Abstract
Polyphosphate (polyP) is an evolutionary ancient inorganic molecule widespread in biology, exerting a broad range of biological activities. The intracellular polymer serves as an energy storage pool and phosphate/calcium ion reservoir with implications for basal cellular functions. Metabolisms of the polymer are well understood in procaryotes and unicellular eukaryotic cells. However, functions, regulation, and association with disease states of the polymer in higher eukaryotic species such as mammalians are just beginning to emerge. The review summarises our current understanding of polyP metabolism, the polymer’s functions, and methods for polyP analysis. In-depth knowledge of the pathways that control polyP turnover will open future perspectives for selective targeting of the polymer.
1. Introduction
Inorganic polyphosphate (polyP) is a linear anionic polymer composed of three to several hundred orthophosphate (Pi) residues that are linked by energy-rich phosphoanhydride bonds [1,2]. In microorganisms, polyP has a variety of cellular functions, including phosphate and energy storage, support of survival in the stationary phase and under nutrient starvation, and, together with polyphosphate kinases, regulation of cell motility, biofilm formation, and virulence [3,4]. Furthermore, the degradation of polyP reduces stress-induced DNA damage in microorganisms and regulates their enzyme activity, which has implications for virulence and sensitivity for antibiotics. In yeast, polyP is crucial for controlling Pi homeostasis but also participates in adaptive processes, including growth and heavy metal ion tolerance [2]. PolyP constitutes a metabolic energy-storing compound in eukaryotes and fuels an array of cellular processes [5,6]. Polyphosphate levels are affected by many pathways in different cells. However, in contrast to unicellular eukaryotes and prokaryotes, little is known about enzymes that regulate polyP in mammalians.
2. Biophysical Characteristics of Polyphosphate
Polyphosphate is an inorganic anionic energy-rich polymer widespread in all living organisms, from bacteria to mammalians [7]. Furthermore, the polymer is also produced synthetically, and synthesised polyP is used in large amounts in an array of applications, e.g., as a water softener, fire extinguisher, or as additives for conservation, pH buffer, and water retention in the food industry [8]. Synthetic polyP molecules are technically produced by condensation of orthophosphoric acid monomers with dehydration under high temperatures. Polymerisation generates energy-rich phosphoanhydrid bonds that form the backbone of linear and branched polymers and supramolecular polyP structures [9]. The nucleotides GTP and ATP share these high-energy phosphoanhydride bonds with polyP. In nature, enzymatic hydrolysis of the terminal high-energy β–γ and α–β phosphoanhydride bonds in ATP to ADP and ADP to AMP, respectively, liberates ~30 kJ/mol of energy [5]. Corresponding to the hydrolysis of the terminal ATP phosphoanhydride bond, the hydrolysis of P-O-P anhydride bonds in linear polyP similarly generates ~30 kJ/mol under standard conditions [10,11]. The kinetics of the Pi-generating hydrolysis reaction is slow. At physiological pH, polyP has a half-life of about 90 min in plasma [12] and several months in aqueous solutions ex vivo. However, it vastly decreases upon increasing temperature and decreasing pH [1,13]. Synthetic polyP molecules are linear, cyclic, or branched and have a chain length from three up to several thousand orthophosphate units [14]. Branched polyP, so-called ultraphosphates, have not been found in living organisms [15]. Their stability depends on pH and the presence or absence of divalent metal cations [16,17]. It is currently unclear whether they are not formed in biological systems or are not found due to their short half-life in aqueous systems, as the branching sites are rapidly hydrolysed [17]. Cyclic polyPs (i.e., metaphosphates) have been synthesised from linear polyP upon solvent removal, and they are comparably stable in aqueous solutions. The linear polyP backbone has a high degree of conformational flexibility with staggered and eclipsed conformations dependent on whether monovalent (e.g., NH4+, Na+, or K+), divalent (e.g., Mg2+, Zn2+, Ca2+), or even trivalent (e.g., Gd3+) cations are complexed to the phosphate backbone [18,19]. Molecular modelling and experimental data revealed that binding Ca2+ ions to polyP molecules leads to aggregate/microparticle formation ex vivo [20]. The length of a polyP molecule in its linear conformation can be estimated from the size of a single phosphate monomer (2.7 Å), indicating that long-chain polyP polymers (around 1000 Pi units) of bacterial and mammalian cell origin is up to ∼300 nm in length. However, the natural polymer is complexed to divalent cations and proteins that compact the polyP within subcellular compartments such as acidocalcisomes or platelet-dense granules. The phosphates in the polyP backbone are strongly acidic (pKa ∼0–3) molecules and are fully deprotonated at physiological pH [21,22]. The negatively charged oxygen atoms from two adjacent phosphate groups complex divalent cations. Thus, polyP provides a scaffold for the local enrichment of Zn2+ in platelet-dense granules and Ca2+ in various intracellular compartments [23]. The complexed cations, at least in part, neutralise the polyanionic polymer backbone [24,25,26] (Figure 1).
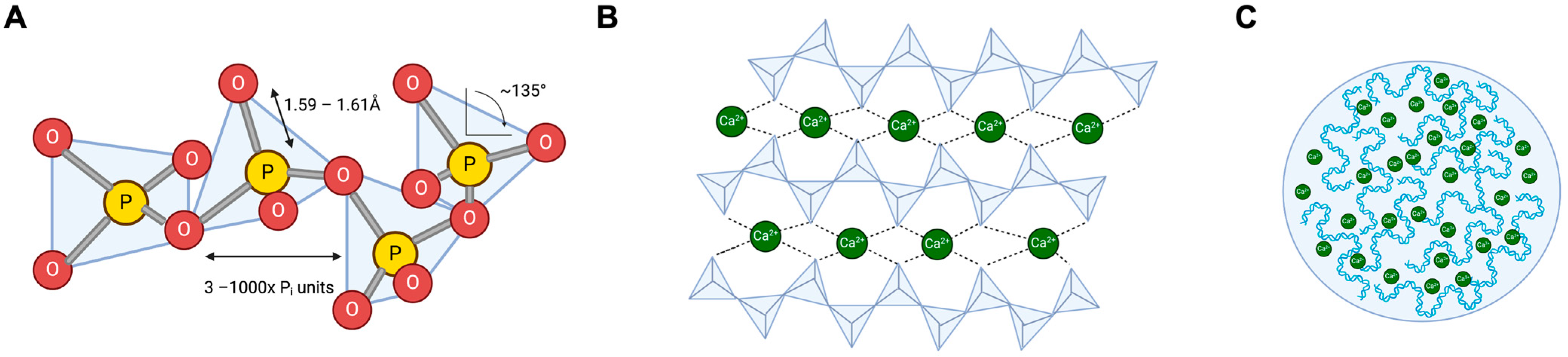
Figure 1.
Gross structures of natural polyP. (A): PolyP is a polymer consisting of 3–1000 tetrahedral phosphate subunits linked to each other via a shared oxygen atom. Most polyP is linear; however, branched forms exist. (B): PolyP binds Ca2+, which binds further polyP and lines up, presumably in a helical secondary structure [27]. (C): Aggregated Ca2+ polyP forms nanoparticles that, in turn, form microsomes within cells.
PolyP in living cells are linear polymers characterised by their mean chain length, distribution of chain length (dispersity), and binding partners, including cations and proteins. PolyP is ubiquitously found in higher organisms, including all mammalian species [28]. However, a comprehensive overview of the organ-specific polymer content and chain length does not currently exist. This is urgently needed to provide the basis for a rational assessment of the (patho)physiological functions of the polymer. Classical studies in rats and mice indicate that polyP content is high in the heart and brain, exceeding polymer levels in the liver by three-fold [5,29]. Based on the high affinity of polyP for binding to Ca2+ (KD in the lower nM range) that exceeds extra- (2.2–2.6 mM) and intracellular (0.1–1.0 mM) Ca2+ levels, the natural polymer is saturated with Ca2+ ions. Ca2+-binding essentially regulates the biophysical properties of the molecule and makes extracellular polyP virtually insoluble in biological fluids [30,31]. Synthetic polyP complexed with divalent cations (Ba2+, Pb2+, Mg2+, or Ca2+) is either insoluble or dissolves in tiny amounts only in aqueous solutions. In contrast, short-chain synthetic polyP complexed with Na+, K+, or NH4+ ions is soluble in water and used as a water softener, e.g., in dishwasher tablets [9,32,33,34]. Both short- and long-chain polymers are virtually insoluble in ethanol, which can be used to precipitate the polymer from biological sources [9,32,33,34]. Precise data on the affinity of polyP for the various complexed cations is not available. Because polyP degrades over time, the total charge of the molecule and binding affinities for multiple cations have remained undefined. Furthermore, no detailed data exist on natural polyP secondary structures, e.g., on cell surfaces or with internal storage pools. However, the fact that intercalating dyes such as DAPI or Sytox Green/Orange stain polyP suggests that at least parts of the polymer have an α-helix-rich structure (reviewed in [35]) similar to DNA [36].
