The Role of Ubiquitination in Osteosarcoma Development and Therapies
Abstract
1. Introduction
2. The UPS
3. E3 Ligases
3.1. RING E3
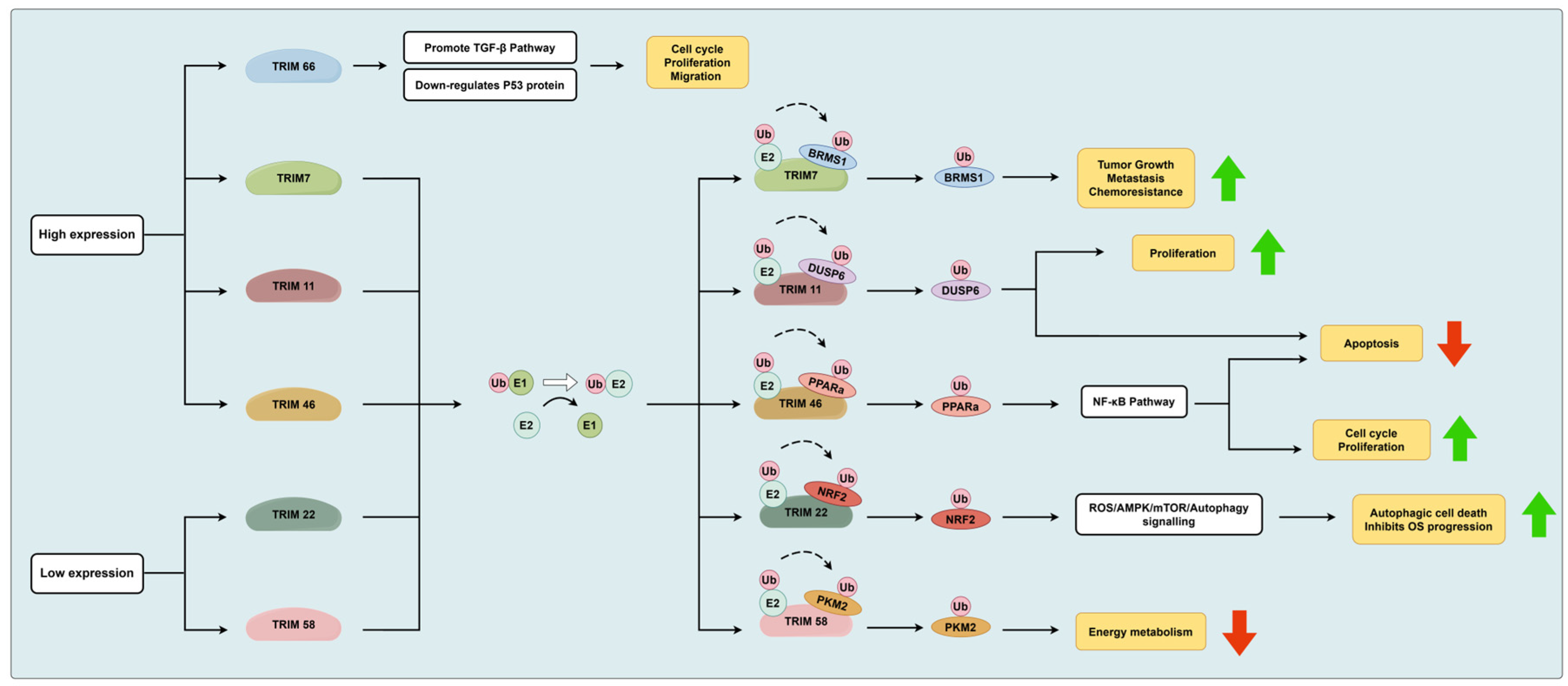
3.1.1. The TRIM Protein Family
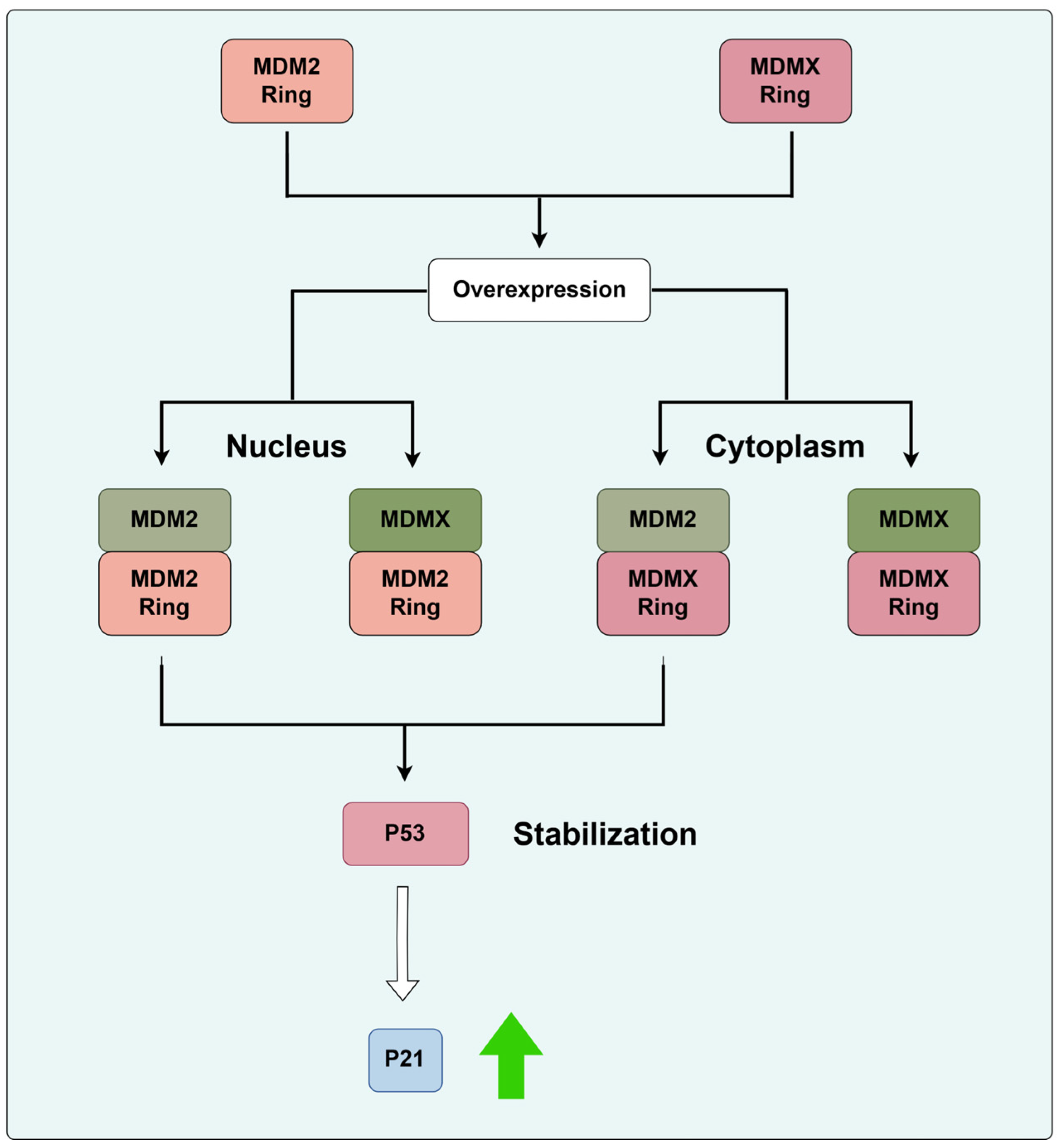
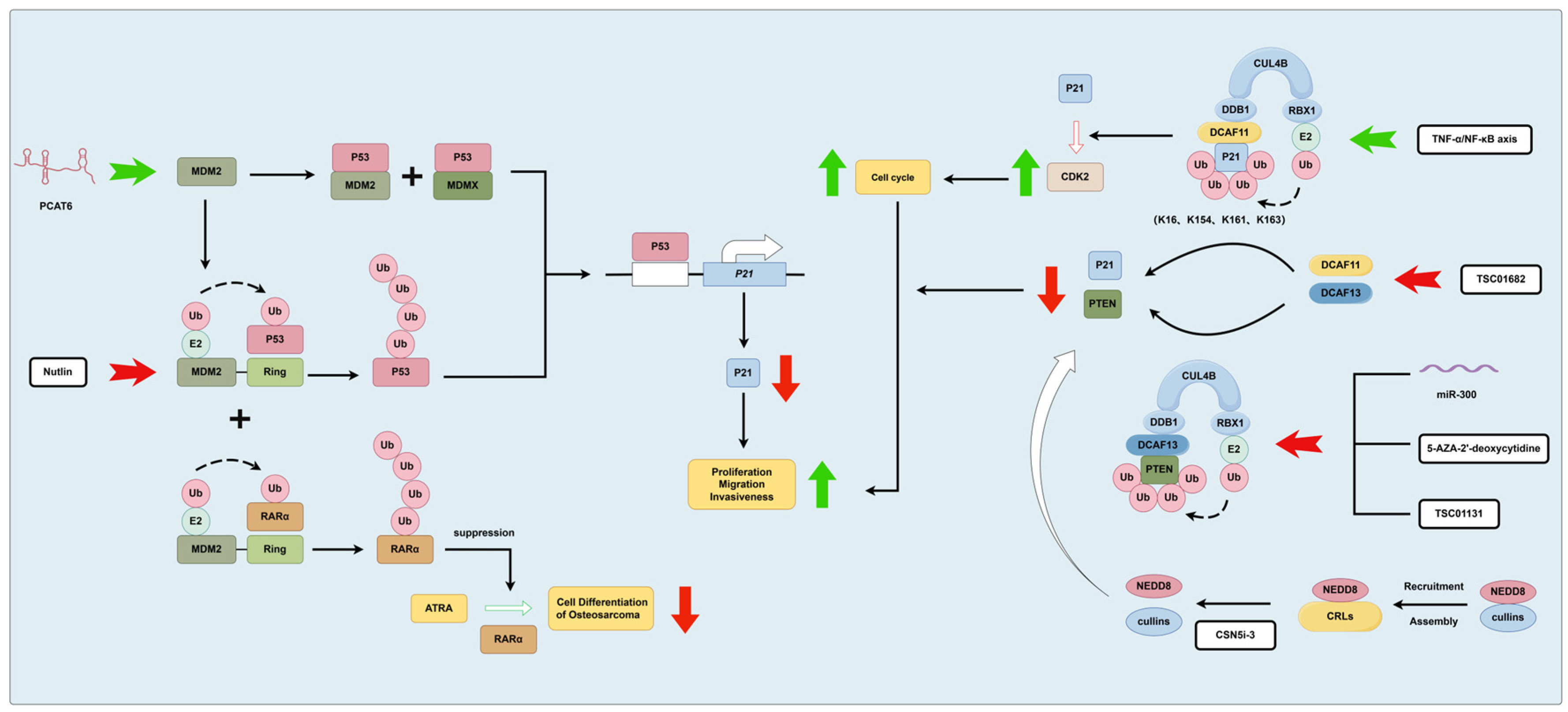
3.1.2. MDM2 and CRLs
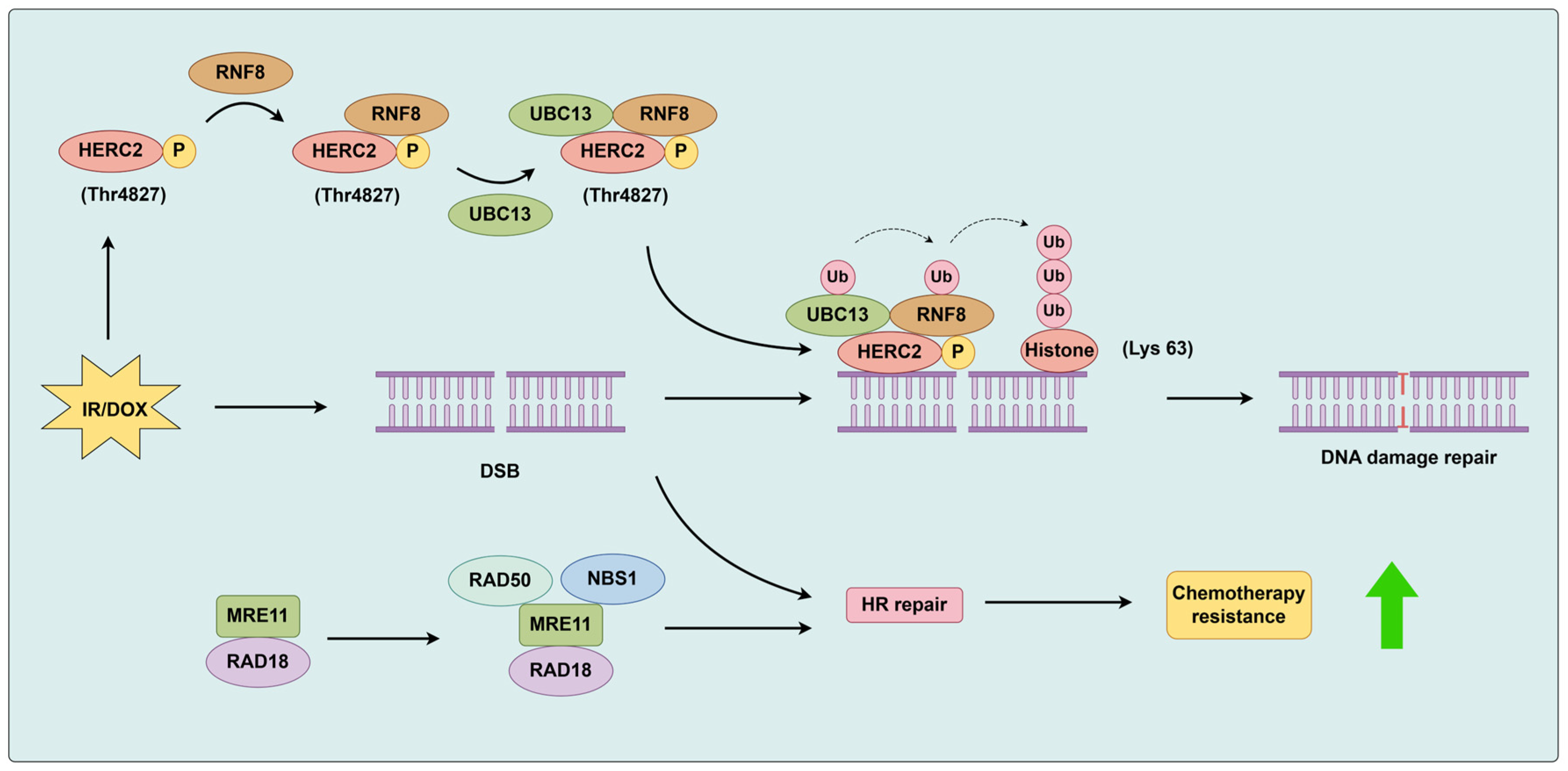
3.1.3. RAD18 and RNF8
3.1.4. SIAH1
3.1.5. Other RING-Type E3 Ligases
3.2. HECT E3
4. E2 Ligases
5. DUBs
5.1. USP1
5.2. USP4/USP17
5.3. USP6/USP27X/USP41/USP43
5.4. USP7
5.5. USP9X
5.6. USP11
5.7. USP22
5.8. USP39
5.9. USP47
5.10. OTUB1
5.11. UCHL1
5.12. BAP1
6. Ubiquilins
7. Signaling Pathways
- p53 pathway
- 2.
- STAT pathway
- 3.
