109, 7257–7262. [Google Scholar] [CrossRef] [PubMed]
- Grygiel-Górniak, B. Peroxisome proliferator-activated receptors and their ligands: Nutritional and clinical implications-a review. Nutr. J. 2014, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Serino, E.; Iannotti, F.A.; Al-Hmadi, H.B.; Caprioglio, D.; Moriello, C.; Masi, F.; Hammami, S.; Appendino, G.; Vitale, R.M.; Taglialatela-Scafati, O. PPARα/γ-targeting amorfrutin phytocannabinoids from aerial parts of Glycyrrhiza foetida. J. Nat. Prod. 2023, 86, 2435–2447. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Lu, J.H. Revision of Glycyrrhiza L. In Taxonomy and Experimental Biology of the Genus Glycyrrhiza L.; Fudan University Press: Shanghai, China, 2015; pp. 3–51. [Google Scholar]
- Ghisalberti, E.L.; Jefferies, P.R.; McAdam, D. Isoprenylated resorcinol derivatives from Glycyrrhiza acanthocarpa. Phytochemistry 1981, 20, 1959–1961. [Google Scholar] [CrossRef]
- Manfredi, K.P.; Vallurupalli, V.; Demidova, M.; Kindscher, K.; Pannell, L.K. Isolation of an anti-HIV diprenylated bibenzyl from Glycyrrhiza lepidota. Phytochemistry 2001, 58, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Jafari, F.; Jafari, M.; Moghadam, A.T.; Emami, S.A.; Jamialahmadi, T.; Mohammadpour, A.H.; Sahebkar, A. A review of Glycyrrhiza glabra (licorice) effects on metabolic syndrome. In Natural Products and Human Diseases: Pharmacology, Molecular Targets, and Therapeutic Benefits; Springer: Berlin/Heidelberg, Germany, 2021; pp. 385–400. [Google Scholar]