3. PolyP Metabolism
It has been established for more than 60 years that polyP is produced in all living organisms [37,38]; nevertheless, little is known about the metabolism, regulation, concentration, chain length, and localisation of the polymer in mammalian cells. There seem to be significant differences between various cell types that most likely reflect diverse but distinct functions of the polymer in subcellular compartments, which are described below and summarised in Table 1.

Table 1.
Major enzymes involved in metabolisms of polyP in different contemporary species.
4. Bacteria
In bacteria, polyP is synthesised and degraded by polyphosphate kinases (PPKs). Two distinct enzymes, PPK1 and PPK2 that differ in their reaction profile [41], reversibly catalyse the polymerisation of the terminal phosphate of ATP into a nascent polyP chain or transfer terminal Pi from the polymer backbone to GDP, synthesising GTP [11,58]. PPK2 is an 80 kDa enzyme bound to the inner leaflet of the plasma membrane and is evolutionarily older than PPK1 [59]. By modulating cellular energy status, polyP protects bacteria from various external stressors, including heat, ultraviolet irradiation, antibiotics, and oxidative stress. PPK1 deficiency by target ablation of ppk1 gene expression impairs growth and biofilm formation and reduces resistance to starvation [60,61]. Moreover, gene ablation of ppk2 in Mycobacterium smegmatis leads to biofilm synthesis and virulence [62,63]. Corynebacterium glutamicum expressed two PPK2 isoenzymes, while Pseudomonas aeruginosa and Sinorhizobium meliloti have three isoenzymes each, and Ralstonia eutropha even expresses five PPK2 variants [64]. Class I PPK2´s phosphorylate nucleoside diphosphates ADP and GDP by transferring the terminal Pi of polyP to the nucleotide diphosphates and vice versa, shuttling Pi to the nascent polyP chain [65]. In contrast, class II PPK2´s catalyse nucleotide monophosphate phosphorylation and were previously called polyP-AMP phosphotransferase (PAP) [66]. Class III PPK2´s phosphorylate mono- or diphosphates in nucleotide to generate ADP or ATP, respectively, using polyP as a phosphate donor [67].
Exopolyphosphatases (PPXs) hydrolyse polyP of chain length > 3 to release the terminal Pi. Based on the primary structure, they are classified into two types, PPX1 and PPX2, respectively. Both enzymes are members of the same protein family, the PPX-GppA phosphatases. Most bacteria, such as Corynebacterium glutamicum, express two distinct exopolyphosphatases. PPX1 and PPX2 from C. glutamicum share low identity on the protein level (25%) and are different from enterobacterial, archaeal, and yeast exopolyphosphatases [57]. X-ray studies of the crystal structure of E. coli PPX1 at 2.2-°A resolution revealed that the molecule is composed of four domains, with two of them mediating binding to polyP > 35-Pi chain length and two harbouring the enzymatic activity, respectively. Bacterial PPX enzymes require both Mg2+ and K+ for maximal activity [68].
5. Unicellular Eukariotes
PolyP presence is evolutionary and not restricted to prokaryotes. The slime mould Dictyostelium discoideum and various fungi, such as the model system Saccharomyces cerevisiae, contain polyP. Eukaryotes express a polyP-modifying enzyme not found in bacteria, endopolyphosphatase1 (PPN1). PPN1 has endo- but also exopolyphosphatase activities and is found in various compartments, including the cytosol, the nuclear and mitochondrial membranes, and within vacuoles [69,70,71]. At these sites, PPN1 colocalises with PPX1, and the activity of both polyP-modifying enzymes is significantly increased by Mg2+ but not by Ca2+ ions [7,25]. The optimal pH for both enzymes is close to 7.0. Similarly to bacterial PPK, the anionic polysaccharide heparin is a competitive inhibitor for yeast endo- and exopolyphosphatases, most likely by competing for the polyanion-binding site in the enzymes [47,72]. Deficient yeast strains in PPX1 and PPN1 still have endopolyphosphatase activity, indicating that other yet unknown enzymes with overlapping substrate specificity exist [69]. Indeed, a yeast strain with vacuole-targeted exopolyphosphatase Ppx1 and concomitantly depletion of the two endopolyphosphatases (ppn1Δppn2Δ, vt-Ppx1) is deficient in polyPase activity [73].
A specific polyP-rich organelle are acidocalcisomes that are membrane-bound vesicular structures initially described in Trypanosoma crucii; however, they exist in other unicellular eukaryotes, bacteria, and human cells [74]. In addition to polyP, acidocalcisomes are rich in Ca2+ and play a role in intracellular calcium signalling and osmoregulation. Acidocalcisomes are acidic, and their specific milieu is maintained by two proton pumps, i.e., vacuolar H+-pyrophosphatase (V-H+-PPase) and H+-ATPase (V-H+-ATPase) and a Ca2+-ATPase, which mediates the transport of H+ into the acidocalcisome lumen in exchange for Ca2+. Ca2+ ions are released from acidocalcisomes into the cytoplasm via an inositol 1,4,5-trisphosphate receptor (IP3 receptor) bound Ca2+ channel [75,76,77]. At the acidocalcisome membrane, the vacuolar transporter chaperone complex (VTC) synthesises polyP from ATP and transports the nascent polymer chain into the organelle lumen [42]. The acidocalcisome polyP is hydrolysed by PPX, and the cleaved Pi is exported via Na+/Pi symporters [78]. Acidocalcisomes contain additional polyP-modifying enzymes such as vacuolar soluble PPase (VSP), PPX, and acid phosphatase. Upon hyperosmotic stress, Trypanosoma crucii produces long-chain polyP, thus lowering Pi and cation levels and consequently reducing osmotic pressure [22]. Based on these data, osmoregulation is considered an essential function of polyP in bacteria and unicellular eukaryotes. However, it is currently unknown whether this fundamental cellular function is conserved in eukaryotes, especially as the intracellular polyP levels are much lower in complex organisms.
6. Mammalians
Mammalian cells contain polyP, although the cellular content of the polymer, its subcellular localisation, and chain length greatly vary [1]. Using [32P]-radiolabeled polymers has shown that polyP content in rat hepatocytes is highest in the nucleus (89 ± 7 µM) followed by plasma membranes (43 ± 3 µM), cytosol (12 ± 2 µM), mitochondria (11 ± 0.6 µM) and lowest in microsomes (4 ± 0.5 µM) [5]. High polyP levels in the cytosol have also been found in various immortalised cultured cell lines, including NIH3T3 fibroblasts, Vero epithelial kidney, and Jurkat CD4+ T cells [5]. In contrast to simpler organisms where polyP metabolising enzymes have been well characterised, the polyP synthesising and degrading machinery in mammalians have remained enigmatic. Surprisingly, and despite the fundamental functions of the polymer, the polyp-metabolising enzymes of simpler organisms are not conserved in mammalians, and knowledge about enzymes involved in polyP turnover is just beginning to emerge [79]. Mammalian phospholipase D shares similarities with bacterial PPK1 and PPK2 but has no polyP-modifying activity [7]. Bacterial PPX1 and human (h)-Prune are functionally equivalent but non-homologous proteins originating from distinct ancestors. PPX1 is a member of the PPX-GppA phosphatases family. At the same time, h-Prune belongs to the DHH phosphoesterases superfamily, which shares the phosphodiesterase activity to hydrolyses short-chain polyP (up to 4 Pi). In contrast to PPX, h-Prune processes long-chain molecules with >25 Pi residues poorly. Consistently, recombinant h-Prune is unable to hydrolyse short- and medium-chain polyP. Conversely, the knockdown of h-Prune expression significantly decreases polyP and ATP content in treated HEK293 cells [52]. Prune is involved in mammalian polyP metabolism, although its exact role and regulation have not been fully elucidated [52,80]. Prune is expressed and functions in polyP metabolism in Drosophila melanogaster, suggesting that some fundamental principles of polyP regulation are conserved throughout evolution. However, the cellular localisation of Prune differs among the species. In Drosophila melanogaster, Prune is predominantly in mitochondria. In contrast, in addition to mitochondria, the polymer is enriched in various other compartments, including ER, cytoplasm, nucleus, and many (secretory) granules and vesicles in mammalian cells [80]. Throughout the species, polyP is enriched in mitochondria and couples phosphate- with ATP-homeostasis. In mammalians, the F0F1-ATP synthase, which generates ATP within the mitochondrial matrix, modulates polyP content and chain length [80,81].
Another group of conserved polyp-metabolising enzymes are alkaline phosphatases (Table 1), ubiquitous membrane-bound glycoproteins that are capable of cleaving phosphoester bonds and additionally have the capacity for degrading polyP [82]. Shrimp alkaline phosphatase efficiently degrades extracellular polyP in plasma, confirming that the enzyme has polyp-metabolising activities [60]. Alkaline phosphatases are similar to yeast cytosolic and nuclear diphosphoinositol polyphosphate phosphohydrolase 1 and 2 (DDP1 and DDP2) [83].
