Pulsed Dipolar EPR for Self-Limited Complexes of Oligonucleotides Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Oligonucleotide Spin Labeling
2.3. Gel Electrophoresis
2.4. Oligonucleotide Complexes Melting Analysis
2.5. EPR Sample Preparation
2.6. CW EPR
2.7. PELDOR Measurements
2.8. Molecular Dynamics
3. Results and Discussion
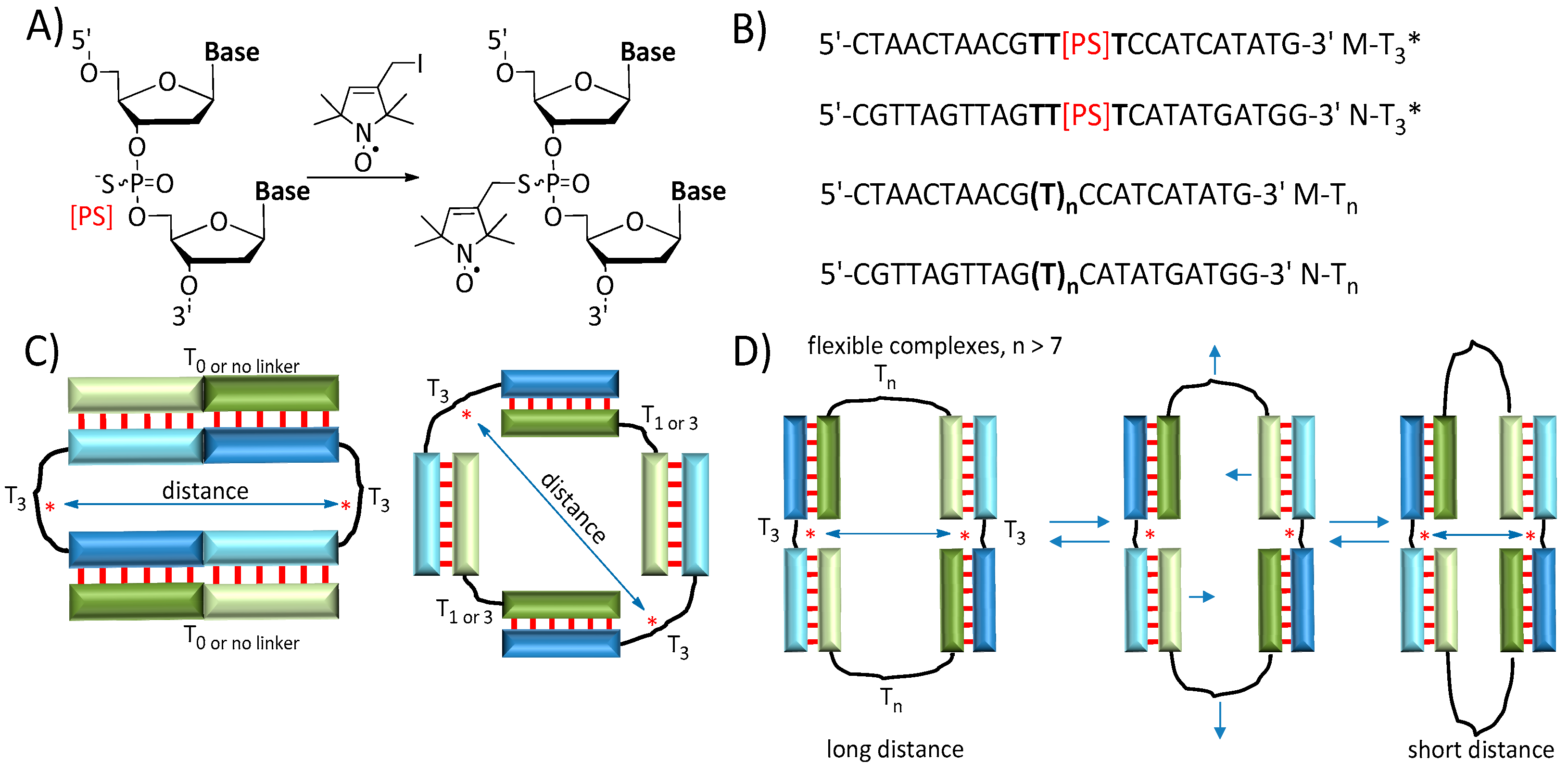
3.1. Oligonucleotide Synthesis
3.2. Electrophoretic and Thermal Denaturation Analysis of Self-Assembled Complexes of Oligonucleotides
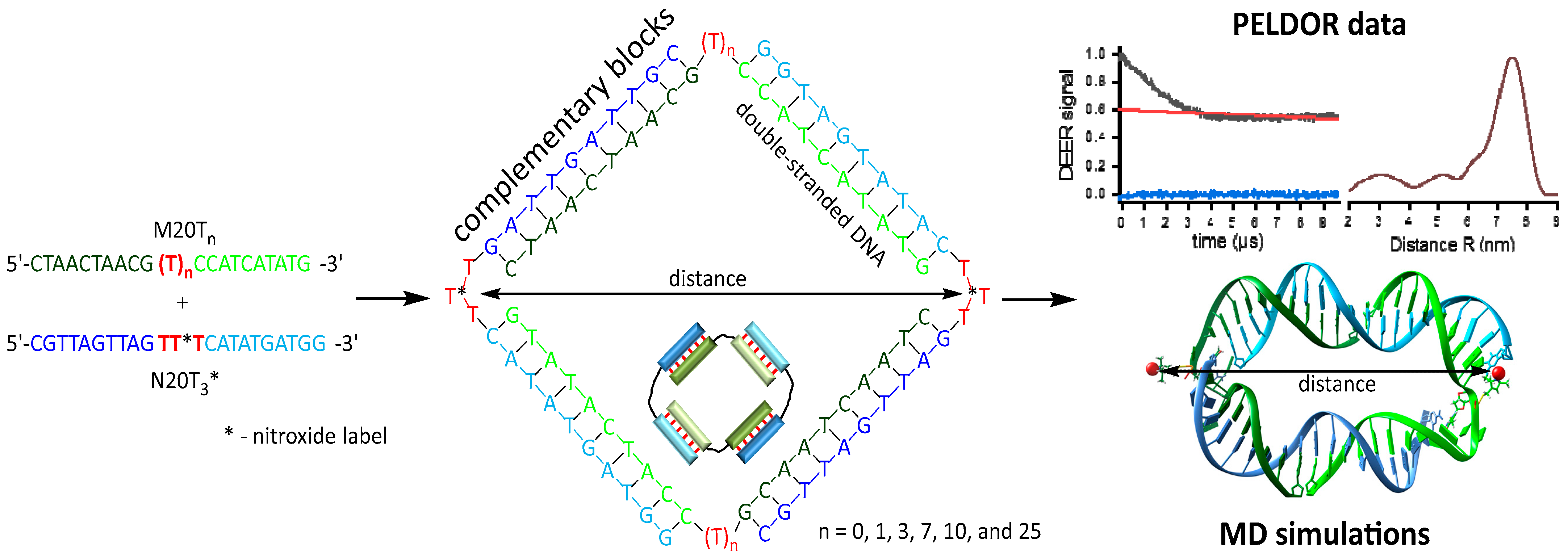
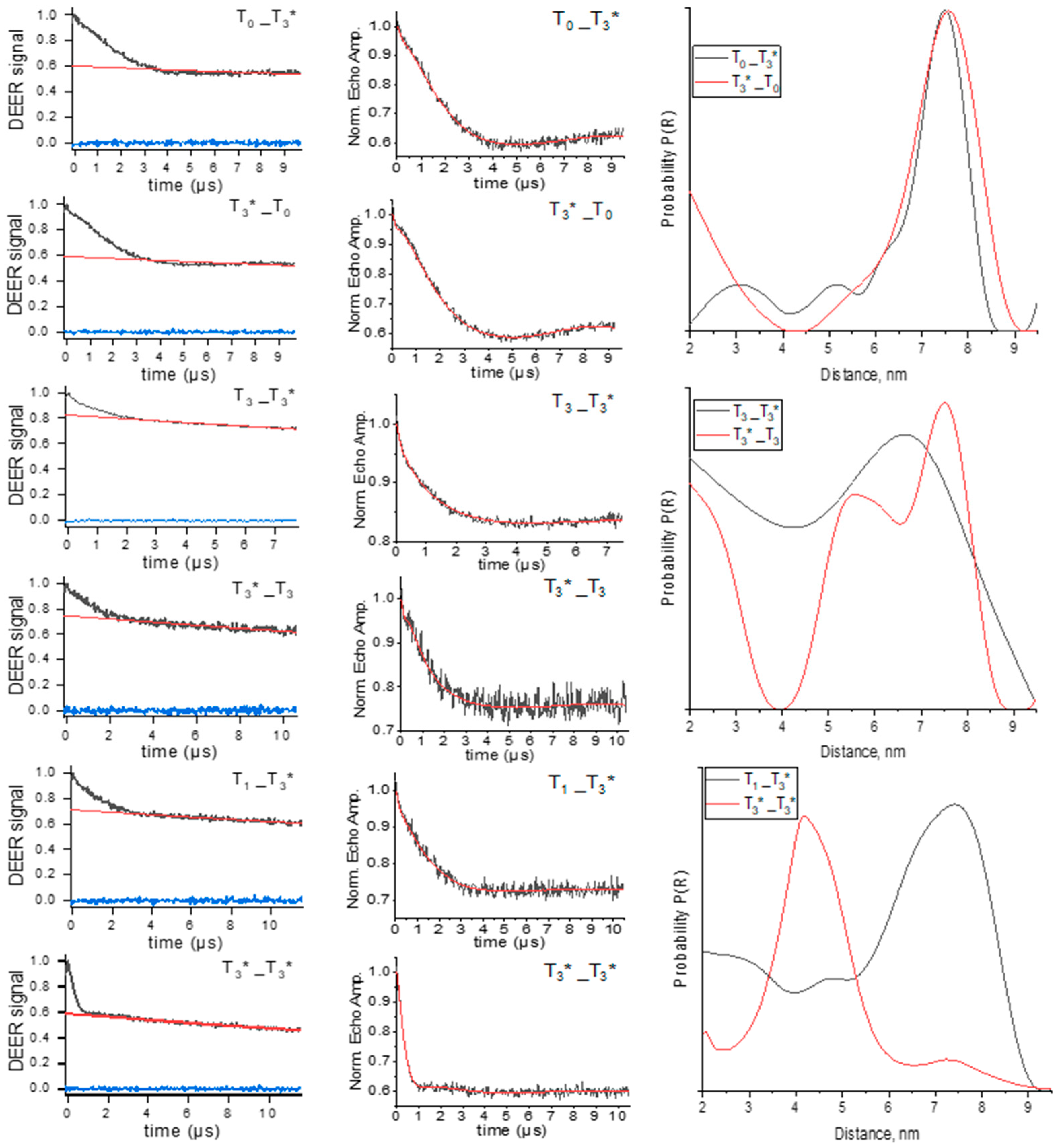
3.3. Distance Measurements by PELDOR
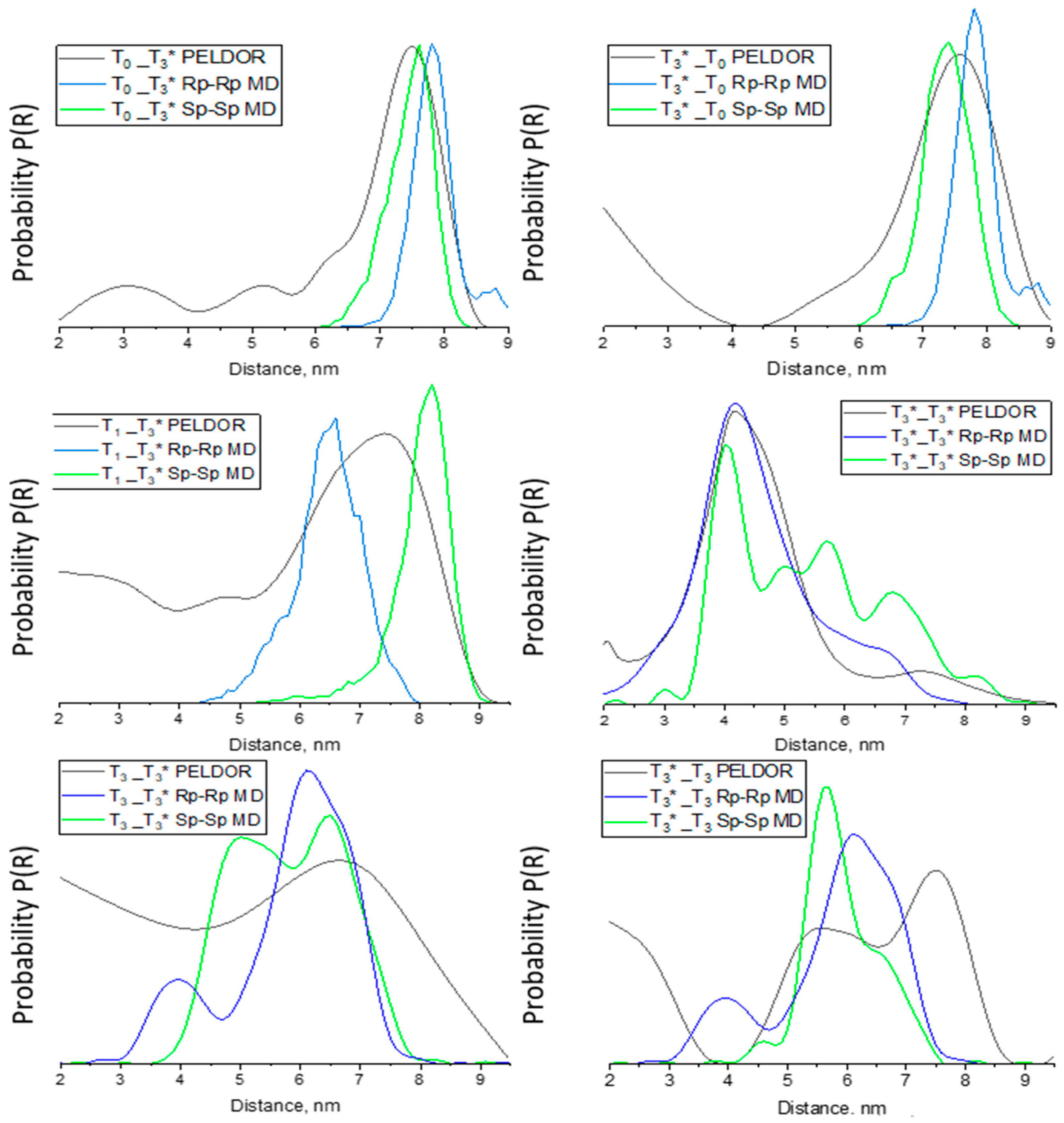
3.4. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, I.; Barinov, N.; Novikov, R.; Severov, V.; Iudin, M.; Vedekhina, T.; Larin, A.; Babenko, V.; Aralov, A.; Gnuchikh, E.; et al. Modeling G4s in chromatin context confirms partial nucleosome exclusion and reveals nucleosome-disrupting effects of the least selective G4 ligands. Biochimie 2023, 204, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Nikitin, M.P. Non-complementary strand commutation as a fundamental alternative for information processing by DNA and gene regulation. Nat. Chem. 2023, 15, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Pavc, D.; Sebastian, N.; Spindler, L.; Drevenšek-Olenik, I.; Podboršek, G.K.; Plavec, J.; Šket, P. Understanding self-assembly at molecular level enables controlled design of DNA G-wires of different properties. Nat. Commun. 2022, 13, 1062. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Narayana, A.; Patel, S.; Sahay, G. Advances in intracellular delivery through supramolecular self-assembly of oligonucleotides and peptides. Theranostics 2019, 9, 3191–3212. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; König, A.; Ghosh, A.; Ghosh, D.; Benabou, S.; Rosu, F.; Gabelica, V. Mass Spectrometry of Nucleic Acid Noncovalent Complexes. Chem. Rev. 2022, 122, 7720–7839. [Google Scholar] [CrossRef]