- PI3K/Akt Pathway
8. Other Related Studies
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Moukengue, B.; Lallier, M.; Marchandet, L.; Baud’huin, M.; Verrecchia, F.; Ory, B.; Lamoureux, F. Origin and Therapies of Osteosarcoma. Cancers 2022, 14, 3503. [Google Scholar] [CrossRef] [PubMed]
- Luetke, A.; Meyers, P.A.; Lewis, I.; Juergens, H. Osteosarcoma treatment—where do we stand? A state of the art review. Cancer Treat. Rev. 2014, 40, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Dhammi, I.K.; Kumar, S. Osteosarcoma: A journey from amputation to limb salvage. Indian J. Orthop. 2014, 48, 233–234. [Google Scholar] [CrossRef] [PubMed]
- D’Arienzo, A.; Ipponi, E.; Ruinato, A.D.; De Franco, S.; Colangeli, S.; Andreani, L.; Capanna, R. Proximal Humerus Reconstruction after Tumor Resection: An Overview of Surgical Management. Adv. Orthop. 2021, 2021, 5559377. [Google Scholar] [CrossRef]
- Fulchignoni, C.; Pietramala, S.; Lopez, I.; Mazzella, G.G.; Comisi, C.; Perisano, C.; Rocchi, L.; Greco, T. Surgical Outcomes and Complications of Custom-Made Prostheses in Upper Limb Oncological Reconstruction: A Systematic Review. J. Funct. Morphol. Kinesiol. 2024, 9, 72. [Google Scholar] [CrossRef]
- Thanindratarn, P.; Dean, D.C.; Nelson, S.D.; Hornicek, F.J.; Duan, Z. Advances in immune checkpoint inhibitors for bone sarcoma therapy. J. Bone Oncol. 2019, 15, 100221. [Google Scholar] [CrossRef] [PubMed]
- Lussier, D.M.; Johnson, J.L.; Hingorani, P.; Blattman, J.N. Combination immunotherapy with alpha-CTLA-4 and alpha-PD-L1 antibody blockade prevents immune escape and leads to complete control of metastatic osteosarcoma. J. Immunother. Cancer 2015, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Mucke, T.; Mitchell, D.A.; Tannapfel, A.; Wolff, K.D.; Loeffelbein, D.J.; Kanatas, A. Effect of neoadjuvant treatment in the management of osteosarcomas of the head and neck. J. Cancer Res. Clin. Oncol. 2014, 140, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Shim, T.; Chillakuru, Y.; Darwish, C.; Chalif, E.; Strum, D.; Benito, D.A.; Mulcahy, C.F.; Monfared, A. Head and neck osteosarcomas: Analysis of treatment trends and survival outcomes in the United States (2004–2016). Head. Neck. 2021, 43, 3294–3305. [Google Scholar] [CrossRef]
- Gill, J.; Gorlick, R. Advancing therapy for osteosarcoma. Nat. Rev. Clin. Oncol. 2021, 18, 609–624. [Google Scholar] [CrossRef]
- Dantuma, N.P.; Bott, L.C. The ubiquitin-proteasome system in neurodegenerative diseases: Precipitating factor, yet part of the solution. Front. Mol. Neurosci. 2014, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.L.; Crews, C.M. Small-molecule control of intracellular protein levels through modulation of the ubiquitin proteasome system. Angew. Chem. Int. Ed. Engl. 2014, 53, 2312–2330. [Google Scholar] [CrossRef] [PubMed]
- Luza, S.; Opazo, C.M.; Bousman, C.A.; Pantelis, C.; Bush, A.I.; Everall, I.P. The ubiquitin proteasome system and schizophrenia. Lancet Psychiatry 2020, 7, 528–537. [Google Scholar] [CrossRef]
- Sun, T.; Liu, Z.; Yang, Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol. Cancer 2020, 19, 146. [Google Scholar] [CrossRef]
- Caba, C.; Mohammadzadeh, A.; Tong, Y. On the Study of Deubiquitinases: Using the Right Tools for the Job. Biomolecules 2022, 12, 703. [Google Scholar] [CrossRef]
- Litwak, S.A.; Wali, J.A.; Pappas, E.G.; Saadi, H.; Stanley, W.J.; Varanasi, L.C.; Kay, T.W.; Thomas, H.E.; Gurzov, E.N. Lipotoxic Stress Induces Pancreatic beta-Cell Apoptosis through Modulation of Bcl-2 Proteins by the Ubiquitin-Proteasome System. J. Diabetes Res. 2015, 2015, 280615. [Google Scholar] [CrossRef]
- Heo, A.J.; Ji, C.H.; Kwon, Y.T. The Cys/N-degron pathway in the ubiquitin-proteasome system and autophagy. Trends. Cell Biol. 2023, 33, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Pohl, C.; Dikic, I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019, 366, 818–822. [Google Scholar] [CrossRef]
- Fasanaro, P.; Capogrossi, M.C.; Martelli, F. Regulation of the endothelial cell cycle by the ubiquitin-proteasome system. Cardiovasc. Res. 2010, 85, 272–280. [Google Scholar] [CrossRef]
- LaPlante, G.; Zhang, W. Targeting the Ubiquitin-Proteasome System for Cancer Therapeutics by Small-Molecule Inhibitors. Cancers 2021, 13, 3079. [Google Scholar] [CrossRef]
- Kulathu, Y.; Komander, D. Atypical ubiquitylation—the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012, 13, 508–523. [Google Scholar] [CrossRef]
- Chen, Y.J.; Wu, H.; Shen, X.Z. The ubiquitin-proteasome system and its potential application in hepatocellular carcinoma therapy. Cancer Lett. 2016, 379, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Wang, Q.; Cai, R.; Yuan, H.; Xu, M. The ubiquitin system: Orchestrating cellular signals in non-small-cell lung cancer. Cell Mol. Biol. Lett. 2020, 25, 1. [Google Scholar] [CrossRef]
- Sahasrabuddhe, A.A.; Elenitoba-Johnson, K.S. Role of the ubiquitin proteasome system in hematologic malignancies. Immunol. Rev. 2015, 263, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Wang, R.; Zhang, Y.; Li, X.; Gan, Y.; Gao, F.; Rong, P.; Wang, W.; Li, W. The role of ubiquitination and deubiquitination in tumor invasion and metastasis. Int. J. Biol. Sci. 2022, 18, 2292–2303. [Google Scholar] [CrossRef]
- Yu, Z.; Li, H.; Zhu, J.; Wang, H.; Jin, X. The roles of E3 ligases in Hepatocellular carcinoma. Am. J. Cancer Res. 2022, 12, 1179–1214. [Google Scholar] [PubMed]
- Cruz Walma, D.A.; Chen, Z.; Bullock, A.N.; Yamada, K.M. Ubiquitin ligases: Guardians of mammalian development. Nat. Rev. Mol. Cell Biol. 2022, 23, 350–367. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ma, L.; Wang, B.; Liu, J.; Wei, W. E3 ubiquitin ligases in cancer and implications for therapies. Cancer Metastasis Rev. 2017, 36, 683–702. [Google Scholar] [CrossRef]
- Knauer, S.K.; Mahendrarajah, N.; Roos, W.P.; Kramer, O.H. The inducible E3 ubiquitin ligases SIAH1 and SIAH2 perform critical roles in breast and prostate cancers. Cytokine Growth. Factor Rev. 2015, 26, 405–413. [Google Scholar] [CrossRef]
- Nguyen, K.M.; Busino, L. Targeting the E3 ubiquitin ligases DCAF15 and cereblon for cancer therapy. Semin. Cancer Biol. 2020, 67, 53–60. [Google Scholar] [CrossRef]
- Venuto, S.; Merla, G. E3 Ubiquitin Ligase TRIM Proteins, Cell Cycle and Mitosis. Cells 2019, 8, 510. [Google Scholar] [CrossRef]
- Williams, F.P.; Haubrich, K.; Perez-Borrajero, C.; Hennig, J. Emerging RNA-binding roles in the TRIM family of ubiquitin ligases. Biol. Chem. 2019, 400, 1443–1464. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guo, Y.; Yang, H.; Shi, G.; Xu, G.; Shi, J.; Yin, N.; Chen, D. TRIM66 overexpresssion contributes to osteosarcoma carcinogenesis and indicates poor survival outcome. Oncotarget 2015, 6, 23708–23719. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhang, Z.; Zhu, X.; Qian, G.; Zhou, Y.; Sun, Y.; Yu, W.; Wang, J.; Lu, H.; Lin, F.; et al. N6-Methyladenosine modification of the TRIM7 positively regulates tumorigenesis and chemoresistance in osteosarcoma through ubiquitination of BRMS1. EBioMedicine 2020, 59, 102955. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xu, X.; Tang, W.; Zhu, Y.; Hu, J.; Zhang, X. Tripartite Motif Containing 11 Interacts with DUSP6 to Promote the Growth of Human Osteosarcoma Cells through Regulating ERK1/2 Pathway. Biomed. Res. Int. 2019, 2019, 9612125. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Cai, X.; Xu, T.; Liu, K.; Yang, D.; Fan, L.; Li, G.; Yu, X. Tripartite Motif-Containing 46 Promotes Viability and Inhibits Apoptosis of Osteosarcoma Cells by Activating NF-B Signaling Through Ubiquitination of PPAR. Oncol. Res. 2020, 28, 409–421. [Google Scholar] [CrossRef]