In addition to polyP synthesis and degradation, the polymer is also regulated by the availability of Pi. The Xenotropic and polytropic retrovirus receptor 1 (XPR1) was initially identified as a docking receptor for retroviruses. However, later studies showed that XPR1 also exports Pi in human and murine platelets [84,85]. XPR1 shares homology with Pho84, Pho87, Pho90, and Pho89 Pi transporters in Saccharomyces cerevisiae [86]. XPR1 is an atypical G-protein-coupled receptor expressed in intracellular organelles and at the plasma membrane. In addition to being regulated by XPR1, polyP itself modulates membrane channels. Primary rat hepatocytes express the mitochondria permeability transition pore (mPTP) at their inner mitochondrial membrane. mPTP is reversibly complexed with the transient receptor potential melastatin 2 (Trpm2) or transient receptor potential channel subfamily A member 1 (TRPA1) [24,87]. PolyP is involved in mPTP activation in murine cardiac myocytes in a polymer chain length-dependent manner. Depleting the polymer protects against Ca2+-induced mPTP opening, suggesting an autocrine regulation of mammalian Ca2+-polyP [88]. mPTP regulates the influx of Ca2+ and, thus, causes mitochondrial swelling, which is regularly associated with cell apoptosis [89].
7. Extracellular PolyP
PolyP has both intracellular and extracellular activities (Table 2). In bacteria and unicellular eukaryotes, polyP is an intracellular polymer. However, when cells disintegrate, the polymer might be exposed to the extracellular environment. In contrast, some eukaryotic cells, including mast cells, astrocytes, and thrombocytes, can actively release the polymer from their secretory storage organelles [30,90].

Table 2.
Gross functions of extracellular and intracellular polyP.
PolyP release and exposure at the cell surface have been identified in thrombocytes [91]. Thrombocytes contain polyP within their dense granules, which appear dark in electron microscopy due to their high Ca2+ and Pi content [92]. The dense granules share similarities with acidocalcisomes in unicellular eukaryotes [93]. However, unlike intracellular acidocalcisomes, dense granules fuse with the cell membrane upon activation and release intracellular polyP to the extracellular compartment [92]. Elegant real-time microscopy studies have visualised polyP “secretion” [20].
In contrast to released soluble mediators from dense granules such as ATP, platelet polyP is a mostly insoluble polymer. It is retained on the surface of procoagulant platelets as Ca2+/Zn2+-rich microparticles [20]. Only small amounts of relatively short-chain polymers of ~70–80 Pi residues are soluble and found in the plasma compartment. However, it is uncertain whether these polymers are in a solution or if they stick to extracellular vesicles or exosomes [94,95]. Polyanion-exchanger-based polyP extraction from platelets showed that platelets, similar to other mammalian cells, mostly contain long-chain polyP with chain lengths of several hundreds of units. However, a small amount of total polyP is in the range of ~80 Pi units [96]. Isolation of platelet polyP using the phenol/chloroform-based extraction method selects for these soluble polymers and results in the purification of short-chain ~80 Pi long polymers [14]. The regulation of platelet polyP remains to be fully elucidated. Nevertheless, intracellular Pi levels have been identified as critical regulators of cellular polymer content [97], and the conditional ablation of platelet XPR1 increased polyP levels and augmented platelet-driven thrombosis with no bleeding abnormalities [85].
Synthetic and cell-purified polyP has modulated an array of reactions in plasma ex vivo [98,99]. However, the in vivo activities of the polymer seem to be mediated chiefly, if not exclusively, via the factor (FXII)-activated plasma contact system [100]. FXII initiates the plasma contact system that drives the formation of bradykinin (BK) via the kallikrein–kinin system and the clots via the intrinsic coagulation pathway, respectively [101]. Binding to negatively charged polyP microparticles on cell surfaces induces a conformational change in FXII zymogen, resulting in an active serine protease, activated FXII (FXIIa) [102,103]. Supporting a direct polyP-mediated FXII activation, immunofluorescent microscopy colocalised FXIIa and polyP on surfaces of cancer cells and cancer-derived exosomes [90]. Injection of polyP into the skin of normal mice induced BK-mediated oedema. In contrast, mice with inherited deficiency in or pharmacologically inhibited expression of FXII and kinin B2 receptors were protected from polyP-induced vascular leakage, indicating that polyP activates FXIIa that drives BK-stimulated vascular leakage [98,104]. Recently, targeting FXIIa has proven to be an efficient therapy for patients with hereditary angioedema (HAE, a life-threatening swelling disease) [103]. An engineered fully human antibody, GaradacimabTM, specifically neutralised FXIIa (originally named 3F7) and potently blocks BK-driven swelling attacks in a prophylactic setting [105]. The exact trigger for FXII activation and kallikrein–kinin-system-mediated BK formation in HAE is not entirely clear; however, clearly, mast cell-derived heparin [106] or polyP (which is a component of mast cell granules [77]) are promising candidates.
Activated platelets and platelet microparticles stimulate thrombin generation in an FXII-contact activation-dependent manner in vivo and human plasma with critical implications for thrombosis in vivo [98,107]. Vice versa, FXII-dependent plasma clotting and thrombus formation are defective in mice and humans with a genetic deficiency in polyP (inositol hexakisphosphate kinase 1 null [Ip6k1−/−] mice [108] and Hermansky–Pudlak syndrome patient platelets [98], respectively). Natural polyP is largely insoluble Ca2+, and Zn2+-rich polyP nanoparticles potently initiate FXII contact activation similarly to kaolin (a silicate commonly used to trigger FXII activation in diagnostic aPTT clotting assays) [94].
Based on the original discovery that FXII-deficient mice are protected from arterial and venous thrombosis but do not bleed excessively [107,109], the FXIIa-triggered contact system has emerged as a promising target for safe anticoagulant drugs. Mimicking FXII/FXIIa inhibitors targeting extracellular polyP have emerged as a novel treatment strategy to dampen thrombosis with preserved haemostasis [103]. Various polycation agents have been used to target extracellular polyP, including spermidine, histone H1, polymyxin B, and cationic polymers [30]. Neutralising the polyP charge interferes with the ability of the polyanion to initiate FXII contact activation and ablates the prothrombotic activities of the polymer while sparing haemostasis [30,103]. Another strategy for interference with polyP prothrombotic activity is based on polymer backbone degradation. Originally, alkaline phosphatase has been shown to interfere with polyP coagulation activity and to block activated platelet-driven thrombosis in mouse models [110,111]. Alkaline phosphatase cleaves the polymer chain; however, it also removes phosphate groups from lipids and proteins, suggesting the use of polyP-specific phosphatases. E. coli exopolyphosphatase (PPX), a member of the PPX-GppA phosphatases family, specifically cleaves polyP of chain length > 35 Pi but no other polyanions such as heparin, DNA/RNA, or synthetic dextran sulphate. PPX efficiently degrades synthetic and natural polyP and ablates the procoagulant activity of the polymer in human plasma. Infusion of PPX into mice confers protection against polyP procoagulant activity and blunts activated platelet-stimulated vascular occlusions in mouse thrombosis models [112,113,114]. Similarly to full-size PPX, the recombinant polyP-binding domains of the enzyme (domains 3 and 4) bind with high affinity to polyP, interfere with procoagulant activities of the polymer, and block polyP-mediated FXII contact activation. Both polyP-degrading PPX and non-degrading PPX_∆12 inhibit thrombus formation in lethal pulmonary embolism without interfering with haemostasis in vivo, reproducing the effects of FXII inhibitors [68,115]. The data show that targeting polyP mimics the selective importance of FXIIa in thrombosis, sparing hemostasis [103,116]. The findings with polyP/FXII also suggest that pharmacologic interference with polyP offers the unique opportunity for blocking thrombosis and inflammation (thrombo-inflammation) without affecting physiologic haemostasis at vascular injury sites (safe anticoagulants) [90,117]. The concept of interference with polyP-triggered coagulation seems to have arisen in blood-sucking insects. Sand flies express proteins in their saliva that allow feeding mammalian blood without triggering brad–kinin-mediated itching and FXIIa-mediated clotting. These proteins, PdSP15a and PdSP15b expose a positively charged helix that binds to the polymer and neutralises polyP procoagulant and proinflammatory activities [118].
The blood circulating histidine-rich glycoprotein (HRG) binds to polyanions such as DNA, RNA, and polyP. HRG has been shown to inhibit polyP procoagulant activity by interfering with FXII contact activation [119]. The underlying mechanism for antagonising FXII activity is a competition between HRG and FXII for binding to polyP. Reduced FXII binding to polyP results in defective FXII contact activation and aPTT prolongation [120,121]. PolyP injection in HRG-deficient (Hrg−/−) mice consistently triggers excess fibrin formation [122,123]. Vice versa, the reconstitution of Hrg−/− mice with HRG restores excess polyP-driven thrombosis in an FXII-dependent manner [119]. The data are consistent with HRG operating as an endogenous FXII regulator by binding to the contact activator polyP.
In addition to procoagulant platelets and activated mast cells, polyP is released from mammalian astrocytes and has a role in neurotransmission. Astrocyte–polyP activates neurons and provokes calcium-mediated signalling [124]. PolyP is increased in human and mouse astrocytes and cerebrospinal fluid amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Targeting secreted polyP in ALS/FTD astrocytes prevents motoneuron death, indicating that polyP released by ALS/FTD astrocytes is a critical factor in neurodegeneration and a potential therapeutic target for interference with the diseases [125]. Intracellular polyP also has chaperone activities in the central nervous system and restores incorrect protein folding, suggesting a role of polyP in amyloidogenic processes and disease states [126,127]. Indeed, polyP improves degenerative neuronal diseases where misfolded protein aggregates seem to mediate the pathology, such as Alzheimer’s, Huntington’s, and Parkinson’s disease [18,128]. Together, the data highlight the importance of polyP in the central nervous system, which is awaiting future studies.