- Liu, K.; Zheng, L.; Ma, C.; Göstl, R.; Herrmann, A. DNA-surfactant complexes: Self-assembly properties and applications. Chem. Soc. Rev. 2017, 46, 5147–5172. [Google Scholar] [CrossRef] [PubMed]
- Zeraati, M.; Langley, D.B.; Schofield, P.; Moye, A.L.; Rouet, R.; Hughes, W.E.; Bryan, T.M.; Dinger, M.E.; Christ, D. I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 2018, 10, 631–637. [Google Scholar] [CrossRef]
- Endeward, B.; Hu, Y.; Bai, G.; Liu, G.; Prisner, T.F.; Fang, X. Long-range distance determination in fully deuterated RNA with pulsed EPR spectroscopy. Biophys. J. 2022, 121, 37–43. [Google Scholar] [CrossRef]
- Spitale, R.C.; Incarnato, D. Probing the dynamic RNA structurome and its functions. Nat. Rev. Genet. 2023, 24, 178–196. [Google Scholar] [CrossRef]
- Collauto, A.; von Bülow, S.; Gophane, D.B.; Saha, S.; Stelzl, L.S.; Hummer, G.; Sigurdsson, S.T.; Prisner, T.F. Compaction of RNA Duplexes in the Cell. Angew. Chemie Int. Ed. 2020, 59, 23025–23029. [Google Scholar] [CrossRef] [PubMed]
- Kristoffersen, E.L.; Coletta, A.; Lund, L.M.; Schiøtt, B.; Birkedal, V. Inhibited complete folding of consecutive human telomeric G-quadruplexes. Nucleic Acids Res. 2023, 51, 1571–1582. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Fan, C.; Gothelf, K.V.; Li, J.; Lin, C.; Liu, L.; Liu, N.; Nijenhuis, M.A.D.; Saccà, B.; Simmel, F.C.; et al. DNA origami. Nat. Rev. Methods Prim. 2021, 1, 13. [Google Scholar] [CrossRef]
- Pan, Q.; Nie, C.; Hu, Y.; Yi, J.; Liu, C.; Zhang, J.; He, M.; He, M.; Chen, T.; Chu, X. Aptamer-Functionalized DNA Origami for Targeted Codelivery of Antisense Oligonucleotides and Doxorubicin to Enhance Therapy in Drug-Resistant Cancer Cells. ACS Appl. Mater. Interfaces 2020, 12, 400–409. [Google Scholar] [CrossRef]