- Petersson, J.; Lonnbro, P.; Herr, A.M.; Morgelin, M.; Gullberg, U.; Drott, K. The human IFN-inducible p53 target gene TRIM22 colocalizes with the centrosome independently of cell cycle phase. Exp. Cell Res. 2010, 316, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhao, Y.; Wang, G.; Feng, S.; Ge, X.; Ye, W.; Wang, Z.; Zhu, Y.; Cai, W.; Bai, J.; et al. TRIM22 inhibits osteosarcoma progression through destabilizing NRF2 and thus activation of ROS/AMPK/mTOR/autophagy signaling. Redox. Biol. 2022, 53, 102344. [Google Scholar] [CrossRef]
- Yuan, P.; Zhou, Y.; Wang, R.; Chen, S.; Wang, Q.; Xu, Z.; Liu, Y.; Yang, H. TRIM58 Interacts with Pyruvate Kinase M2 to Inhibit Tumorigenicity in Human Osteosarcoma Cells. Biomed. Res. Int. 2020, 2020, 8450606. [Google Scholar] [CrossRef]
- Wang, W.; Qin, J.J.; Rajaei, M.; Li, X.; Yu, X.; Hunt, C.; Zhang, R. Targeting MDM2 for novel molecular therapy: Beyond oncology. Med. Res. Rev. 2020, 40, 856–880. [Google Scholar] [CrossRef]
- Karni-Schmidt, O.; Lokshin, M.; Prives, C. The Roles of MDM2 and MDMX in Cancer. Annu. Rev. Pathol. 2016, 11, 617–644. [Google Scholar] [CrossRef]
- Egorova, O.; Lau, H.H.; McGraphery, K.; Sheng, Y. Mdm2 and MdmX RING Domains Play Distinct Roles in the Regulation of p53 Responses: A Comparative Study of Mdm2 and MdmX RING Domains in U2OS Cells. Int. J. Mol. Sci. 2020, 21, 1309. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Xu, Y.; Zheng, J. Long non-coding RNA PCAT6 promotes the development of osteosarcoma by increasing MDM2 expression. Oncol. Rep. 2020, 44, 2465–2474. [Google Scholar] [CrossRef] [PubMed]
- Ying, M.; Zhang, L.; Zhou, Q.; Shao, X.; Cao, J.; Zhang, N.; Li, W.; Zhu, H.; Yang, B.; He, Q. The E3 ubiquitin protein ligase MDM2 dictates all-trans retinoic acid-induced osteoblastic differentiation of osteosarcoma cells by modulating the degradation of RARalpha. Oncogene 2016, 35, 4358–4367. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Gao, H.; Ji, Y.; Zhou, Q.; Du, Z.; Tian, L.; Jiang, Y.; Yao, K.; Zhou, Z. Targeting p53-MDM2 interaction by small-molecule inhibitors: Learning from MDM2 inhibitors in clinical trials. J. Hematol. Oncol. 2022, 15, 91. [Google Scholar] [CrossRef] [PubMed]
- Henningsen, K.M.; Manzini, V.; Magerhans, A.; Gerber, S.; Dobbelstein, M. MDM2-Driven Ubiquitination Rapidly Removes p53 from Its Cognate Promoters. Biomolecules 2021, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S.; et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, K.; Hou, C.; Jiang, K.; Chen, B.; Chen, J.; Lao, L.; Qian, L.; Zhong, G.; Liu, Z.; et al. CRL4B(DCAF11) E3 ligase targets p21 for degradation to control cell cycle progression in human osteosarcoma cells. Sci. Rep. 2017, 7, 1175. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, B.; Jiang, K.; Lao, L.; Shen, H.; Chen, Z. Activation of TNF-alpha/NF-kappaB axis enhances CRL4B(DCAF)(11) E3 ligase activity and regulates cell cycle progression in human osteosarcoma cells. Mol. Oncol. 2018, 12, 476–494. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, W.; Jiang, K.; Chen, B.; Wang, K.; Lao, L.; Hou, C.; Wang, F.; Zhang, C.; Shen, H. MicroRNA-300 Regulates the Ubiquitination of PTEN through the CRL4B(DCAF13) E3 Ligase in Osteosarcoma Cells. Mol. Ther. Nucleic. Acids. 2018, 10, 254–268. [Google Scholar] [CrossRef]
- Chen, B.; Feng, Y.; Zhang, M.; Cheng, G.; Chen, B.; Wang, H. Small molecule TSC01682 inhibits osteosarcoma cell growth by specifically disrupting the CUL4B-DDB1 interaction and decreasing the ubiquitination of CRL4B E3 ligase substrates. Am. J. Cancer Res. 2019, 9, 1857–1870. [Google Scholar]
- Samsa, W.E.; Mamidi, M.K.; Bashur, L.A.; Elliott, R.; Miron, A.; Chen, Y.; Lee, B.; Greenfield, E.M.; Chan, R.; Danielpour, D.; et al. The crucial p53-dependent oncogenic role of JAB1 in osteosarcoma in vivo. Oncogene 2020, 39, 4581–4591. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Pei, H.; Lu, S.J.; Liu, Z.J.; Yan, L.; Zhao, X.M.; Hu, B.; Lu, H.G. SPOP suppresses osteosarcoma invasion via PI3K/AKT/NF-kappaB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Suzuki, M.; Kawai, H.; Suzuki, F.; Kamiya, K. Asymmetric nature of two subunits of RAD18, a RING-type ubiquitin ligase E3, in the human RAD6A-RAD18 ternary complex. Nucleic. Acids. Res. 2012, 40, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Gu, J.; Liu, C.; Liu, N.; Yu, Z.; Zhou, C.; Heng, W.; Cao, Z.; Wei, F.; Zhu, K.; et al. Genome-wide CRISPR screen identified Rad18 as a determinant of doxorubicin sensitivity in osteosarcoma. J. Exp. Clin. Cancer Res. 2022, 41, 154. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Poe, J.C.; Yang, L.; Fedoriw, A.; Desai, S.; Magnuson, T.; Li, Z.; Fedoriw, Y.; Araki, K.; Gao, Y.; et al. Rad18 confers hematopoietic progenitor cell DNA damage tolerance independently of the Fanconi Anemia pathway in vivo. Nucleic. Acids. Res. 2016, 44, 4174–4188. [Google Scholar] [CrossRef]
- Zhou, T.; Yi, F.; Wang, Z.; Guo, Q.; Liu, J.; Bai, N.; Li, X.; Dong, X.; Ren, L.; Cao, L.; et al. The Functions of DNA Damage Factor RNF8 in the Pathogenesis and Progression of Cancer. Int. J. Biol. Sci. 2019, 15, 909–918. [Google Scholar] [CrossRef]
- Bekker-Jensen, S.; Rendtlew Danielsen, J.; Fugger, K.; Gromova, I.; Nerstedt, A.; Lukas, C.; Bartek, J.; Lukas, J.; Mailand, N. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 2010, 12, 80–86. [Google Scholar] [CrossRef]
- Hodge, C.D.; Ismail, I.H.; Edwards, R.A.; Hura, G.L.; Xiao, A.T.; Tainer, J.A.; Hendzel, M.J.; Glover, J.N. RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment. J. Biol. Chem. 2016, 291, 9396–9410. [Google Scholar] [CrossRef]
- Chen, X.; He, Z.; Fu, M.; Wang, Y.; Wu, H.; Li, X.; Cao, H.; Zheng, S.J. The E3 Ubiquitin Ligase Siah-1 Suppresses Avian Reovirus Infection by Targeting p10 for Degradation. J. Virol. 2018, 92, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Loh, K.M.; van Amerongen, R.; Nusse, R. Generating Cellular Diversity and Spatial Form: Wnt Signaling and the Evolution of Multicellular Animals. Dev. Cell 2016, 38, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, S.; Kishida, S.; Yamamoto, H.; Murai, H.; Koyama, S.; Kikuchi, A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 1998, 17, 1371–1384. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Jiang, B.; Jiang, X.; Charlat, O.; Chen, A.; Mickanin, C.; Bauer, A.; Xu, W.; Yan, X.; Cong, F. The SIAH E3 ubiquitin ligases promote Wnt/beta-catenin signaling through mediating Wnt-induced Axin degradation. Genes Dev. 2017, 31, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Zhang, J. Inactivation of carboxyl terminus of Hsc70-interacting protein prevents hypoxia-induced pulmonary arterial smooth muscle cells proliferation by reducing intracellular Ca(2+) concentration. Pulm. Circ. 2019, 9, 2045894019875343. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qin, G.; Liang, X.; Wang, W.; Wang, Z.; Liao, D.; Zhong, L.; Zhang, R.; Zeng, Y.X.; Wu, Y.; et al. Targeting the CK1alpha/CBX4 axis for metastasis in osteosarcoma. Nat. Commun. 2020, 11, 1141. [Google Scholar] [CrossRef] [PubMed]