8. PolyP Signalling
Established functions of polyP in prokaryotic cells include storage for energy, Pi and Ca2+, biofilm formation, biomineralisation, and bone formation [2,32,129,130] (Table 2). Importantly, intracellularly, polyP activates mTOR and, thus, promotes cell proliferation and growth [131]. The exopolyphosphatase h-prune provides another link of polyP and mitogenic activity. The binding of h-prune to nm23-h1, a metastasis suppressor, inhibits h-prone phosphatase activity [52]. Defective phosphatase activity, at least in yeast, increases polyP content and might provide a rationale for h-prune expression in cancer associated with poor prognosis via augmented metastasis and tumour growth [52,80]. The precise function of polyP in cancer is likely complex and might be cell-type specific. In myeloma cancer cells, the intracellular polyP level is significantly higher than that of “healthy” primary plasma cells. The addition of synthetic polyP initiates apoptosis of myeloma cells [132]. However, the underlying mechanisms are unclear and might include a change in pH, complexation of surface-bound ions, activation of specific signalling pathways, or local increase in Pi. In addition to regulating basal cellular functions, it has been suggested that polyP is covalently attached to target proteins [133], e.g., nuclear signal recognition 1 (Nsr1) and its interacting partner, topoisomerase 1 (Top1; polyphosphorylation), on lysine residues within conserved N-terminal polyacidic serine and lysin- (PASK) or histidine-rich clusters [134,135]. However, recent studies have challenged the hypothesis of polyP being a novel noncovalently attached post-translational modification [136,137]. Lysine and histidine residues are positively charged at physiological pH, and these amino acids bind tightly electrostatically (but not covalently) to the negatively charged polyP backbone. The conserved histidine a-helical domain (CHAD) is an example of a polyP-binding protein found in several procaryotic cells [138].
Extracellular polyP has emerged as a procoagulant and proinflammatory mediator. Due to its high biological activity and link to disease states, the polymer has potential diagnostic implications as a (predictive) biomarker with multiple methods for polymer analysis (Table 3). Intercalating dyes can visualise polyP in histology upon separation in agarose and urea gels and individual cells by flow cytometry (FACS) [1]. Specificity for polyP upon UREA-PAGE separation is increased by photobleaching (negative DAPI staining), where polyP appears dark and DNA or proteoglycans turn whitish [36,77,132]. DAPI binds both to polyP and DNA but differs in its emission wavelength. DAPI bound to DNA emits a greenish colour around 461 nm, while DAPI/polyP is bluish at 525 nm [139]. As the dyes bind to helical polyP portions, the polymer backbone needs to exceed >15 Pi to allow for detection. Dyes JC-D7 and JC-D8 differ in emission spectrum intensity depending on the polyP chain length with >15 Pi [140]. Elegant work using FITC-labelled polyP has shown that cells endocytose polyP microparticles. Using FITC-polyP allows for intracellular polyP staining and the analyses of transport and enrichment of (labelled) polyP to distinct cellular compartments depending on cell activation, metabolic state, and environment [141]. For flow cytometry and histology, neutral red was initially used to visualise polyP [142]. The polyP-binding domain of PPX (PPBD, synonymous to PPX_∆12) selectively binds to polyP with chain length > 38 Pi units [142]. PPBD [143] has been used in histology and FACS to visualise and quantify the polymer in an experimental setting and might provide a lead structure for a diagnostic polyP probe with clinical implications. Furthermore, CHADs bind to polyP-rich granules in vivo and may provide an alternative investigation for polyP [144]. Biophysical methods such as small-angle X-ray and 31P NMR have been established to analyse the structure of the polymer dependent on bound cations in dry form or solution, respectively. The analysis of polyP in clinical settings, its association with FXII activation states, and its role in diseases requires future research.

Table 3.
Methods for analysing polyphosphate.
9. Future Perspectives
What are the most urgent needs for the field? So far, a comprehensive overview of polyP amount and chain length in all mammalian organs is missing. This “polyP atlas” would provide the basis for analysis of changes in the polymer associated with disease states. Additionally, and as a next step, the amount and composition of polyP in all cellular sub-compartments require attention. Furthermore, the polyp-metabolising enzymes (if existing) need to be identified and fully characterised to allow for selective and specific modulation of polyP content, chain length, and, thus, activities.
Author Contributions
R.S., M.W., H.J.J. and T.R. wrote and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the P06/KFO306 and INST 152/876-1 FUGG grants of the DFG (to T.R.).
Acknowledgments
We would like to thank Verena Horneffer-van der Sluis for proofreading.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Desfougeres, Y.; Saiardi, A.; Azevedo, C. Inorganic polyphosphate in mammals: Where’s Wally? Biochem. Soc. Trans. 2020, 48, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Rao, N.N.; Gomez-Garcia, M.R.; Kornberg, A. Inorganic polyphosphate: Essential for growth and survival. Annu. Rev. Biochem. 2009, 78, 605–647. [Google Scholar] [CrossRef]
- Wang, L.; Yan, J.; Wise, M.J.; Liu, Q.; Asenso, J.; Huang, Y.; Dai, S.; Liu, Z.; Du, Y.; Tang, D. Distribution Patterns of Polyphosphate Metabolism Pathway and Its Relationships With Bacterial Durability and Virulence. Front. Microbiol. 2018, 9, 782. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.N.; Docampo, R. Polyphosphate and its diverse functions in host cells and pathogens. PLoS Pathog. 2013, 9, e1003230. [Google Scholar] [CrossRef] [PubMed]
- Kumble, K.D.; Kornberg, A. Inorganic polyphosphate in mammalian cells and tissues. J. Biol. Chem. 1995, 270, 5818–5822. [Google Scholar] [CrossRef] [PubMed]
- Lynn, W.S.; Brown, R.H. Synthesis of Polyphosphate by Rat Liver Mitochondria. Biochem. Biophys. Res. Commun. 1963, 11, 367–371. [Google Scholar] [CrossRef]
- Kulakovskaya, T.; Kulaev, I. Enzymes of inorganic polyphosphate metabolism. Prog. Mol. Subcell. Biol. 2013, 54, 39–63. [Google Scholar] [CrossRef]
- Hirokado, M.; Shimamura, Y.; Nakajima, K.; Ozawa, H.; Kimura, K.; Yasuda, K.; Nishijima, M. Methods for determination of milt protein and epsilon-polylysine in food additive preparations and processed foods by capillary zone electrophoresis. Shokuhin Eiseigaku Zasshi 2001, 42, 79–83. [Google Scholar] [CrossRef]
- Thilo, E. Die kondensierten Phosphate. Angew. Chem. 1955, 67, 141–145. [Google Scholar] [CrossRef]
- Achbergerova, L.; Nahalka, J. Polyphosphate—An ancient energy source and active metabolic regulator. Microb. Cell Fact. 2011, 10, 63. [Google Scholar] [CrossRef]
- Curnutt, S.G.; Schmidt, R.R. Possible Mechanisms Controlling the Intracellular Level of Inorganic Polyphosphate during Synchronous Growth of Chlorella Pyrenoidosa. Ii. Atp/Adp Ratio. Biochim. Biophys. Acta 1964, 86, 201–203. [Google Scholar] [CrossRef]
- Smith, S.A.; Mutch, N.J.; Baskar, D.; Rohloff, P.; Docampo, R.; Morrissey, J.H. Polyphosphate modulates blood coagulation and fibrinolysis. Proc. Natl. Acad. Sci. USA 2006, 103, 903–908. [Google Scholar] [CrossRef]
- Dassa, E.; Boquet, P.L. Is the acid phosphatase of Escherichia coli with pH optimum of 2.5 A polyphosphate depolymerase? FEBS Lett. 1981, 135, 148–150. [Google Scholar] [CrossRef]