- Xu, T.; Yang, M.; Li, Y.; Chen, X.; Wang, Q.; Deng, W.; Pang, X.; Yu, K.; Jiang, B.; Guan, S.; et al. An integrated exact mass spectrometric strategy for comprehensive and rapid characterization of phenolic compounds in licorice. Rapid Commun. Mass Spectr. 2013, 27, 2297–2309. [Google Scholar] [CrossRef] [PubMed]
- Errichiello, F.; D’Amato, M.; Gambuti, A.; Moio, L.; Pastore, A.; Hekmat, A.H.; Stornaiuolo, M.; Serino, E.; Taglialatela-Scafati, O.; Forino, M. Oleanolic acid: A promising antidiabetic metabolite detected in Aglianico grape pomace. J. Funct. Foods 2023, 104, 105548. [Google Scholar] [CrossRef]
- Vasconcelos, J.M.; Silva, A.M.; Cavaleiro, J.A. Chromones and flavanones from Artemisia campestris subsp. maritima. Phytochemistry 1998, 49, 1421–1424. [Google Scholar] [CrossRef]
- Chen, R.; Liu, X.; Zou, J.; Yin, Y.; Ou, B.; Li, J.; Wang, R.; Xie, D.; Zhang, P.; Dai, J. Regio-and stereospecific prenylation of flavonoids by Sophora flavescens prenyltransferase. Adv. Synth. Catal. 2013, 355, 1817–1828. [Google Scholar] [CrossRef]
- Iinuma, M.; Yokoyama, J.; Ohyama, M.; Tanaka, T.; Ruangrungsi, N. Eight phenolic compounds in root of Sophora exigua. Phytochemistry 1994, 35, 785–789. [Google Scholar] [CrossRef]
- Roussis, V.; Ampofo, S.A.; Wiemer, D.F. Flavanones from Lonchocarpus minimiflorus. Phytochemistry 1987, 26, 2371–2375. [Google Scholar] [CrossRef]