- Ryu, Y.; Hong, C.A.; Song, Y.; Beak, J.; Seo, B.A.; Lee, J.J.; Kim, H.S. Modular protein-DNA hybrid nanostructures as a drug delivery platform. Nanoscale 2020, 12, 4975–4981. [Google Scholar] [CrossRef]
- Vázquez-gonzález, M.; Willner, I. Aptamer-functionalized hybrid nanostructures for sensing, drug delivery, catalysis and mechanical applications. Int. J. Mol. Sci. 2021, 22, 1803. [Google Scholar] [CrossRef]
- Li, M.; Zheng, M.; Wu, S.; Tian, C.; Liu, D.; Weizmann, Y.; Jiang, W.; Wang, G.; Mao, C. In vivo production of RNA nanostructures via programmed folding of single-stranded RNAs. Nat. Commun. 2018, 9, 2196. [Google Scholar] [CrossRef] [PubMed]
- Qu, N.; Ying, Y.; Qin, J.; Chen, A.K. Rational design of self-assembled RNA nanostructures for HIV-1 virus assembly blockade. Nucleic Acids Res. 2022, 50, E44. [Google Scholar] [CrossRef]
- van Dyck, J.F.; Burns, J.R.; Le Huray, K.I.P.; Konijnenberg, A.; Howorka, S.; Sobott, F. Sizing up DNA nanostructure assembly with native mass spectrometry and ion mobility. Nat. Commun. 2022, 13, 3610. [Google Scholar] [CrossRef]
- Shi, X.; Pumm, A.; Maffeo, C.; Kohler, F.; Feigl, E.; Zhao, W.; Verschueren, D.; Golestanian, R.; Aksimentiev, A.; Dietz, H.; et al. A DNA turbine powered by a transmembrane potential across a nanopore. Nat. Nanotechnol. 2023, 19, 338–344. [Google Scholar] [CrossRef]
- Rothenbühler, S.; Iacovache, I.; Langenegger, S.M.; Zuber, B.; Häner, R. Complex DNA Architectonics─Self-Assembly of Amphiphilic Oligonucleotides into Ribbons, Vesicles, and Asterosomes. Bioconjug. Chem. 2023, 34, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Sontakke, V.A.; Yokobayashi, Y. Programmable Macroscopic Self-Assembly of DNA-Decorated Hydrogels. J. Am. Chem. Soc. 2022, 144, 2149–2155. [Google Scholar] [CrossRef]