- Bru, S.; Jimenez, J.; Canadell, D.; Arino, J.; Clotet, J. Improvement of biochemical methods of polyP quantification. Microb. Cell 2016, 4, 6–15. [Google Scholar] [CrossRef]
- Christ, J.J.; Willbold, S.; Blank, L.M. Methods for the Analysis of Polyphosphate in the Life Sciences. Anal. Chem. 2020, 92, 4167–4176. [Google Scholar] [CrossRef]
- Jessen, H.J.; Dürr-Mayer, T.; Haas, T.M.; Ripp, A.; Cummins, C.C. Lost in Condensation: Poly-, Cyclo-, and Ultraphosphates. Acc. Chem. Res. 2021, 54, 4036–4050. [Google Scholar] [CrossRef]
- Dürr-Mayer, T.; Qiu, D.; Eisenbeis, V.B.; Steck, N.; Häner, M.; Hofer, A.; Mayer, A.; Siegel, J.S.; Baldridge, K.K.; Jessen, H.J. The chemistry of branched condensed phosphates. Nat. Commun. 2021, 12, 5368. [Google Scholar] [CrossRef]
- Kornberg, A. Inorganic polyphosphate: A molecule of many functions. Prog. Mol. Subcell. Biol. 1999, 23, 1–18. [Google Scholar] [CrossRef]
- Vanwazer, J.R.; Campanella, D.A. Structure and Properties of the Condensed Phosphates.4. Complex Ion Formation in Polyphosphate Solutions. J. Am. Chem. Soc. 1950, 72, 655–663. [Google Scholar] [CrossRef]
- Verhoef, J.J.; Barendrecht, A.D.; Nickel, K.F.; Dijkxhoorn, K.; Kenne, E.; Labberton, L.; McCarty, O.J.; Schiffelers, R.; Heijnen, H.F.; Hendrickx, A.P.; et al. Polyphosphate nanoparticles on the platelet surface trigger contact system activation. Blood 2017, 129, 1707–1717. [Google Scholar] [CrossRef]
- Greenfield, N.J.; Hussain, M.; Lenard, J. Effects of Growth-State and Amines on Cytoplasmic and Vacuolar Ph, Phosphate and Polyphosphate Levels in Saccharomyces-Cerevisiae—A P-31-Nuclear Magnetic-Resonance Study. Biochim. Biophys. Acta 1987, 926, 205–213. [Google Scholar] [CrossRef]
- Ruiz, F.A.; Rodrigues, C.O.; Docampo, R. Rapid changes in polyphosphate content within acidocalcisomes in response to cell growth, differentiation, and environmental stress in Trypanosoma cruzi. J. Biol. Chem. 2001, 276, 26114–26121. [Google Scholar] [CrossRef]
- Foley, B.; Johnson, S.A.; Hackley, B.; Smith, J.C., Jr.; Halsted, J.A. Zinc content of human platelets. Proc. Soc. Exp. Biol. Med. 1968, 128, 265–269. [Google Scholar] [CrossRef]
- Elustondo, P.A.; Nichols, M.; Negoda, A.; Thirumaran, A.; Zakharian, E.; Robertson, G.S.; Pavlov, E.V. Mitochondrial permeability transition pore induction is linked to formation of the complex of ATPase C-subunit, polyhydroxybutyrate and inorganic polyphosphate. Cell Death Discov. 2016, 2, 16070. [Google Scholar] [CrossRef]
- Kulakovskaya, T.V.; Lichko, L.P.; Vagabov, V.M.; Kulaev, I.S. Inorganic polyphosphates in mitochondria. Biochem. Biokhimiia 2010, 75, 825–831. [Google Scholar] [CrossRef]
- Pestov, N.A.; Kulakovskaya, T.V.; Kulaev, I.S. Inorganic polyphosphate in mitochondria of Saccharomyces cerevisiae at phosphate limitation and phosphate excess. FEMS Yeast Res. 2004, 4, 643–648. [Google Scholar] [CrossRef][Green Version]
- Jackson, L.E.; Kariuki, B.M.; Smith, M.E.; Barralet, J.E.; Wright, A.J. Synthesis and structure of a calcium polyphosphate with a unique criss-cross arrangement of helical phosphate chains. Chem. Mater. 2005, 17, 4642–4646. [Google Scholar] [CrossRef]
- Goh, S.H.; Wright, J.A.; LeJohn, H.B. Possible regulation of macromolecular biosynthesis in mammalian cells by a novel dinucleoside polyphosphate (HS3) produced during step-down growth conditions. J. Cell Physiol. 1977, 93, 353–362. [Google Scholar] [CrossRef]
- Jankowski, J.; Jankowski, V.; Laufer, U.; van der Giet, M.; Henning, L.; Tepel, M.; Zidek, W.; Schluter, H. Identification and quantification of diadenosine polyphosphate concentrations in human plasma. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 1231–1238. [Google Scholar] [CrossRef]
- Mailer, R.K.W.; Hanel, L.; Allende, M.; Renne, T. Polyphosphate as a Target for Interference With Inflammation and Thrombosis. Front. Med. 2019, 6, 76. [Google Scholar] [CrossRef]
- Muller, W.E.G.; Schroder, H.C.; Wang, X. Inorganic Polyphosphates As Storage for and Generator of Metabolic Energy in the Extracellular Matrix. Chem. Rev. 2019, 119, 12337–12374. [Google Scholar] [CrossRef]
- Leyhausen, G.; Lorenz, B.; Zhu, H.; Geurtsen, W.; Bohnensack, R.; Muller, W.E.; Schroder, H.C. Inorganic polyphosphate in human osteoblast-like cells. J. Bone Miner. Res. 1998, 13, 803–812. [Google Scholar] [CrossRef]
- Taylor, A.W.; Frazier, A.W.; Gurney, E.L. Solubility products of magnesium ammonium and magnesium potassium phosphates. Trans. Faraday Soc. 1963, 59, 1585–1589. [Google Scholar] [CrossRef]
- Gornagy, S.; Kaldi, P.; Besan, J.; Szanto, A. Solubility of Micronutrients in Ammonium Polyphosphate Solutions. Hung. J. Ind. Chem. 1978, 6, 259–274. [Google Scholar]
- Chiti, A. A new member in the EJNMMI family: The European Journal of Hybrid Imaging. Eur. J. Hybrid. Imag. 2017, 1, 6. [Google Scholar] [CrossRef]
- Smith, S.A.; Morrissey, J.H. Sensitive fluorescence detection of polyphosphate in polyacrylamide gels using 4’,6-diamidino-2-phenylindol. Electrophoresis 2007, 28, 3461–3465. [Google Scholar] [CrossRef]
- Griffin, J.B.; Davidian, N.M.; Penniall, R. Studies of phosphorus metabolism by isolated nuclei. VII. Identification of polyphosphate as a product. J. Biol. Chem. 1965, 240, 4427–4434. [Google Scholar] [CrossRef]
- Ebel, J.P.; Muller, S. Cytochemical study of inorganic polyphosphates in living organisms. I. Study of metachromatic reactions and coloration with methyl green and pyronine as a function of the length of the polyphosphate chain. Exp. Cell Res. 1958, 15, 21–28. [Google Scholar] [CrossRef]
- Ahn, K.; Kornberg, A. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J. Biol. Chem. 1990, 265, 11734–11739. [Google Scholar] [CrossRef]
- Zhu, Y.; Huang, W.; Lee, S.S.; Xu, W. Crystal structure of a polyphosphate kinase and its implications for polyphosphate synthesis. EMBO Rep. 2005, 6, 681–687. [Google Scholar] [CrossRef]
- Zhang, H.; Ishige, K.; Kornberg, A. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. USA 2002, 99, 16678–16683. [Google Scholar] [CrossRef]
- Hothorn, M.; Neumann, H.; Lenherr, E.D.; Wehner, M.; Rybin, V.; Hassa, P.O.; Uttenweiler, A.; Reinhardt, M.; Schmidt, A.; Seiler, J.; et al. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 2009, 324, 513–516. [Google Scholar] [CrossRef]
- Gomez-Garcia, M.R.; Kornberg, A. Formation of an actin-like filament concurrent with the enzymatic synthesis of inorganic polyphosphate. Proc. Natl. Acad. Sci. USA 2004, 101, 15876–15880. [Google Scholar] [CrossRef]
- Liu, W.; Wang, J.; Comte-Miserez, V.; Zhang, M.; Yu, X.; Chen, Q.; Jessen, H.J.; Mayer, A.; Wu, S.; Ye, S. Cryo-EM structure of the polyphosphate polymerase VTC reveals coupling of polymer synthesis to membrane transit. EMBO J. 2023, 42, e113320. [Google Scholar] [CrossRef]
- Lonetti, A.; Szijgyarto, Z.; Bosch, D.; Loss, O.; Azevedo, C.; Saiardi, A. Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 2011, 286, 31966–31974. [Google Scholar] [CrossRef]
- Kumble, K.D.; Kornberg, A. Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. J. Biol. Chem. 1996, 271, 27146–27151. [Google Scholar] [CrossRef]
- Andreeva, N.A.; Kulakovskaya, T.V.; Kulakovskaya, E.V.; Kulaev, I.S. Polyphosphates and exopolyphosphatases in cytosol and mitochondria of Saccharomyces cerevisiae during growth on glucose or ethanol under phosphate surplus. Biochem. Biokhimiia 2008, 73, 65–69. [Google Scholar] [CrossRef]
- Gerasimaite, R.; Sharma, S.; Desfougeres, Y.; Schmidt, A.; Mayer, A. Coupled synthesis and translocation restrains polyphosphate to acidocalcisome-like vacuoles and prevents its toxicity. J. Cell Sci. 2014, 127, 5093–5104. [Google Scholar] [CrossRef]