- Ostendorff, H.P.; Peirano, R.I.; Peters, M.A.; Schluter, A.; Bossenz, M.; Scheffner, M.; Bach, I. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature 2002, 416, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, Y.; Gao, R.; Yang, X.; Yan, X.; Wang, C.; Jiang, S.; Yu, L. RLIM interacts with Smurf2 and promotes TGF-beta induced U2OS cell migration. Biochem. Biophys. Res. Commun. 2011, 414, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shen, J.; Li, X.; Wang, X.; Long, M.; Lin, F.; Wei, J.; Yang, L.; Yang, C.; Dong, K.; et al. Rlim, an E3 ubiquitin ligase, influences the stability of Stathmin protein in human osteosarcoma cells. Cell Signal. 2014, 26, 1532–1538. [Google Scholar] [CrossRef] [PubMed]
- Liyasova, M.S.; Ma, K.; Lipkowitz, S. Molecular pathways: Cbl proteins in tumorigenesis and antitumor immunity-opportunities for cancer treatment. Clin. Cancer Res. 2015, 21, 1789–1794. [Google Scholar] [CrossRef]
- Severe, N.; Marie, P. Implication of the ubiquitin ligase c-Cbl in bone formation and tumorigenesis. Med. Sci. 2012, 28, 970–975. [Google Scholar] [CrossRef]
- Severe, N.; Dieudonne, F.X.; Marty, C.; Modrowski, D.; Patino-Garcia, A.; Lecanda, F.; Fromigue, O.; Marie, P.J. Targeting the E3 ubiquitin casitas B-lineage lymphoma decreases osteosarcoma cell growth and survival and reduces tumorigenesis. J. Bone Min. Res. 2012, 27, 2108–2117. [Google Scholar] [CrossRef]
- Araki, T.; Milbrandt, J. ZNRF proteins constitute a family of presynaptic E3 ubiquitin ligases. J. Neurosci. 2003, 23, 9385–9394. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Yang, Y.; An, Q.; Qi, Y. MicroRNA-100 suppresses human osteosarcoma cell proliferation and chemo-resistance via ZNRF2. Oncotarget 2017, 8, 34678–34686. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zhong, G.; Li, J.; Pan, J.; Zhao, Y.; Song, H.; Sun, W.; Jin, X.; Li, Y.; Du, R.; et al. Targeting E3 Ubiquitin Ligase WWP1 Prevents Cardiac Hypertrophy Through Destabilizing DVL2 via Inhibition of K27-Linked Ubiquitination. Circulation 2021, 144, 694–711. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zan, P.; Li, S.; Liu, J.; Wang, J.; Chen, D.; Wang, H.; Qian, Y.; Luo, L.; Huang, X. Knockdown of WWP1 inhibits growth and invasion, but induces apoptosis of osteosarcoma cells. Int. J. Clin. Exp. Pathol. 2015, 8, 7869–7877. [Google Scholar] [PubMed]
- Zhang, W.; Zhuang, Y.; Zhang, Y.; Yang, X.; Zhang, H.; Wang, G.; Yin, W.; Wang, R.; Zhang, Z.; Xiao, W. Uev1A facilitates osteosarcoma differentiation by promoting Smurf1-mediated Smad1 ubiquitination and degradation. Cell Death Dis. 2017, 8, e2974. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.T.; Li, D.Z.; Li, J.M.; Fang, J.; Li, H.Z.; Tong, P.J.; Liu, F.C. Lentivirus-mediated RNA interference targeting UbcH10 reduces cell growth and invasion of human osteosarcoma cells via inhibition of Ki-67 and matrix metalloproteinases. Oncol. Lett. 2015, 9, 2171–2176. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Leng, H.; Chen, H.; Wang, L.; Jiang, N.; Huo, X.; Yu, B. Knockdown of UBE2T Inhibits Osteosarcoma Cell Proliferation, Migration, and Invasion by Suppressing the PI3K/Akt Signaling Pathway. Oncol. Res. 2016, 24, 361–369. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, C.C.; Zhang, H.P.; Li, G.Q.; Li, S.S. MLN4924 suppresses neddylation and induces cell cycle arrest, senescence, and apoptosis in human osteosarcoma. Oncotarget 2016, 7, 45263–45274. [Google Scholar] [CrossRef]
- Chen, S.; Yang, J.; Zhang, Y.; Duan, C.; Liu, Q.; Huang, Z.; Xu, Y.; Zhou, L.; Xu, G. Ubiquitin-conjugating enzyme UBE2O regulates cellular clock function by promoting the degradation of the transcription factor BMAL1. J. Biol. Chem. 2018, 293, 11296–11309. [Google Scholar] [CrossRef]
- He, M.; Zhou, Z.; Wu, G.; Chen, Q.; Wan, Y. Emerging role of DUBs in tumor metastasis and apoptosis: Therapeutic implication. Pharmacol. Ther. 2017, 177, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.A.; Maecker, H.L.; French, D.M.; Liu, J.; Gregg, A.; Silverstein, L.B.; Cao, T.C.; Carano, R.A.; Dixit, V.M. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 2011, 146, 918–930. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, H.; Zhong, N.; Jiang, Z.; Xu, L.; Deng, Y.; Jiang, Z.; Wang, H.; Wang, J. Gene silencing of USP1 by lentivirus effectively inhibits proliferation and invasion of human osteosarcoma cells. Int. J. Oncol. 2016, 49, 2549–2557. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Feng, Z.; Huang, H.; Wang, G.; Chen, Z.; Xu, G.; Xie, Z.; Jie, Z.; Zhao, X.; Ma, Q.; et al. USP1 inhibition suppresses the progression of osteosarcoma via destabilizing TAZ. Int. J. Biol. Sci. 2022, 18, 3122–3136. [Google Scholar] [CrossRef]
- Liang, Q.; Dexheimer, T.S.; Zhang, P.; Rosenthal, A.S.; Villamil, M.A.; You, C.; Zhang, Q.; Chen, J.; Ott, C.A.; Sun, H.; et al. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 2014, 10, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Xiong, M.; Dai, G.; Yu, L.; Zhang, Z.; Chen, J.; Guo, W. MicroRNA-192-5p suppresses the initiation and progression of osteosarcoma by targeting USP1. Oncol. Lett. 2018, 15, 6947–6956. [Google Scholar] [CrossRef] [PubMed]
- Sarri, N.; Wang, K.; Tsioumpekou, M.; Castillejo-Lopez, C.; Lennartsson, J.; Heldin, C.H.; Papadopoulos, N. Deubiquitinating enzymes USP4 and USP17 finetune the trafficking of PDGFRbeta and affect PDGF-BB-induced STAT3 signalling. Cell Mol. Life Sci. 2022, 79, 85. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Liu, W.; Li, J. USP17 is upregulated in osteosarcoma and promotes cell proliferation, metastasis, and epithelial-mesenchymal transition through stabilizing SMAD4. Tumour. Biol. 2017, 39, 1010428317717138. [Google Scholar] [CrossRef]
- Wang, Q.L.; Wang, L.; Li, Q.Y.; Li, H.Y.; Lin, L.; Wei, D.; Xu, J.Y.; Luo, X.J. Micafungin exerts antitumor effect on breast cancer and osteosarcoma through preventing EMT in tumor cells in an USP7/AKT/GSK-3beta pathway-dependent manner. Naunyn. Schmiedebergs. Arch. Pharmacol. 2023, 397, 4447–4459. [Google Scholar] [CrossRef]
- Yang, Z.; Yu, W.; Xu, A.; Liu, B.; Jin, L.; Tao, H.; Wang, D. mTORC1 accelerates osteosarcoma progression via m(6)A-dependent stabilization of USP7 mRNA. Cell Death Discov. 2024, 10, 127. [Google Scholar] [CrossRef]
- Zeng, Q.; Li, Z.; Zhao, X.; Guo, L.; Yu, C.; Qin, J.; Zhang, S.; Zhang, Y.; Yang, X. Ubiquitin-specific protease 7 promotes osteosarcoma cell metastasis by inducing epithelial-mesenchymal transition. Oncol. Rep. 2019, 41, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, X.; Cai, H.; Yang, W.; Lei, H.; Xu, H.; Wang, W.; Zhu, Q.; Kang, J.; Yin, T.; et al. Targeting USP9x/SOX2 axis contributes to the anti-osteosarcoma effect of neogambogic acid. Cancer Lett. 2020, 469, 277–286. [Google Scholar] [CrossRef]
- Zheng, W.; Li, S.; Huang, J.; Dong, Y.; Zhang, H.; Zheng, J. Down-Regulation of Ubiquitin-Specific Peptidase 9X Inhibited Proliferation, Migration and Invasion of Osteosarcoma via ERK1/2 and PI3K/Akt Signaling Pathways. Biol. Pharm. Bull. 2022, 45, 1283–1290. [Google Scholar] [CrossRef]
- Wiltshire, T.D.; Lovejoy, C.A.; Wang, T.; Xia, F.; O’Connor, M.J.; Cortez, D. Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J. Biol. Chem. 2010, 285, 14565–14571. [Google Scholar] [CrossRef]
- Stockum, A.; Snijders, A.P.; Maertens, G.N. USP11 deubiquitinates RAE1 and plays a key role in bipolar spindle formation. PLoS ONE 2018, 13, e0190513. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, D.; Liu, L.; Wang, L.; Yan, M. miR-140 inhibits osteosarcoma progression by impairing USP22-mediated LSD1 stabilization and promoting p21 expression. Mol. Ther. Nucleic. Acids. 2021, 24, 436–448. [Google Scholar] [CrossRef]