- Zamoskovtseva, A.A.; Golyshev, V.M.; Kizilova, V.A.; Shevelev, G.Y.; Pyshnyi, D.V.; Lomzov, A.A. Pairing nanoarchitectonics of oligodeoxyribonucleotides with complex diversity: Concatemers and self-limited complexes. RSC Adv. 2022, 12, 6416–6431. [Google Scholar] [CrossRef] [PubMed]
- Stelzl, L.S.; Erlenbach, N.; Heinz, M.; Prisner, T.F.; Hummer, G. Resolving the Conformational Dynamics of DNA with Ångstrom Resolution by Pulsed Electron-Electron Double Resonance and Molecular Dynamics. J. Am. Chem. Soc. 2017, 139, 11674–11677. [Google Scholar] [CrossRef]
- Ketter, S.; Joseph, B. Gd3+-Trityl-Nitroxide Triple Labeling and Distance Measurements in the Heterooligomeric Cobalamin Transport Complex in the Native Lipid Bilayers. J. Am. Chem. Soc. 2023, 145, 960–966. [Google Scholar] [CrossRef]
- Krumkacheva, O.A.; Timofeev, I.O.; Politanskaya, L.V.; Polienko, Y.F.; Tretyakov, E.V.; Rogozhnikova, O.Y.; Trukhin, D.V.; Tormyshev, V.M.; Chubarov, A.S.; Bagryanskaya, E.G.; et al. Triplet Fullerenes as Prospective Spin Labels for Nanoscale Distance Measurements by Pulsed Dipolar EPR. Angew. Chemie Int. Ed. 2019, 58, 13271–13275. [Google Scholar] [CrossRef] [PubMed]
- Ketter, S.; Dajka, M.; Rogozhnikova, O.; Dobrynin, S.A.; Tormyshev, V.M.; Bagryanskaya, E.G.; Joseph, B. In situ distance measurements in a membrane transporter using maleimide functionalized orthogonal spin labels and 5-pulse electron double resonance spectroscopy. bioRxiv 2022. [Google Scholar] [CrossRef]
- Bulygin, K.N.; Timofeev, I.O.; Malygin, A.A.; Graifer, D.M.; Meschaninova, M.I.; Venyaminova, A.G.; Krumkacheva, O.A.; Fedin, M.V.; Frolova, Y.L.; Karpova, G.G.; et al. Two alternative conformations of mRNA in the human ribosome during elongation and termination of translation as revealed by EPR spectroscopy. Comput. Struct. Biotechnol. J. 2021, 19, 4702–4710. [Google Scholar] [CrossRef]