- Solesio, M.E.; Garcia Del Molino, L.C.; Elustondo, P.A.; Diao, C.; Chang, J.C.; Pavlov, E.V. Inorganic polyphosphate is required for sustained free mitochondrial calcium elevation, following calcium uptake. Cell Calcium 2020, 86, 102127. [Google Scholar] [CrossRef]
- Christ, J.J.; Blank, L.M. Analytical polyphosphate extraction from Saccharomyces cerevisiae. Anal. Biochem. 2018, 563, 71–78. [Google Scholar] [CrossRef]
- Samper-Martin, B.; Sarrias, A.; Lazaro, B.; Perez-Montero, M.; Rodriguez-Rodriguez, R.; Ribeiro, M.P.C.; Banon, A.; Wolfgeher, D.; Jessen, H.J.; Alsina, B.; et al. Polyphosphate degradation by Nudt3-Zn2+ mediates oxidative stress response. Cell Rep. 2021, 37, 110004. [Google Scholar] [CrossRef] [PubMed]
- Tammenkoski, M.; Koivula, K.; Cusanelli, E.; Zollo, M.; Steegborn, C.; Baykov, A.A.; Lahti, R. Human metastasis regulator protein H-prune is a short-chain exopolyphosphatase. Biochemistry 2008, 47, 9707–9713. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem. Sci. 1998, 23, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Ugochukwu, E.; Lovering, A.L.; Mather, O.C.; Young, T.W.; White, S.A. The crystal structure of the cytosolic exopolyphosphatase from Saccharomyces cerevisiae reveals the basis for substrate specificity. J. Mol. Biol. 2007, 371, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- Hurlimann, H.C.; Stadler-Waibel, M.; Werner, T.P.; Freimoser, F.M. Pho91 Is a vacuolar phosphate transporter that regulates phosphate and polyphosphate metabolism in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 4438–4445. [Google Scholar] [CrossRef] [PubMed]
- Gerasimaitė, R.; Mayer, A. Ppn2, a novel Zn2+-dependent polyphosphatase in the acidocalcisome-like yeast vacuole. J. Cell Sci. 2017, 130, 1625–1636. [Google Scholar] [CrossRef] [PubMed]
- Lindner, S.N.; Knebel, S.; Wesseling, H.; Schoberth, S.M.; Wendisch, V.F. Exopolyphosphatases PPX1 and PPX2 from Corynebacterium glutamicum. Appl. Environ. Microbiol. 2009, 75, 3161–3170. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Suzuki, H.; Kaneko, T.; Ikeda, Y. Properties of polyphosphate kinase prepared from mycobacterium smegmatis. Biochim. Biophys. Acta 1972, 268, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Achbergerová, L.; Nahálka, J. PPK1 and PPK2—Which polyphosphate kinase is older? Biologia 2014, 6, 263–269. [Google Scholar] [CrossRef]
- Rashid, M.H.; Rumbaugh, K.; Passador, L.; Davies, D.G.; Hamood, A.N.; Iglewski, B.H.; Kornberg, A. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2000, 97, 9636–9641. [Google Scholar] [CrossRef]
- Tan, S.; Fraley, C.D.; Zhang, M.; Dailidiene, D.; Kornberg, A.; Berg, D.E. Diverse phenotypes resulting from polyphosphate kinase gene (ppk1) inactivation in different strains of Helicobacter pylori. J. Bacteriol. 2005, 187, 7687–7695. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, A.; De Piano, C.; Gelman, E.; McKinney, J.D.; Dhar, N. Elucidating the role of (p)ppGpp in mycobacterial persistence against antibiotics. IUBMB Life 2018, 70, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.M.; Dutta, N.K.; Hung, C.F.; Wu, T.C.; Rubin, H.; Karakousis, P.C. Stringent Response Factors PPX1 and PPK2 Play an Important Role in Mycobacterium tuberculosis Metabolism, Biofilm Formation, and Sensitivity to Isoniazid In Vivo. Antimicrob. Agents Chemother. 2016, 60, 6460–6470. [Google Scholar] [CrossRef] [PubMed]
- Neville, N.; Roberge, N.; Jia, Z. Polyphosphate Kinase 2 (PPK2) Enzymes: Structure, Function, and Roles in Bacterial Physiology and Virulence. Int. J. Mol. Sci. 2022, 23, 670. [Google Scholar] [CrossRef] [PubMed]
- Racki, L.R.; Tocheva, E.I.; Dieterle, M.G.; Sullivan, M.C.; Jensen, G.J.; Newman, D.K. Polyphosphate granule biogenesis is temporally and functionally tied to cell cycle exit during starvation in. Proc. Natl. Acad. Sci. USA 2017, 114, E2440–E2449. [Google Scholar] [CrossRef] [PubMed]
- Bonting, C.F.C.; Kortstee, G.J.J.; Zehnder, A.J.B. Properties of Polyphosphate—Amp Phosphotransferase of Acinetobacter Strain-210a. J. Bacteriol. 1991, 173, 6484–6488. [Google Scholar] [CrossRef] [PubMed]
- Motomura, K.; Hirota, R.; Okada, M.; Ikeda, T.; Ishida, T.; Kuroda, A. A New Subfamily of Polyphosphate Kinase 2 (Class III PPK2) Catalyses both Nucleoside Monophosphate Phosphorylation and Nucleoside Diphosphate Phosphorylation. Appl. Environ. Microb. 2014, 80, 2602–2608. [Google Scholar] [CrossRef] [PubMed]
- Rangarajan, E.S.; Nadeau, G.; Li, Y.; Wagner, J.; Hung, M.N.; Schrag, J.D.; Cygler, M.; Matte, A. The structure of the exopolyphosphatase (PPX) from Escherichia coli O157:H7 suggests a binding mode for long polyphosphate chains. J. Mol. Biol. 2006, 359, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Lichko, L.P.; Kulakovskaya, T.V.; Kulakovskaya, E.V.; Kulaev, I.S. Inactivation of PPX1 and PPN1 genes encoding exopolyphosphatases of Saccharomyces cerevisiae does not prevent utilisation of polyphosphates as phosphate reserve. Biochem. Biokhimiia 2008, 73, 985–989. [Google Scholar] [CrossRef]
- Sethuraman, A.; Rao, N.N.; Kornberg, A. The endopolyphosphatase gene: Essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2001, 98, 8542–8547. [Google Scholar] [CrossRef]
- Lichko, L.P.; Kulakovskaya, T.V.; Kulaev, I.S. Inorganic polyphosphates and exopolyphosphatases in different cell compartments of Saccharomyces cerevisiae. Biochem. Biokhimiia 2006, 71, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, N.A.; Kulakovskaia, T.V.; Kulaev, I.S. Characteristics of polyphosphatase activity of Saccharomyces cerevisiae cytosol. Biokhimiia 1994, 59, 1882–1891. [Google Scholar] [PubMed]
- Azevedo, C.; Desfougères, Y.; Jiramongkol, Y.; Partington, H.; Trakansuebkul, S.; Singh, J.; Steck, N.; Jessen, H.J.; Saiardi, A. Development of a yeast model to study the contribution of vacuolar polyphosphate metabolism to lysine polyphosphorylation. J. Biol. Chem. 2020, 295, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Lander, N.; Cordeiro, C.; Huang, G.; Docampo, R. Polyphosphate and acidocalcisomes. Biochem. Soc. Trans. 2016, 44, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Docampo, R.; Moreno, S.N. Acidocalcisomes. Cell Calcium 2011, 50, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Docampo, R.; de Souza, W.; Miranda, K.; Rohloff, P.; Moreno, S.N. Acidocalcisomes—Conserved from bacteria to man. Nat. Rev. Microbiol. 2005, 3, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Sanchez, D.; Hernandez-Ruiz, L.; Ruiz, F.A.; Docampo, R. Polyphosphate is a novel proinflammatory regulator of mast cells and is located in acidocalcisomes. J. Biol. Chem. 2012, 287, 28435–28444. [Google Scholar] [CrossRef]
- Gerasimaite, R.; Pavlovic, I.; Capolicchio, S.; Hofer, A.; Schmidt, A.; Jessen, H.J.; Mayer, A. Inositol Pyrophosphate Specificity of the SPX-Dependent Polyphosphate Polymerase VTC. ACS Chem. Biol. 2017, 12, 648–653. [Google Scholar] [CrossRef]
- Kornberg, A.; Rao, N.N.; Ault-Riche, D. Inorganic polyphosphate: A molecule of many functions. Annu. Rev. Biochem. 1999, 68, 89–125. [Google Scholar] [CrossRef] [PubMed]
- Scoma, E.R.; Da Costa, R.T.; Leung, H.H.; Urquiza, P.; Guitart-Mampel, M.; Hambardikar, V.; Riggs, L.M.; Wong, C.O.; Solesio, M.E. Human Prune Regulates the Metabolism of Mammalian Inorganic Polyphosphate and Bioenergetics. Int. J. Mol. Sci. 2023, 24, 13859. [Google Scholar] [CrossRef]