- Ito, C.; Sato, K.; Oka, T.; Inoue, M.; Ju-Ichi, M.; Omura, M.; Furukawa, H. Two flavanones from Citrus species. Phytochemistry 1989, 28, 3562–3564. [Google Scholar] [CrossRef]
- Chang, Y.C.; Nair, M.G.; Santell, R.C.; Helferich, W.G. Microwave-mediated synthesis of anticarcinogenic isoflavones from soybeans. J. Agric. Food Chem. 1994, 42, 1869–1871. [Google Scholar] [CrossRef]
- Xu, K.; Deng, R.; Lü, H. Preparative isolation, and purification of phlorizin from apple tree bark by high-speed counter-current chromatography. Asian J. Chem. 2011, 23, 3807–3810. [Google Scholar]
- Gurbuz, P.; Baran, M.Y.; Demirezer, L.O.; Guvenalp, Z.; Kuruuzum-Uz, A. Phenylacylated-flavonoids from Peucedanum chryseum. Rev. Bras. Farmacogn. 2018, 28, 228–230. [Google Scholar] [CrossRef]
- Ko, R.K.; Kang, M.C.; Kim, B.S.; Han, J.H.; Kim, G.O.; Lee, N.H. A new phloroglucinol glycoside from Aster subulatus Michx. Bull. Kor. Chem. Soc. 2009, 30, 1167–1169. [Google Scholar]
- Seigler, D.S.; Pauli, G.F.; Nahrstedt, A.; Leen, R. Cyanogenic allosides and glucosides from Passiflora edulis and Carica papaya. Phytochemistry 2002, 60, 873–882. [Google Scholar] [CrossRef]
- Monteiro, L.B.; Davanzo, G.G.; de Aguiar, C.F.; Moraes-Vieira, P.M.M. Using flow cytometry for mitochondrial assays. MethodsX 2020, 7, 100938. [Google Scholar] [CrossRef]