- Peter, M.F.; Gebhardt, C.; Mächtel, R.; Muñoz, G.G.M.; Glaenzer, J.; Narducci, A.; Thomas, G.H.; Cordes, T.; Hagelueken, G. Cross-validation of distance measurements in proteins by PELDOR/DEER and single-molecule FRET. Nat. Commun. 2022, 13, 4396. [Google Scholar] [CrossRef]
- Gopinath, A.; Joseph, B. Conformational Flexibility of the Protein Insertase BamA in the Native Asymmetric Bilayer Elucidated by ESR Spectroscopy. Angew. Chem. Int. Ed. 2022, 61, e202113448. [Google Scholar] [CrossRef]
- Sannikova, N.E.; Kolokolov, M.I.; Khlynova, T.A.; Chubarov, A.S.; Polienko, Y.F.; Fedin, M.V.; Krumkacheva, O.A. Revealing Light-Induced Structural Shifts in G-Quadruplex-Porphyrin Complexes: A Pulsed Dipolar EPR Study. Phys. Chem. Chem. Phys. 2023, 25, 22455–22466. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, G. DEER Distance Measurements on Proteins. Annu. Rev. Phys. Chem. 2012, 63, 419–446. [Google Scholar] [CrossRef] [PubMed]
- Depmeier, H.; Hoffmann, E.; Bornewasser, L.; Kath-Schorr, S. Strategies for Covalent Labeling of Long RNAs. ChemBioChem 2021, 22, 2826–2847. [Google Scholar] [CrossRef] [PubMed]
- Torricella, F.; Pierro, A.; Mileo, E.; Belle, V.; Bonucci, A. Nitroxide spin labels and EPR spectroscopy: A powerful association for protein dynamics studies. Biochim. Biophys. Acta-Proteins Proteom. 2021, 1869, 140653. [Google Scholar] [CrossRef] [PubMed]
- Saran, R.; Huang, Z.; Liu, J. Phosphorothioate nucleic acids for probing metal binding, biosensing and nanotechnology. Coord. Chem. Rev. 2021, 428, 213624. [Google Scholar] [CrossRef]