- Zhang, D.; Jiang, F.; Wang, X.; Li, G. Downregulation of Ubiquitin-Specific Protease 22 Inhibits Proliferation, Invasion, and Epithelial-Mesenchymal Transition in Osteosarcoma Cells. Oncol. Res. 2017, 25, 743–751. [Google Scholar] [CrossRef]
- Yadav, P.; Subbarayalu, P.; Medina, D.; Nirzhor, S.; Timilsina, S.; Rajamanickam, S.; Eedunuri, V.K.; Gupta, Y.; Zheng, S.; Abdelfattah, N.; et al. M6A RNA Methylation Regulates Histone Ubiquitination to Support Cancer Growth and Progression. Cancer Res. 2022, 82, 1872–1889. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Liu, J.; Zhang, H.; Li, J.; Shang, G. Long Intergenic Noncoding RNA00265 Enhances Cell Viability and Metastasis via Targeting miR-485-5p/USP22 Axis in Osteosarcoma. Front. Oncol. 2022, 12, 907472. [Google Scholar] [CrossRef] [PubMed]
- Gan, Z.; Han, K.; Lin, S.; Hu, H.; Shen, Z.; Min, D. Knockdown of ubiquitin-specific peptidase 39 inhibited the growth of osteosarcoma cells and induced apoptosis in vitro. Biol. Res. 2017, 50, 15. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, B.; Li, X.; Jin, W.; Han, C.; Wang, L.; Wang, H. MiR-1281, a p53-responsive microRNA, impairs the survival of human osteosarcoma cells upon ER stress via targeting USP39. Am. J. Cancer Res. 2018, 8, 1764–1774. [Google Scholar]
- Zhang, S.; Ding, L.; Gao, F.; Fan, H. Long non-coding RNA DSCAM-AS1 upregulates USP47 expression through sponging miR-101-3p to accelerate osteosarcoma progression. Biochem. Cell Biol. 2020, 98, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Herhaus, L.; Perez-Oliva, A.B.; Cozza, G.; Gourlay, R.; Weidlich, S.; Campbell, D.G.; Pinna, L.A.; Sapkota, G.P. Casein kinase 2 (CK2) phosphorylates the deubiquitylase OTUB1 at Ser16 to trigger its nuclear localization. Sci. Signal. 2015, 8, ra35. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Qiao, G.; Min, D.; Zhang, Z.; Lin, F.; Yang, Q.; Feng, T.; Tang, L.; Sun, Y.; Zhao, H.; et al. Heterogeneous expression and biological function of ubiquitin carboxy-terminal hydrolase-L1 in osteosarcoma. Cancer Lett. 2015, 359, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Sun, H.; Cheng, C.; Wang, G. BRCA1-Associated Protein-1 Suppresses Osteosarcoma Cell Proliferation and Migration Through Regulation PI3K/Akt Pathway. DNA Cell Biol. 2017, 36, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Chang, N.; Yoon, Y.; Yang, H.; Cho, H.; Kim, E.; Shin, Y.; Kang, W.; Oh, Y.T.; Mun, G.I.; et al. USP1 targeting impedes GBM growth by inhibiting stem cell maintenance and radioresistance. Neuro Oncol. 2016, 18, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Li, K.Y.; Su, Y.; Shen, Y.; Lei, M.Z.; Zhang, F.; Yin, M.; Chen, Z.J.; Wen, W.Y.; Hu, W.G.; et al. Diet high in branched-chain amino acid promotes PDAC development by USP1-mediated BCAT2 stabilization. Natl. Sci. Rev. 2022, 9, nwab212. [Google Scholar] [CrossRef]
- Lim, K.S.; Li, H.; Roberts, E.A.; Gaudiano, E.F.; Clairmont, C.; Sambel, L.A.; Ponnienselvan, K.; Liu, J.C.; Yang, C.; Kozono, D.; et al. USP1 Is Required for Replication Fork Protection in BRCA1-Deficient Tumors. Mol. Cell 2018, 72, 925–941.e924. [Google Scholar] [CrossRef]
- Soboleva, T.A.; Jans, D.A.; Johnson-Saliba, M.; Baker, R.T. Nuclear-cytoplasmic shuttling of the oncogenic mouse UNP/USP4 deubiquitylating enzyme. J. Biol. Chem. 2005, 280, 745–752. [Google Scholar] [CrossRef]
- Hu, B.; Zhang, D.; Zhao, K.; Wang, Y.; Pei, L.; Fu, Q.; Ma, X. Spotlight on USP4: Structure, Function, and Regulation. Front. Cell Dev. Biol. 2021, 9, 595159. [Google Scholar] [CrossRef]
- Ducker, C.; Shaw, P.E. USP17-mediated de-ubiquitination and cancer: Clients cluster around the cell cycle. Int. J. Biochem. Cell Biol. 2021, 130, 105886. [Google Scholar] [CrossRef]
- Mehic, M.; de Sa, V.K.; Hebestreit, S.; Heldin, C.H.; Heldin, P. The deubiquitinating enzymes USP4 and USP17 target hyaluronan synthase 2 and differentially affect its function. Oncogenesis 2017, 6, e348. [Google Scholar] [CrossRef] [PubMed]
- Lavaud, M.; Mullard, M.; Tesfaye, R.; Amiaud, J.; Legrand, M.; Danieau, G.; Brion, R.; Morice, S.; Regnier, L.; Dupuy, M.; et al. Overexpression of the Ubiquitin Specific Proteases USP43, USP41, USP27x and USP6 in Osteosarcoma Cell Lines: Inhibition of Osteosarcoma Tumor Growth and Lung Metastasis Development by the USP Antagonist PR619. Cells 2021, 10, 2268. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, M.; Jolly, L.A.; Gecz, J.; Wood, S.A. La FAM fatale: USP9X in development and disease. Cell Mol. Life Sci. 2015, 72, 2075–2089. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Yang, M.; Wang, K.; Wang, Y.; Zhong, B.; Jiang, N. Deubiquitinating enzyme OTUB1 in immunity and cancer: Good player or bad actor? Cancer Lett. 2022, 526, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Bishop, P.; Rocca, D.; Henley, J.M. Ubiquitin C-terminal hydrolase L1 (UCH-L1): Structure, distribution and roles in brain function and dysfunction. Biochem. J. 2016, 473, 2453–2462. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, S.; Shimada, K.; Honoki, K.; Kido, A.; Akahane, M.; Tanaka, Y.; Konishi, N. Ubiquilin 2 enhances osteosarcoma progression through resistance to hypoxic stress. Oncol. Rep. 2015, 33, 1799–1806. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, W.; Qiao, R.H.; Wang, D.M.; Huang, X.W.; Li, B.; Wang, D. UHRF1 promotes human osteosarcoma cell invasion by downregulating the expression of E-cadherin in an Rb1-dependent manner. Mol. Med. Rep. 2016, 13, 315–320. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Deng, X.; Deng, J.; Yi, X.; Zou, Y.; Liu, H.; Li, C.; Deng, B.; Fan, H.; Hao, L. Ubiquitin-like protein FAT10 promotes osteosarcoma glycolysis and growth by upregulating PFKFB3 via stabilization of EGFR. Am. J. Cancer Res. 2020, 10, 2066–2082. [Google Scholar]
- Ma, C.; Zhang, Z.; Cui, Y.; Yuan, H.; Wang, F. Silencing FAT10 inhibits metastasis of osteosarcoma. Int. J. Oncol. 2016, 49, 666–674. [Google Scholar] [CrossRef]
- Yi, X.; Deng, X.; Zhao, Y.; Deng, B.; Deng, J.; Fan, H.; Du, Y.; Hao, L. Ubiquitin-like protein FAT10 promotes osteosarcoma growth by modifying the ubiquitination and degradation of YAP1. Exp. Cell Res. 2020, 387, 111804. [Google Scholar] [CrossRef]
- Pei, H.; Chen, L.; Liao, Q.M.; Wang, K.J.; Chen, S.G.; Liu, Z.J.; Zhang, Z.C. SUMO-specific protease 2 (SENP2) functions as a tumor suppressor in osteosarcoma via SOX9 degradation. Exp. Ther. Med. 2018, 16, 5359–5365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, J.; Deng, Z.; Liu, H.; Mao, W.; Jiang, F.; Xia, Z.; Li, J.D. Circadian clock components RORalpha and Bmal1 mediate the anti-proliferative effect of MLN4924 in osteosarcoma cells. Oncotarget 2016, 7, 66087–66099. [Google Scholar] [CrossRef] [PubMed]
- Heessen, S.; Leonchiks, A.; Issaeva, N.; Sharipo, A.; Selivanova, G.; Masucci, M.G.; Dantuma, N.P. Functional p53 chimeras containing the Epstein-Barr virus Gly-Ala repeat are protected from Mdm2- and HPV-E6-induced proteolysis. Proc. Natl. Acad. Sci. USA 2002, 99, 1532–1537. [Google Scholar] [CrossRef] [PubMed]
- Rajendra, R.; Malegaonkar, D.; Pungaliya, P.; Marshall, H.; Rasheed, Z.; Brownell, J.; Liu, L.F.; Lutzker, S.; Saleem, A.; Rubin, E.H. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J. Biol. Chem. 2004, 279, 36440–36444. [Google Scholar] [CrossRef]