- Esterbauer, H.; Oberkofler, H.; Krempler, F.; Patsch, W. Human peroxisome proliferator activated receptor gamma coactivator 1 (PPARGC1) gene: cDNA sequence, genomic organization, chromosomal localization, and tissue expression. Genomics 2000, 62, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Bala, M.; Gupta, P.; Gupta, S.; Dua, A.; Injeti, E.; Mittal, A. Efficient and modified 2-NBDG assay to measure glucose uptake in cultured myotubes. J. Pharmacol. Toxicol. Methods 2021, 109, 107069. [Google Scholar] [CrossRef] [PubMed]
- Ehrenkranz, J.R.; Lewis, N.G.; Kahn, C.R.; Roth, J. Phlorizin: A review. Diabetes Metab. Res. Rev. 2005, 21, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Blaschek, W. Natural products as lead compounds for sodium glucose cotransporter (SGLT) inhibitors. Planta Med. 2017, 83, 985–993. [Google Scholar] [CrossRef]
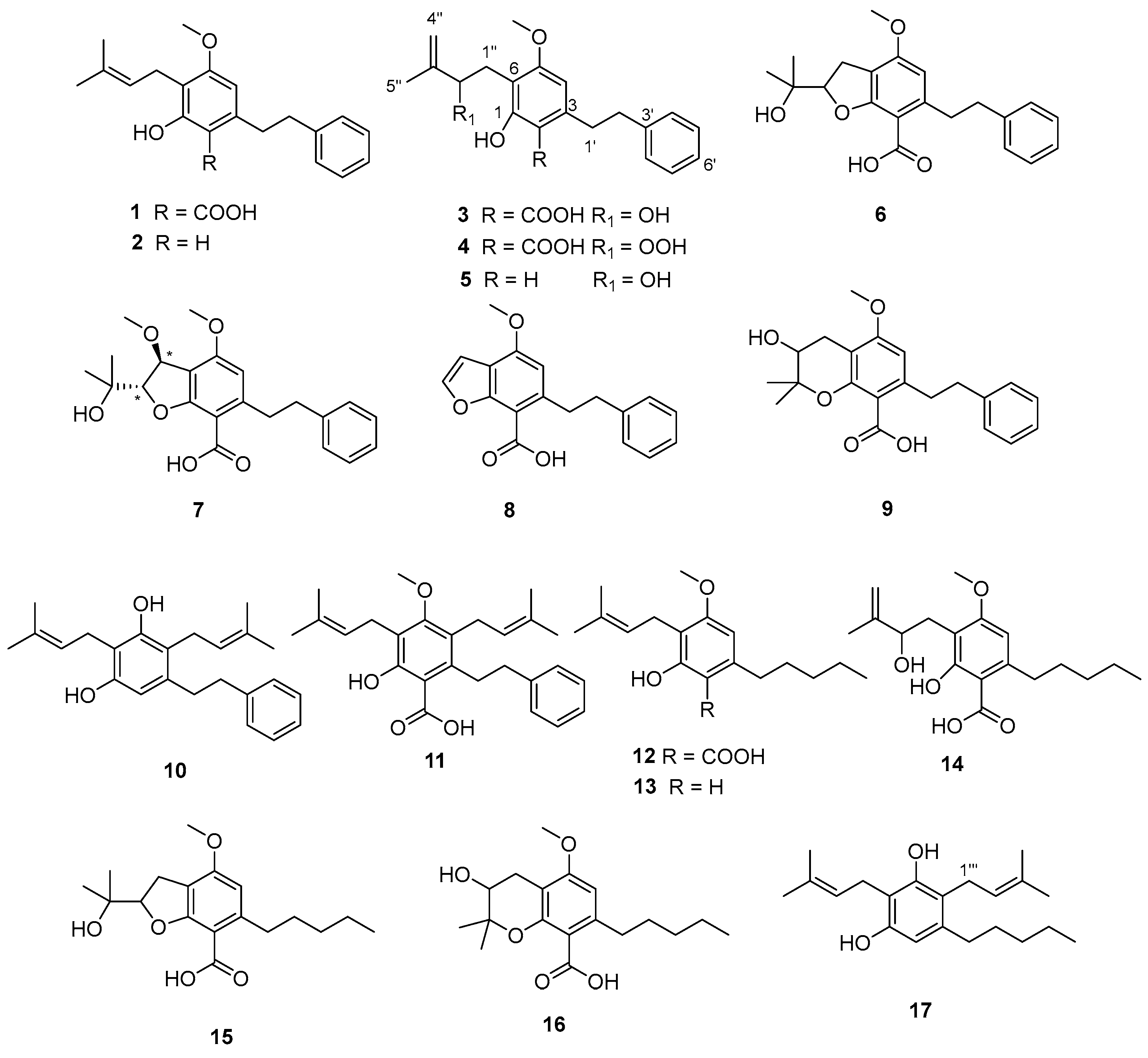
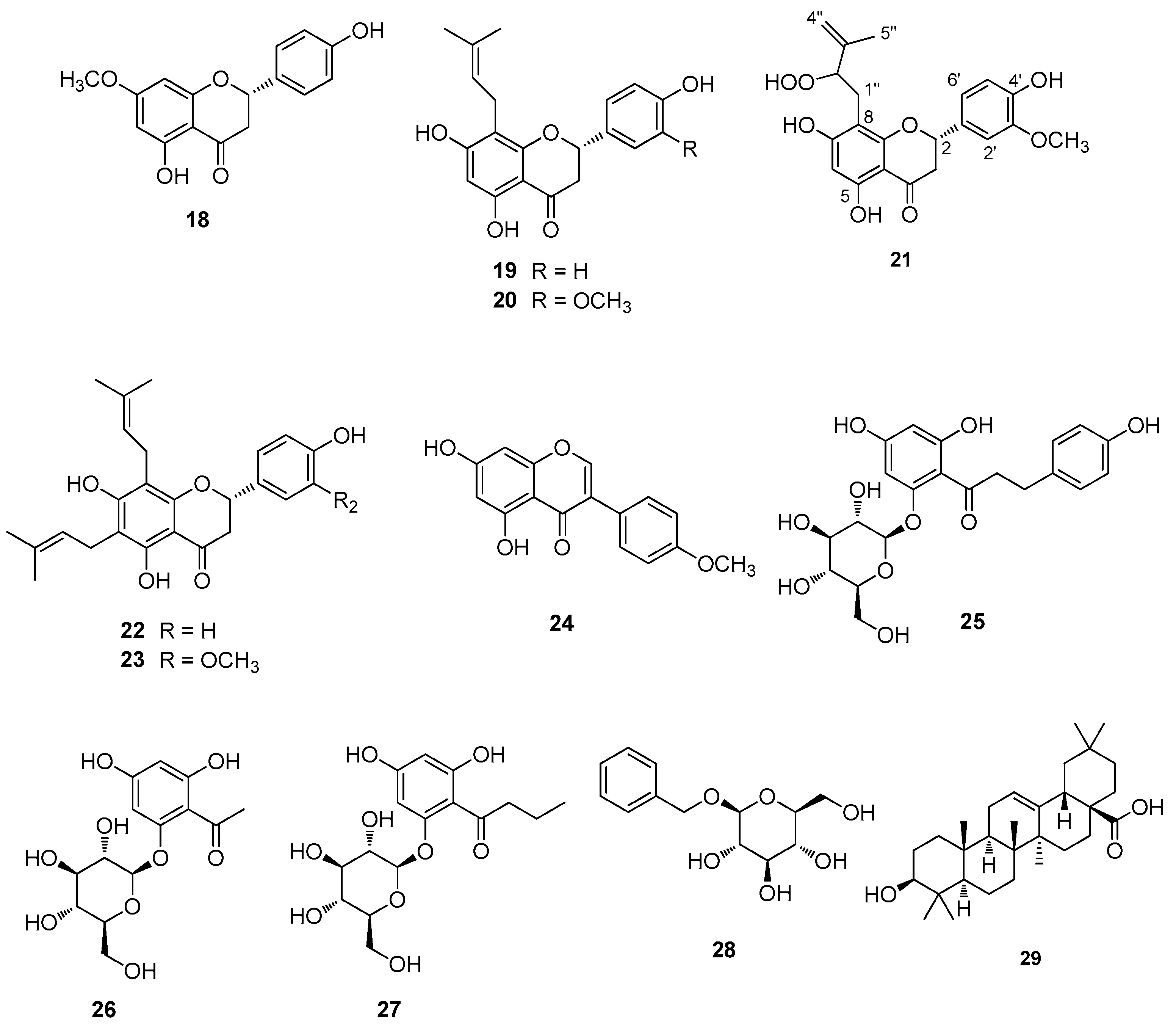
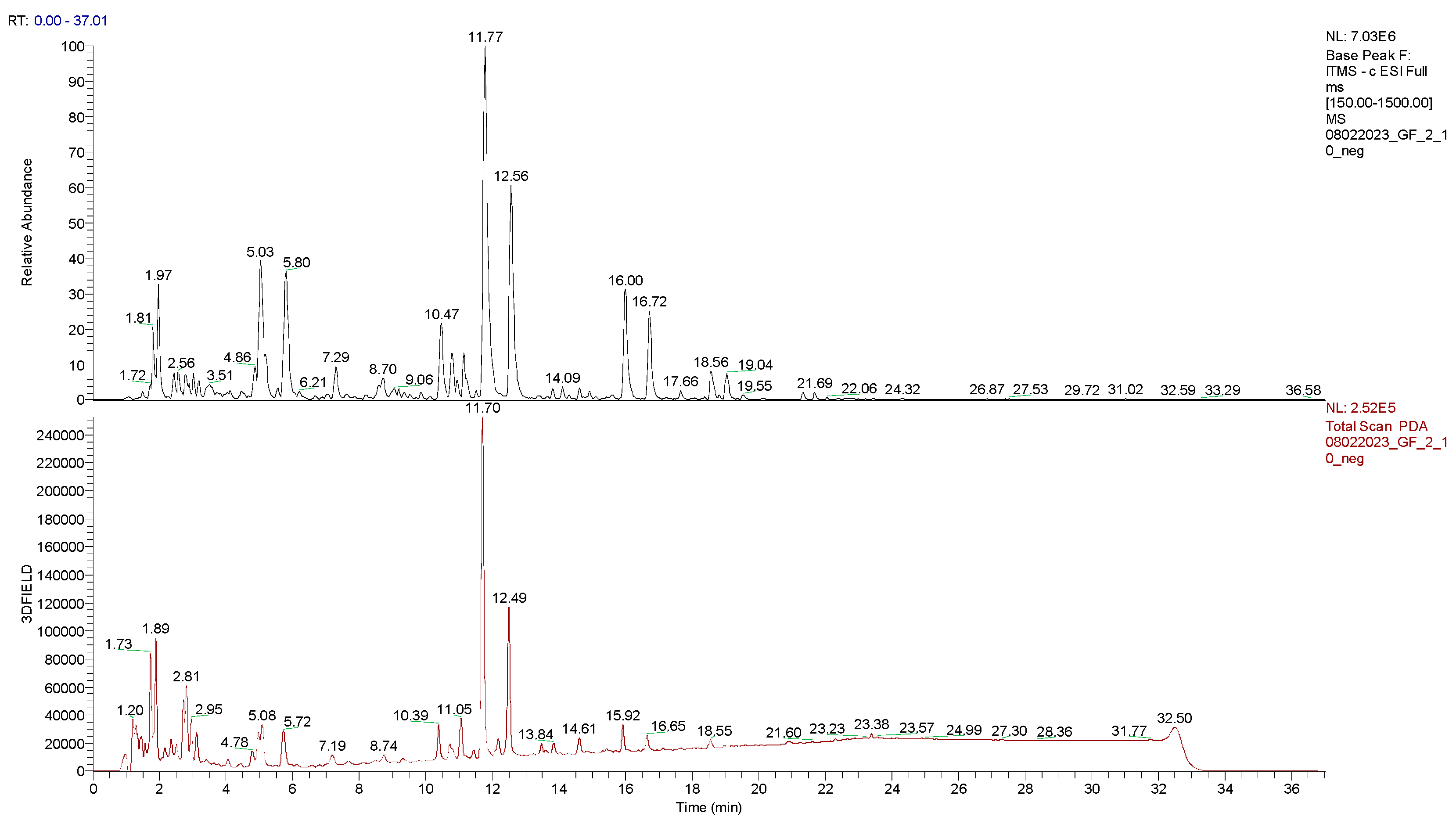

| Assignment | Rt (min) | Precursor ion (m/z) | Fragments | UV (λmax in nm) |
|---|---|---|---|---|
| Amorfrutin 3 | 3.02 | 355.30 | 311.28; 293.24; | 220.00; 260.00; 300.00 |
| Biochanin A | 3.17 | 283.28 | 268.06; 265.20; 240.29 | 220.00; 260.00; |
| Exiguaflavanone K | 9.06 | 369.33 | 339.12; 325.18; 293.23 284.17; | 220.00; 265.00; 290.00; 375.00 |
| Lonchocarpol A | 10.47 | 407.33 | 398.22; 301.11; 313.21; 287.24; 261.22; 243.18 | 220.00; 295.00; 345.00 |
| Hiravanone | 10.77 | 437.43 | 422.15; 313.20; 301.18; 287.11; 261.25; 243.23 | 220.00; 295.00; 345.00 (220; 290; 335; 375; 425) |