- Qin, P.Z.; Haworth, I.S.; Cai, Q.; Kusnetzow, A.K.; Grant, G.P.; Price, E.A.; Sowa, G.Z.; Popova, A.; Herreros, B.; He, H. Measuring nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nat. Protoc. 2007, 2, 2354–2365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, C.X.; Di Felice, R.; Sponer, J.; Islam, B.; Stadlbauer, P.; Ding, Y.; Mao, L.; Mao, Z.W.; Qin, P.Z. Conformations of Human Telomeric G-Quadruplex Studied Using a Nucleotide-Independent Nitroxide Label. Biochemistry 2016, 55, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Hankovszky, H.O.; Hideg, K.; Lex, L. Nitroxyls; VII1. Synthesis and Reactions of Highly Reactive 1-Oxyl-2,2,5,5-tetramethyl-2,5-dihydropyrrole-3-ylmethyl Sulfonates. Synthesis 1980, 1980, 914–916. [Google Scholar] [CrossRef]
- Pannier, M.; Veit, S.; Godt, A.; Jeschke, G.; Spiess, H.W. Dead-Time Free Measurement of Dipole—Dipole Interactions between Electron Spins. J. Magn. Reson. 2000, 142, 331–340. [Google Scholar] [CrossRef]
- Schmidt, R.R.; Martin, R.E.; Pannier, M.; Diederich, F.; Gramlich, V. Determination of End-to-End Distances in a Series of TEMPO Diradicals of up to 2.8 nm Length with a New Four-Pulse Double Electron Electron Resonance Experiment. Angew. Chemie Int. Ed. 1998, 37, 2833–2837. [Google Scholar]
- Jeschke, G.; Chechik, V.; Ionita, P.; Godt, A.; Zimmermann, H.; Banham, J.; Timmel, C.R.; Hilger, D.; Jung, H. DeerAnalysis2006—A comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006, 30, 473–498. [Google Scholar] [CrossRef]
- Russell, H.; Cura, R.; Lovett, J.E. DEER Data Analysis Software: A Comparative Guide. Front. Mol. Biosci. 2022, 9, 915167. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cerutti, D.S.; Cheatham, T.E.; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Greene, D.; Homeyer, N.; et al. AMBER 14; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Dupradeau, F.