- Saini, H.; Sharma, H.; Mukherjee, S.; Chowdhury, S.; Chowdhury, R. Verteporfin disrupts multiple steps of autophagy and regulates p53 to sensitize osteosarcoma cells. Cancer Cell Int. 2021, 21, 52. [Google Scholar] [CrossRef]
- Fossey, S.L.; Bear, M.D.; Lin, J.; Li, C.; Schwartz, E.B.; Li, P.K.; Fuchs, J.R.; Fenger, J.; Kisseberth, W.C.; London, C.A. The novel curcumin analog FLLL32 decreases STAT3 DNA binding activity and expression, and induces apoptosis in osteosarcoma cell lines. BMC Cancer 2011, 11, 112. [Google Scholar] [CrossRef]
- Lei, Z.; Duan, H.; Zhao, T.; Zhang, Y.; Li, G.; Meng, J.; Zhang, S.; Yan, W. PARK2 inhibits osteosarcoma cell growth through the JAK2/STAT3/VEGF signaling pathway. Cell Death Dis. 2018, 9, 375. [Google Scholar] [CrossRef]
- Jiang, L.; Liao, J.; Liu, J.; Wei, Q.; Wang, Y. Geranylgeranylacetone promotes human osteosarcoma cell apoptosis by inducing the degradation of PRMT1 through the E3 ubiquitin ligase CHIP. J. Cell Mol. Med. 2021, 25, 7961–7972. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z.; Yang, X.; Lu, W.; Chen, Y.; Lin, Y.; Wang, J.; Lin, S.; Yun, J.P. H3K27 acetylation activated-COL6A1 promotes osteosarcoma lung metastasis by repressing STAT1 and activating pulmonary cancer-associated fibroblasts. Theranostics 2021, 11, 1473–1492. [Google Scholar] [CrossRef]
- Wan, Z.; Huang, S.; Mo, F.; Yao, Y.; Liu, G.; Han, Z.; Chen, M.; Zhiyun, L. CSN5 controls the growth of osteosarcoma via modulating the EGFR/PI3K/Akt axis. Exp. Cell Res. 2019, 384, 111646. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wu, W.; Deng, S.; Shao, Z.; Jin, X. TRAIP modulates the IGFBP3/AKT pathway to enhance the invasion and proliferation of osteosarcoma by promoting KANK1 degradation. Cell Death Dis. 2021, 12, 767. [Google Scholar] [CrossRef]
- Li, X.; Zhong, L.; Wang, Z.; Chen, H.; Liao, D.; Zhang, R.; Zhang, H.; Kang, T. Phosphorylation of IRS4 by CK1gamma2 promotes its degradation by CHIP through the ubiquitin/lysosome pathway. Theranostics 2018, 8, 3643–3653. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, S.R.; Kishi, C.; Ishihara, N.; Mizushima, N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J. Biol. Chem. 2011, 286, 19630–19640. [Google Scholar] [CrossRef]
- Bartek, J.; Hodny, Z. PARK2 orchestrates cyclins to avoid cancer. Nat. Genet. 2014, 46, 527–528. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Wang, H.; Wang, X.; Zhao, Y.; Zhao, D.; Wang, C.; Li, Y.; Yang, Z.; Lu, S.; Zeng, Q.; et al. Molecular mechanisms of Polyphyllin I-induced apoptosis and reversal of the epithelial-mesenchymal transition in human osteosarcoma cells. J. Ethnopharmacol. 2015, 170, 117–127. [Google Scholar] [CrossRef]
- Vanarotti, M.; Grace, C.R.; Miller, D.J.; Actis, M.L.; Inoue, A.; Evison, B.J.; Vaithiyalingam, S.; Singh, A.P.; McDonald, E.T.; Fujii, N. Structures of REV1 UBM2 Domain Complex with Ubiquitin and with a Small-Molecule that Inhibits the REV1 UBM2-Ubiquitin Interaction. J. Mol. Biol. 2018, 430, 2857–2872. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.E.; Xu, H.Y.; Hao, J.; Huo, W.W.; Qian, Y.; Hou, B.L. The Ubiquitination of Spinal MrgC Alleviates Bone Cancer Pain and Reduces Intracellular Calcium Concentration in Spinal Neurons in Mice. Neurochem. Res. 2019, 44, 2527–2535. [Google Scholar] [CrossRef]
- Campanero, M.R.; Flemington, E.K. Regulation of E2F through ubiquitin-proteasome-dependent degradation: Stabilization by the pRB tumor suppressor protein. Proc. Natl. Acad. Sci. USA 1997, 94, 2221–2226. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, Q.; Zhang, N.; Li, W.; Ying, M.; Ding, W.; Yang, B.; He, Q. E2F1 impairs all-trans retinoic acid-induced osteogenic differentiation of osteosarcoma via promoting ubiquitination-mediated degradation of RARalpha. Cell Cycle 2014, 13, 1277–1287. [Google Scholar] [CrossRef]
- Luo, J.; Xia, Y.; Luo, J.; Li, J.; Zhang, C.; Zhang, H.; Ma, T.; Yang, L.; Kong, L. GRP78 inhibition enhances ATF4-induced cell death by the deubiquitination and stabilization of CHOP in human osteosarcoma. Cancer Lett. 2017, 410, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Jing, J.; Hu, X.; Yu, S.; Yao, F.; Li, Z.; Cheng, L. Gankyrin activates the hedgehog signalling to drive metastasis in osteosarcoma. J. Cell Mol. Med. 2021, 25, 6232–6241. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, D.; Qin, P.; Zhang, J.; Cui, X.; Gao, J.; Wang, J.; Li, J. Long non-coding RNA EPIC1 inhibits viability and invasion of osteosarcoma cells by promoting MEF2D ubiquitylation. Int. J. Biol. Macromol. 2019, 128, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Wagle, S.; Park, S.H.; Kim, K.M.; Moon, Y.J.; Bae, J.S.; Kwon, K.S.; Park, H.S.; Lee, H.; Moon, W.S.; Kim, J.R.; et al. DBC1/CCAR2 is involved in the stabilization of androgen receptor and the progression of osteosarcoma. Sci. Rep. 2015, 5, 13144. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Qin, P.; Zhang, D.; Cui, X.; Gao, J.; Yu, Z.; Chai, Y.; Wang, J.; Li, J. Long non-coding RNA PVT1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing ERG and sponging miR-183-5p. Aging 2019, 11, 9581–9596. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Yi, X.; Deng, J.; Zou, Y.; Wang, S.; Shan, W.; Liu, P.; Zhang, Z.; Chen, L.; Hao, L. ROCK2 promotes osteosarcoma growth and metastasis by modifying PFKFB3 ubiquitination and degradation. Exp. Cell Res. 2019, 385, 111689. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Yi, X.; Huang, D.; Liu, P.; Chen, L.; Du, Y.; Hao, L. ROCK2 mediates osteosarcoma progression and TRAIL resistance by modulating O-GlcNAc transferase degradation. Am. J. Cancer Res. 2020, 10, 781–798. [Google Scholar] [PubMed]
- Cao, K.; Wang, H.; Fang, Y.; Wang, Y.; Wei, L.; Chen, X.; Jiang, Z.; Wei, X.; Hu, Y. Histone Deacetylase 4 Promotes Osteosarcoma Cell Proliferation and Invasion by Regulating Expression of Proliferating Cell Nuclear Antigen. Front. Oncol. 2019, 9, 870. [Google Scholar] [CrossRef]
- Kim, K.M.; Hussein, U.K.; Park, S.H.; Kang, M.A.; Moon, Y.J.; Zhang, Z.; Song, Y.; Park, H.S.; Bae, J.S.; Park, B.H.; et al. FAM83H is involved in stabilization of beta-catenin and progression of osteosarcomas. J. Exp. Clin. Cancer Res. 2019, 38, 267. [Google Scholar] [CrossRef]
- Zhang, Q.; Yin, X.; Zhang, Y. MicroRNA-221 Promotes Cell Proliferation and Inhibits Apoptosis in Osteosarcoma Cells by Directly Targeting FBXW11 and Regulating Wnt Signaling. Arch. Med. Res. 2021, 52, 191–199. [Google Scholar] [CrossRef]
- Shen, S.; Yao, T.; Xu, Y.; Zhang, D.; Fan, S.; Ma, J. CircECE1 activates energy metabolism in osteosarcoma by stabilizing c-Myc. Mol. Cancer 2020, 19, 151. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Tan, Y.; Zang, C.; Zhao, F.; Cai, C.; Kong, L.; Deng, H.; Chao, F.; Xia, R.; Xie, M.; et al. LAMTOR5-AS1 regulates chemotherapy-induced oxidative stress by controlling the expression level and transcriptional activity of NRF2 in osteosarcoma cells. Cell Death Dis. 2021, 12, 1125. [Google Scholar] [CrossRef] [PubMed]
- Anninga, J.K.; Gelderblom, H.; Fiocco, M.; Kroep, J.R.; Taminiau, A.H.; Hogendoorn, P.C.; Egeler, R.M. Chemotherapeutic adjuvant treatment for osteosarcoma: Where do we stand? Eur. J. Cancer 2011, 47, 2431–2445. [Google Scholar] [CrossRef] [PubMed]
- Meyers, P.A. Systemic therapy for osteosarcoma and Ewing sarcoma. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, e644–e647. [Google Scholar] [CrossRef]