- Baev, A.Y.; Angelova, P.R.; Abramov, A.Y. Inorganic polyphosphate is produced and hydrolysed in F0F1-ATP synthase of mammalian mitochondria. Biochem. J. 2020, 477, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, D.J.; Rogers, C.E.; Harris, H. Expression of alkaline phosphatase loci in mammalian tissues. Proc. Natl. Acad. Sci. USA 1980, 77, 2857–2860. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.K.; Fonzi, W.A.; Wilkerson, J.; Opheim, D.J. A particulate form of alkaline phosphatase in the yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta 1981, 657, 482–494. [Google Scholar] [CrossRef]
- Giovannini, D.; Touhami, J.; Charnet, P.; Sitbon, M.; Battini, J.L. Inorganic phosphate export by the retrovirus receptor XPR1 in metazoans. Cell Rep. 2013, 3, 1866–1873. [Google Scholar] [CrossRef] [PubMed]
- Mailer, R.K.; Allende, M.; Heestermans, M.; Schweizer, M.; Deppermann, C.; Frye, M.; Pula, G.; Odeberg, J.; Gelderblom, M.; Rose-John, S.; et al. Xenotropic and polytropic retrovirus receptor 1 regulates procoagulant platelet polyphosphate. Blood 2021, 137, 1392–1405. [Google Scholar] [CrossRef] [PubMed]
- Battini, J.L.; Rasko, J.E.; Miller, A.D. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: Possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. USA 1999, 96, 1385–1390. [Google Scholar] [CrossRef] [PubMed]
- Seidlmayer, L.K.; Juettner, V.V.; Kettlewell, S.; Pavlov, E.V.; Blatter, L.A.; Dedkova, E.N. Distinct mPTP activation mechanisms in ischaemia-reperfusion: Contributions of Ca2+, ROS, pH, and inorganic polyphosphate. Cardiovasc. Res. 2015, 106, 237–248. [Google Scholar] [CrossRef]
- Seidlmayer, L.K.; Gomez-Garcia, M.R.; Shiba, T.; Porter, G.A., Jr.; Pavlov, E.V.; Bers, D.M.; Dedkova, E.N. Dual role of inorganic polyphosphate in cardiac myocytes: The importance of polyP chain length for energy metabolism and mPTP activation. Arch. Biochem. Biophys. 2019, 662, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Lemasters, J.J.; Theruvath, T.P.; Zhong, Z.; Nieminen, A.L. Mitochondrial calcium and the permeability transition in cell death. Biochim. Biophys. Acta (BBA) Bioenerg. 2009, 1787, 1395–1401. [Google Scholar] [CrossRef]
- Nickel, K.F.; Labberton, L.; Long, A.T.; Langer, F.; Fuchs, T.A.; Stavrou, E.X.; Butler, L.M.; Renne, T. The polyphosphate/factor XII pathway in cancer-associated thrombosis: Novel perspectives for safe anticoagulation in patients with malignancies. Thromb. Res. 2016, 141 (Suppl. S2), S4–S7. [Google Scholar] [CrossRef]
- Caen, J.; Wu, Q. Hageman factor, platelets and polyphosphates: Early history and recent connection. J. Thromb. Haemost. 2010, 8, 1670–1674. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, F.A.; Lea, C.R.; Oldfield, E.; Docampo, R. Human platelet dense granules contain polyphosphate and are similar to acidocalcisomes of bacteria and unicellular eukaryotes. J. Biol. Chem. 2004, 279, 44250–44257. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.N.; Docampo, R. The role of acidocalcisomes in parasitic protists. J. Eukaryot. Microbiol. 2009, 56, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Nickel, K.F.; Ronquist, G.; Langer, F.; Labberton, L.; Fuchs, T.A.; Bokemeyer, C.; Sauter, G.; Graefen, M.; Mackman, N.; Stavrou, E.X.; et al. The polyphosphate-factor XII pathway drives coagulation in prostate cancer-associated thrombosis. Blood 2015, 126, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Santi, M.J.; Montilla, M.; Carroza, M.A.; Ruiz, F.A. Novel assay for prothrombotic polyphosphates in plasma reveals their correlation with obesity. Thromb. Res. 2016, 144, 53–55. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.J.; Smith, S.A.; Morrissey, J.H. Polyphosphate in thrombosis, hemostasis, and inflammation. Res. Pract. Thromb. Haemost. 2019, 3, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Bondy-Chorney, E.; Abramchuk, I.; Nasser, R.; Holinier, C.; Denoncourt, A.; Baijal, K.; McCarthy, L.; Khacho, M.; Lavallee-Adam, M.; Downey, M. A Broad Response to Intracellular Long-Chain Polyphosphate in Human Cells. Cell Rep. 2020, 33, 108318. [Google Scholar] [CrossRef] [PubMed]
- Muller, F.; Mutch, N.J.; Schenk, W.A.; Smith, S.A.; Esterl, L.; Spronk, H.M.; Schmidbauer, S.; Gahl, W.A.; Morrissey, J.H.; Renne, T. Platelet polyphosphates are proinflammatory and procoagulant mediators in vivo. Cell 2009, 139, 1143–1156. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, J.H.; Smith, S.A. Polyphosphate as modulator of hemostasis, thrombosis, and inflammation. J. Thromb. Haemost. 2015, 13 (Suppl. S1), S92–S97. [Google Scholar] [CrossRef]
- Maas, C.; Renne, T. Coagulation factor XII in thrombosis and inflammation. Blood 2018, 131, 1903–1909. [Google Scholar] [CrossRef]
- Bjorkqvist, J.; Nickel, K.F.; Stavrou, E.; Renne, T. In vivo activation and functions of the protease factor XII. Thromb. Haemost. 2014, 112, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Konrath, S.; Mailer, R.K.; Renne, T. Mechanism, Functions, and Diagnostic Relevance of FXII Activation by Foreign Surfaces. Hamostaseologie 2021, 41, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Kenne, E.; Nickel, K.F.; Long, A.T.; Fuchs, T.A.; Stavrou, E.X.; Stahl, F.R.; Renne, T. Factor XII: A novel target for safe prevention of thrombosis and inflammation. J. Intern. Med. 2015, 278, 571–585. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, G.; Revenko, A.S.; Crosby, J.R.; May, C.; Gao, D.; Zhao, C.; Monia, B.P.; MacLeod, A.R. Inhibition of vascular permeability by antisense-mediated inhibition of plasma kallikrein and coagulation factor 12. Nucleic Acid. Ther. 2013, 23, 175–187. [Google Scholar] [CrossRef]
- Larsson, M.; Rayzman, V.; Nolte, M.W.; Nickel, K.F.; Björkqvist, J.; Jämsä, A.; Hardy, M.P.; Fries, M.; Schmidbauer, S.; Hedenqvist, P.; et al. A Factor XIIa Inhibitory Antibody Provides Thromboprotection in Extracorporeal Circulation Without Increasing Bleeding Risk. Sci. Transl. Med. 2014, 6, 222ra217. [Google Scholar] [CrossRef] [PubMed]
- Oschatz, C.; Maas, C.; Lecher, B.; Jansen, T.; Bjorkqvist, J.; Tradler, T.; Sedlmeier, R.; Burfeind, P.; Cichon, S.; Hammerschmidt, S.; et al. Mast cells increase vascular permeability by heparin-initiated bradykinin formation in vivo. Immunity 2011, 34, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Tucker, E.I.; Pine, M.S.; Sisler, I.; Matafonov, A.; Sun, M.F.; White-Adams, T.C.; Smith, S.A.; Hanson, S.R.; McCarty, O.J.; et al. A role for factor XIIa-mediated factor XI activation in thrombus formation in vivo. Blood 2010, 116, 3981–3989. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Shukla, D.; Suman, K.; Lakshmi, B.J.; Manorama, R.; Kumar, S.; Bhandari, R. Inositol hexakisphosphate kinase 1 maintains hemostasis in mice by regulating platelet polyphosphate levels. Blood 2013, 122, 1478–1486. [Google Scholar] [CrossRef] [PubMed]
- Iwaki, T.; Castellino, F.J. Plasma levels of bradykinin are suppressed in factor XII-deficient mice. Thromb. Haemost. 2006, 95, 1003–1010. [Google Scholar] [CrossRef]
- Smith, S.A.; Choi, S.H.; Collins, J.N.; Travers, R.J.; Cooley, B.C.; Morrissey, J.H. Inhibition of polyphosphate as a novel strategy for preventing thrombosis and inflammation. Blood 2012, 120, 5103–5110. [Google Scholar] [CrossRef]
- Smith, S.A.; Gajsiewicz, J.M.; Morrissey, J.H. Ability of Polyphosphate and Nucleic Acids to Trigger Blood Clotting: Some Observations and Caveats. Front. Med. 2018, 5, 107. [Google Scholar] [CrossRef]
- Akiyama, M.; Crooke, E.; Kornberg, A. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J. Biol. Chem. 1993, 268, 633–639. [Google Scholar] [CrossRef]
- Labberton, L.; Kenne, E.; Long, A.T.; Nickel, K.F.; Di Gennaro, A.; Rigg, R.A.; Hernandez, J.S.; Butler, L.; Maas, C.; Stavrou, E.X.; et al. Neutralising blood-borne polyphosphate in vivo provides safe thromboprotection. Nat. Commun. 2016, 7, 12616. [Google Scholar] [CrossRef] [PubMed]