-Y.; Pigache, A.; Zaffran, T.; Savineau, C.; Lelong, R.; Grivel, N.; Lelong, D.; Rosanski, W.; Cieplak, C. The R.E.D. tools: Advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 2010, 77, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Ivani, I.; Dans, P.D.; Noy, A.; Pérez, A.; Faustino, I.; Hospital, A.; Walther, J.; Andrio, P.; Goñi, R.; Balaceanu, A.; et al. Parmbsc1: A refined force field for DNA simulations. Nat. Methods 2016, 13, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Joung, I.S.; Cheatham, T.E. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B 2008, 112, 9020–9041. [Google Scholar] [CrossRef] [PubMed]
- Chubarov, A.S.; Oscorbin, I.P.; Filipenko, M.L.; Lomzov, A.A.; Pyshnyi, D.V. Allele-specific PCR for KRAS mutation detection using phosphoryl guanidine modified primers. Diagnostics 2020, 10, 872. [Google Scholar] [CrossRef] [PubMed]
- Hett, T.; Zbik, T.; Mukherjee, S.; Matsuoka, H.; Bönigk, W.; Klose, D.; Rouillon, C.; Brenner, N.; Peuker, S.; Klement, R.; et al. Spatiotemporal Resolution of Conformational Changes in Biomolecules by Combining Pulsed Electron-Electron Double Resonance Spectroscopy with Microsecond Freeze-Hyperquenching. J. Am. Chem. Soc. 2021, 143, 6981–6989. [Google Scholar] [CrossRef] [PubMed]
- Worswick, S.G.; Spencer, J.A.; Jeschke, G.; Kuprov, I. Deep neural network processing of DEER data. Sci. Adv. 2018, 4, eaat5218. [Google Scholar] [CrossRef]
- Schiemann, O.; Heubach, C.A.; Abdullin, D.; Ackermann, K.; Azarkh, M.; Bagryanskaya, E.G.; Drescher, M.; Endeward, B.; Freed, J.H.; Galazzo, L.; et al. Benchmark Test and Guidelines for DEER/PELDOR Experiments on Nitroxide-Labeled Biomolecules. J. Am. Chem. Soc. 2021, 143, 17875–17890. [Google Scholar] [CrossRef]
- Islam, M.A.; Fujisaka, A.; Kawakami, J.; Yamaguchi, T.; Obika, S. Different reactivity of Sp and Rp isomers of phosphorothioate-modified oligonucleotides in a duplex structure. Bioorganic Med. Chem. Lett. 2020, 30, 127166. [Google Scholar] [CrossRef] [PubMed]