- Marina, N.M.; Smeland, S.; Bielack, S.S.; Bernstein, M.; Jovic, G.; Krailo, M.D.; Hook, J.M.; Arndt, C.; van den Berg, H.; Brennan, B.; et al. Comparison of MAPIE versus MAP in patients with a poor response to preoperative chemotherapy for newly diagnosed high-grade osteosarcoma (EURAMOS-1): An open-label, international, randomised controlled trial. Lancet Oncol. 2016, 17, 1396–1408. [Google Scholar] [CrossRef] [PubMed]
- Corre, I.; Verrecchia, F.; Crenn, V.; Redini, F.; Trichet, V. The Osteosarcoma Microenvironment: A Complex But Targetable Ecosystem. Cells 2020, 9, 976. [Google Scholar] [CrossRef]
- Hu, Z.; Wen, S.; Huo, Z.; Wang, Q.; Zhao, J.; Wang, Z.; Chen, Y.; Zhang, L.; Zhou, F.; Guo, Z.; et al. Current Status and Prospects of Targeted Therapy for Osteosarcoma. Cells 2022, 11, 3507. [Google Scholar] [CrossRef]
| DUBs | Participating Process | Regulatory Axis |
|---|---|---|
| USP1 | Maintaining osteosarcoma stem cell state [82]; Promoting osteosarcoma growth and invasion [83,84]; Involved in DNA damage response [85]. | USP1/IDs [82]; USP1/TAZ/Hippo pathway [84]; miR-192-5p/USP1 [86]; USP1-UAF1 [85]. |
| USP4/USP17 | Promoting osteosarcoma cell proliferation, migration, and invasion [87,88]. | (USP4/USP17)/PDGFRβ/STAT3 pathway [87]; USP17/SMAD4 [88]. |
| USP7 | Promoting osteosarcoma cell proliferation, migration, and invasion [89,90,91]. | USP7/Wnt-β-catenin pathway [91]; USP7/AKT/GSK-3β/EMT [89]; E2/mTORC1/USP7/NLRP3 pathway [90]. |
| USP9X | Promoting osteosarcoma cell proliferation, migration, and invasion [92,93]. | USP9X-SOX2 [92]; USP9X/ERK1/2 [93]; USP9X/PI3K/Akt pathway [93]. |
| USP11 | Involved in DNA damage response, promoting HR repair [94]; Involved in bipolar spindle formation [95]. | USP11/RAE1 [95]. |
| USP22 | Promoting osteosarcoma cell proliferation, migration, and invasion [96,97]. | USP22/PI3K/Akt pathway [97]; USP22/EMT [97]; USP22/LSD1 [96]; MiR-140/USP22 [96] ALKBH5/USP22(m6A) [98]; linc00265/miR-485-5p/USP22 [99]. |
| USP39 | Promoting apoptosis in osteosarcoma cells and participating in cell cycle regulation [100,101]. | USP39/p21 [100]; USP39/PARP [100]; miR-1281/USP39/ER stress [101]. |
| USP47 | Promoting osteosarcoma cell proliferation, migration, and invasion [102]. | USP47/Wnt-β-catenin pathway [102]; USP47/AKT-mTOR pathway [102]; DSCAM-AS1/miR-101-3p/USP47 [102]. |
| OTUB1 | Involved in DNA damage repair [103]. | CK2/OTUB1/53BP1 [103]. |
| UCHL1 | Promoting osteosarcoma cell proliferation and invasion, while inhibiting apoptosis [104]. | UCHL1/p-Akt [104]; UCHL1/p-ERK [104]. |
| BAP1 | Inhibiting proliferation, migration, and invasion of osteosarcoma cells [105]. | BAP1/PI3K-Akt pathway [105]; miR-125/BAP1 [105]. |
| Pathways | Molecules and Drugs | Regulation of Pathways by Molecules or Drugs | References |
|---|---|---|---|
| P53 pathway | MDM2 | Negative regulation. | [42] |
| P53 chimeras | Positive regulation. | [124] | |
| Topors | Negative regulation. | [125] | |
| VP+MG132 | Negative regulation. | [126] | |
| STAT pathway | USP4/USP17 | Positive regulation (STAT3). | [87] |
| FLLL32 | Negative regulation (STAT3). | [127] | |
| PARK2 | Negative regulation (STAT3). | [128] | |
| GGA | Negative regulation (STAT3). | [129] | |
| COL6A1 | Negative regulation (STAT1). | [130] | |
| PI3K/Akt pathway | UBE2T | Negative regulation. | [78] |
| SPOP | Negative regulation. | [53] | |
| USP9X | Downregulation of USP9X inhibits the activation of the PI3K/Akt signaling pathway. | [93] | |
| USP22 | Downregulation of USP22 inhibits the activation of the PI3K/Akt signaling pathway. | [97] | |
| BAP1 | Negative regulation. | [105] | |
| CSN5 | Positive regulation. | [131] | |
| TRAIP | Positive regulation. | [132] | |
| IRS4 | Positive regulation. | [133] | |
| Wnt-β-catenin pathway | SIAH1 | Positive regulation. | [63] |
| USP7 | Positive regulation. | [91] | |
| USP47 | Downregulation of USP47 results in the inactivation of the Wnt/β-catenin signaling pathway. | [102] |
| Molecules | Processes | Mechanism | References |
|---|---|---|---|
| pRB | Regulation of the cell cycle. | Preventing the degradation of transcription factor E2F1 via the ubiquitin–proteasome pathway. | [139] |
| E2F1 | Regulation of cell differentiation. | Specifically binding to RARα, promoting its ubiquitin-mediated degradation. | [140] |
| GRP78 | Regulation of endoplasmic reticulum stress. | Directly interacting with the N-terminal domain of CHOP and promoting CHOP ubiquitination in a p300-dependent manner. | [141] |
| Gankyrin | Promoting migration and invasion. | “Interacting with Gli1 to inhibit ubiquitin-proteasome-mediated degradation of Gli1 and regulating the expression of stemness factors.” | [142] |
| EPIC1 | Inhibiting invasion. | A long non-coding RNA that negatively regulates the expression of MEF2D protein by increasing its ubiquitination levels. | [143] |
| DBC1 and AR | Involved in proliferation and proliferation-related signaling pathways. | DBC1 negatively regulates AR protein expression by inhibiting AR polyubiquitination–proteasome pathway degradation. | [144] |
| PVT1 | Promoting proliferation and migration. | By inhibiting the ubiquitination of ERG and competitively binding with miR-183-5p, leading to upregulation of ERG expression. | [145] |
| ROCK2 | Promoting proliferation, migration, and invasion; Participating in TRAIL resistance and inhibiting apoptosis. | ROCK2 interacts directly with PFKFB3, reducing the ubiquitination level of PFKFB3 and increasing its stability; ROCK2 inhibits TRAIL activation and osteosarcoma cell apoptosis by ubiquitin–proteasome degradation of OGT. | [146,147] |
| HDAC4 and PCNA | Promoting proliferation and invasion while inhibiting apoptosis. | HDAC4 directly binds to PCNA and inhibits PCNA ubiquitination–proteasome degradation, positively regulating PCNA protein expression. | [148] |
| FAM83H | Promoting proliferation and invasive capacity. | FAM83H inhibits the ubiquitination–proteasome degradation of β-catenin by interacting with it. | [149] |
| miR-221 | Promoting proliferation and inhibiting cell apoptosis. | Inhibiting the Wnt signaling pathway by targeting the ubiquitin–proteasome system component FBXW11. | [150] |
| CircECE1 | Promoting proliferation and metastasis. | Interacting with c-Myc, preventing SPOP-mediated c-Myc ubiquitination and degradation, thereby activating the Warburg effect through TXNIP transcription. | [151] |
| LAMTOR5-AS1 | Inhibiting proliferation and multidrug resistance | Mediating the interaction between NRF2 and KEAP1 inhibits the ubiquitination degradation of NRF2, upregulating the expression of HO-1. | [152] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, P.; Feng, Z.; Liu, Y.; Zhang, K.; Zhao, G.; Lei, Z.; Di, T.; Zhang, H. The Role of Ubiquitination in Osteosarcoma Development and Therapies. Biomolecules 2024, 14, 791. https://doi.org/10.3390/biom14070791
Mao P, Feng Z, Liu Y, Zhang K, Zhao G, Lei Z, Di T, Zhang H. The Role of Ubiquitination in Osteosarcoma Development and Therapies. Biomolecules. 2024; 14(7):791. https://doi.org/10.3390/biom14070791
Chicago/Turabian StyleMao, Peng, Zuxi Feng, Yong Liu, Kai Zhang, Guanghai Zhao, Zeyuan Lei, Tianning Di, and Haihong Zhang. 2024. "The Role of Ubiquitination in Osteosarcoma Development and Therapies" Biomolecules 14, no. 7: 791. https://doi.org/10.3390/biom14070791
APA StyleMao, P., Feng, Z., Liu, Y., Zhang, K., Zhao, G., Lei, Z., Di, T., & Zhang, H. (2024). The Role of Ubiquitination in Osteosarcoma Development and Therapies. Biomolecules, 14(7), 791. https://doi.org/10.3390/biom14070791