| Amorfrutin A | 11.77 | 339.28 | 295.15; 225.19 | 225.00; 270.00; 305.00; 375.00; 425.00 |
| Amorfrutin 2 | 12.56 | 305.24 | 290.24; 275.25; 261.18; 247.22 | 220.00; 270.00; 305.00; 375.00; 420.00 |
| Amorfrutin C | 16.00 | 407.33 | 363.30; 348.25; 331.27; 305.12; 285.29 | 220.00; 315.00; 375.00; 410.00 |
| Position | δH, Mult., J in Hz | δC, Mult. |
|---|---|---|
| 1 | 155.8, C | |
| 2 | 6.26, s | 103.5, CH |
| 3 | 142.7, C | |
| 4 | 6.49, s | 110.3, CH |
| 5 | 158.2, C | |
| 6 | 112.6, C | |
| 1′ | 2.85, m | 37.9, CH2 |
| 2′ | 2.91, m | 37.8, CH2 |
| 3′ | 141.5, C | |
| 4′ | 7.20, overlapped | 128.4, CH |
| 5′ | 7.28, overlapped | 128.6, CH |
| 6′ | 7.20, overlapped | 125.9, CH |
| 7′ | 7.28, overlapped | 128.6, CH |
| 8′ | 7.20, overlapped | 128.4, CH |
| 1a″ 1b″ | 3.10, dd, 14.6, 2.3 2.78, dd, 14.6, 8.0 | 30.8, CH2 |
| 2″ | 4.29, dd, 8.0, 2.3 | 76.8, CH |
| 3″ | 146.9, C | |
| 4a″ 4b″ | 4.86, s 4.99, s | 110.3, CH2 |
| 5″ | 1.84, s | 18.1, CH3 |
| -OMe | 3.74, s | 55.5, CH3 |
| Position | δH, Mult., J in Hz | δC, Mult. |
|---|---|---|
| 2 | 5.32, m | 79.2, CH |
| 3a 3b | 3.09, m 2.79, m | 43.4, CH2 |
| 4 | 196.0, C | |
| 5 | 161.9, C | |
| 6 | 6.07, s | 96.4, CH |
| 7 | 163.7, C | |
| 8 | 105.3, C | |
| 9 | 163.5, C | |
| 10 | 102.8, C | |
| 1′ | 135.9, C | |
| 2′ | 6.96, overlapped | 108.8, CH |
| 3′ | 146.4, C | |
| 4′ | 146.3, C | |
| 5′ | 6.94, overlapped | 114.4, CH |
| 6′ | 6.93, overlapped | 119.6, CH |