| Oligonucleotide 1 | Abbreviation | Complex Type 2 | Melting Temperature Tm (°C) | |
|---|---|---|---|---|
| No 1 | No 2 | |||
| M | N-T3* | T0_T3* | t, h | 43.0 ± 0.6 |
| M-T1 | N-T3* | T1_T3* | t, (h) | 38.5 ± 1.2 |
| M-T3 | N-T3* | T3_T3* | d, t | 39.2 ± 0.8 |
| M-T7 | N-T3* | T7_T3* | d, (t) | 41.0 ± 0.7 |
| M-T10 | N-T3* | T10_T3* | d, (t) | 42.5 ± 0.8 |
| M-T25 | N-T3* | T25_T3* | d, t | 43.0 ± 0.8 |
| M-T3* | N-T3* | T3*_T3* | d, t | 39.0 ± 1.0 |
| M-T3* | N | T3*_T0 | t, h | 42.7 ± 0.6 |
| M-T3* | N-T3 | T3*_T3 | d, t | 39.0 ± 1.0 |
| M-T3* | N-T25 | T3*_T25 | d, t | 43.5 ± 0.8 |
| Sample | Distance MD, nm | PELDOR Distance, nm | |
|---|---|---|---|
| Sp-Sp | Rp-Rp | ||
| T0_T3* | 7.2 | 8.0 | 7.5 |
| T3*_T0 | 7.3 | 7.4 | 7.5 |
| T1_T3* | 8.3 | 6.5 | 7.4 |
| T3_T3* | 4.2 | 5.1 | 6.5–6.8 |
| T3*_T3 | 5.9 | 6.4 | 5.6, 7.7 |
| T3*_T3* | 4.0, 5.3, 5.5, 5.7, 6.9, 7.4 | 3.4, 3.8, 3.9, 4.1, 5.0, 5.6 | 4.3 |
| T7_T3* | 8.5 | 8.0 | - |
| T10_T3* | 8.0 | 9.4 | - |
| T25_T3* | 10.1 | 8.6 | - |
| T3*_T25 | 13.3 | 13.0 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chubarov, A.S.; Endeward, B.; Kanarskaya, M.A.; Polienko, Y.F.; Prisner, T.F.; Lomzov, A.A. Pulsed Dipolar EPR for Self-Limited Complexes of Oligonucleotides Studies. Biomolecules 2024, 14, 887. https://doi.org/10.3390/biom14080887
Chubarov AS, Endeward B, Kanarskaya MA, Polienko YF, Prisner TF, Lomzov AA. Pulsed Dipolar EPR for Self-Limited Complexes of Oligonucleotides Studies. Biomolecules. 2024; 14(8):887. https://doi.org/10.3390/biom14080887
Chicago/Turabian StyleChubarov, Alexey S., Burkhard Endeward, Maria A. Kanarskaya, Yuliya F. Polienko, Thomas F. Prisner, and Alexander A. Lomzov. 2024. "Pulsed Dipolar EPR for Self-Limited Complexes of Oligonucleotides Studies" Biomolecules 14, no. 8: 887. https://doi.org/10.3390/biom14080887