| OMe | 3.93, s | 55.9, CH3 |
| 1″a 1″b | 3.08, m 2.79, m | 23.8, CH2 |
| 2″ | 4.48, m | 88.8, CH |
| 3″ | 143.4, C | |
| 4″ | 5.07, s | 113.5, CH2 |
| 5″ | 1.87, s | 18.9, CH3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Hmadi, H.B.; Serino, E.; Pastore, A.; Chianese, G.; Hammami, S.; Stornaiuolo, M.; Taglialatela-Scafati, O. Metabolites from Aerial Parts of Glycyrrhiza foetida as Modulators of Targets Related to Metabolic Syndrome. Biomolecules 2024, 14, 467. https://doi.org/10.3390/biom14040467
Al-Hmadi HB, Serino E, Pastore A, Chianese G, Hammami S, Stornaiuolo M, Taglialatela-Scafati O. Metabolites from Aerial Parts of Glycyrrhiza foetida as Modulators of Targets Related to Metabolic Syndrome. Biomolecules. 2024; 14(4):467. https://doi.org/10.3390/biom14040467
Chicago/Turabian StyleAl-Hmadi, Hekmat B., Elena Serino, Arianna Pastore, Giuseppina Chianese, Saoussen Hammami, Mariano Stornaiuolo, and Orazio Taglialatela-Scafati. 2024. "Metabolites from Aerial Parts of Glycyrrhiza foetida as Modulators of Targets Related to Metabolic Syndrome" Biomolecules 14, no. 4: 467. https://doi.org/10.3390/biom14040467
APA StyleAl-Hmadi, H. B., Serino, E., Pastore, A., Chianese, G., Hammami, S., Stornaiuolo, M., & Taglialatela-Scafati, O. (2024). Metabolites from Aerial Parts of Glycyrrhiza foetida as Modulators of Targets Related to Metabolic Syndrome. Biomolecules, 14(4), 467. https://doi.org/10.3390/biom14040467