- Niebuhr, K.; Ebel, F.; Frank, R.; Reinhard, M.; Domann, E.; Carl, U.D.; Walter, U.; Gertler, F.B.; Wehland, J.; Chakraborty, T. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 1997, 16, 5433–5444. [Google Scholar] [CrossRef] [PubMed]
- Bolesch, D.G.; Keasling, J.D. Polyphosphate binding and chain length recognition of Escherichia coli exopolyphosphatase. J. Biol. Chem. 2000, 275, 33814–33819. [Google Scholar] [CrossRef]
- Kleinschnitz, C.; Stoll, G.; Bendszus, M.; Schuh, K.; Pauer, H.U.; Burfeind, P.; Renne, C.; Gailani, D.; Nieswandt, B.; Renne, T. Targeting coagulation factor XII provides protection from pathological thrombosis in cerebral ischemia without interfering with hemostasis. J. Exp. Med. 2006, 203, 513–518. [Google Scholar] [CrossRef]
- Nickel, K.F.; Long, A.T.; Fuchs, T.A.; Butler, L.M.; Renne, T. Factor XII as a Therapeutic Target in Thromboembolic and Inflammatory Diseases. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, P.H.; Xu, X.; Oliveira, F.; Chagas, A.C.; Nascimento, C.R.; Francischetti, I.M.; Juliano, M.A.; Juliano, L.; Scharfstein, J.; Valenzuela, J.G.; et al. Novel family of insect salivary inhibitors blocks contact pathway activation by binding to polyphosphate, heparin, and dextran sulfate. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2759–2770. [Google Scholar] [CrossRef]
- Malik, R.A.; Zhou, J.; Fredenburgh, J.C.; Truong, T.K.; Crosby, J.R.; Revenko, A.S.; Weitz, J.I. Polyphosphate-induced thrombosis in mice is factor XII dependent and is attenuated by histidine-rich glycoprotein. Blood Adv. 2021, 5, 3540–3551. [Google Scholar] [CrossRef] [PubMed]
- MacQuarrie, J.L.; Stafford, A.R.; Yau, J.W.; Leslie, B.A.; Vu, T.T.; Fredenburgh, J.C.; Weitz, J.I. Histidine-rich glycoprotein binds factor XIIa with high affinity and inhibits contact-initiated coagulation. Blood 2011, 117, 4134–4141. [Google Scholar] [CrossRef]
- Malik, R.A.; Liao, P.; Zhou, J.; Hussain, R.; Fredenburgh, J.C.; Hettrick, L.; Revenko, A.S.; Weitz, J.I. Histidine-rich glycoprotein attenuates catheter thrombosis. Blood Adv. 2023, 7, 5651–5660. [Google Scholar] [CrossRef] [PubMed]
- Banno, F.; Kita, T.; Fernandez, J.A.; Yanamoto, H.; Tashima, Y.; Kokame, K.; Griffin, J.H.; Miyata, T. Exacerbated venous thromboembolism in mice carrying a protein S K196E mutation. Blood 2015, 126, 2247–2253. [Google Scholar] [CrossRef] [PubMed]
- Zilberman-Rudenko, J.; Reitsma, S.E.; Puy, C.; Rigg, R.A.; Smith, S.A.; Tucker, E.I.; Silasi, R.; Merkulova, A.; McCrae, K.R.; Maas, C.; et al. Factor XII Activation Promotes Platelet Consumption in the Presence of Bacterial-Type Long-Chain Polyphosphate In Vitro and In Vivo. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 1748–1760. [Google Scholar] [CrossRef] [PubMed]
- Holmstrom, K.M.; Marina, N.; Baev, A.Y.; Wood, N.W.; Gourine, A.V.; Abramov, A.Y. Signalling properties of inorganic polyphosphate in the mammalian brain. Nat. Commun. 2013, 4, 1362. [Google Scholar] [CrossRef] [PubMed]
- Arredondo, C.; Cefaliello, C.; Dyrda, A.; Jury, N.; Martinez, P.; Díaz, I.; Amaro, A.; Tran, H.; Morales, D.; Pertusa, M.; et al. Excessive release of inorganic polyphosphate by ALS/FTD astrocytes causes non-cell-autonomous toxicity to motoneurons. Neuron 2022, 110, 1656–1670.e1612. [Google Scholar] [CrossRef]
- Gray, M.J.; Wholey, W.Y.; Wagner, N.O.; Cremers, C.M.; Mueller-Schickert, A.; Hock, N.T.; Krieger, A.G.; Smith, E.M.; Bender, R.A.; Bardwell, J.C.; et al. Polyphosphate is a primordial chaperone. Mol. Cell 2014, 53, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Jakob, U. Inorganic polyphosphate, a multifunctional polyanionic protein scaffold. J. Biol. Chem. 2019, 294, 2180–2190. [Google Scholar] [CrossRef]
- Angelova, P.R.; Iversen, K.Z.; Teschemacher, A.G.; Kasparov, S.; Gourine, A.V.; Abramov, A.Y. Signal transduction in astrocytes: Localisation and release of inorganic polyphosphate. Glia 2018, 66, 2126–2136. [Google Scholar] [CrossRef]
- Brown, M.R.; Kornberg, A. Inorganic polyphosphate in the origin and survival of species. Proc. Natl. Acad. Sci. USA 2004, 101, 16085–16087. [Google Scholar] [CrossRef]
- Omelon, S.; Georgiou, J.; Henneman, Z.J.; Wise, L.M.; Sukhu, B.; Hunt, T.; Wynnyckyj, C.; Holmyard, D.; Bielecki, R.; Grynpas, M.D. Control of vertebrate skeletal mineralisation by polyphosphates. PLoS ONE 2009, 4, e5634. [Google Scholar] [CrossRef]
- Wang, L.; Fraley, C.D.; Faridi, J.; Kornberg, A.; Roth, R.A. Inorganic polyphosphate stimulates mammalian TOR, a kinase involved in the proliferation of mammary cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11249–11254. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Ruiz, L.; Gonzalez-Garcia, I.; Castro, C.; Brieva, J.A.; Ruiz, F.A. Inorganic polyphosphate and specific induction of apoptosis in human plasma cells. Haematologica 2006, 91, 1180–1186. [Google Scholar] [PubMed]
- Azevedo, C.; Singh, J.; Steck, N.; Hofer, A.; Ruiz, F.A.; Singh, T.; Jessen, H.J.; Saiardi, A. Screening a Protein Array with Synthetic Biotinylated Inorganic Polyphosphate To Define the Human PolyP-ome. ACS Chem. Biol. 2018, 13, 1958–1963. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Livermore, T.; Saiardi, A. Protein polyphosphorylation of lysine residues by inorganic polyphosphate. Mol. Cell 2015, 58, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Neville, N.; Lehotsky, K.; Yang, Z.; Klupt, K.A.; Denoncourt, A.; Downey, M.; Jia, Z. Modification of histidine repeat proteins by inorganic polyphosphate. Cell Rep. 2023, 42, 113082. [Google Scholar] [CrossRef] [PubMed]
- Neville, N.; Lehotsky, K.; Klupt, K.A.; Downey, M.; Jia, Z. Polyphosphate attachment to lysine repeats is a non-covalent protein modification. Mol. Cell 2024, 84, 1802–1810.e4. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Borghi, F.; Su, X.B.; Saiardi, A. On the covalent nature of lysine polyphosphorylation. Mol. Cell 2024, 84, 1811–1815.e3. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Orts, L.; Hohmann, U.; Zhu, J.S.; Hothorn, M. Molecular characterisation of CHAD domains as inorganic polyphosphate-binding modules. Life Sci. Alliance 2019, 2, e201900385. [Google Scholar] [CrossRef] [PubMed]
- Aschar-Sobbi, R.; Abramov, A.Y.; Diao, C.; Kargacin, M.E.; Kargacin, G.J.; French, R.J.; Pavlov, E. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J. Fluoresc. 2008, 18, 859–866. [Google Scholar] [CrossRef]
- Angelova, P.R.; Agrawalla, B.K.; Elustondo, P.A.; Gordon, J.; Shiba, T.; Abramov, A.Y.; Chang, Y.T.; Pavlov, E.V. In situ investigation of mammalian inorganic polyphosphate localisation using novel selective fluorescent probes JC-D7 and JC-D8. ACS Chem. Biol. 2014, 9, 2101–2110. [Google Scholar] [CrossRef]
- Fernandes-Cunha, G.M.; McKinlay, C.J.; Vargas, J.R.; Jessen, H.J.; Waymouth, R.M.; Wender, P.A. Delivery of Inorganic Polyphosphate into Cells Using Amphipathic Oligocarbonate Transporters. ACS Central Sci. 2018, 4, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Labberton, L.; Long, A.T.; Gendler, S.J.; Snozek, C.L.; Stavrou, E.X.; Nickel, K.F.; Maas, C.; Blankenberg, S.; Hernandez, J.S.; Renne, T. A Flow Cytometry-Based Assay for Procoagulant Platelet Polyphosphate. Cytom. B Clin. Cytom. 2018, 94, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Ohtomo, R.; Kuga-Uetake, Y.; Aono, T.; Saito, M. Direct Labeling of Polyphosphate at the Ultrastructural Level in Saccharomyces cerevisiae by Using the Affinity of the Polyphosphate Binding Domain of Escherichia coli Exopolyphosphatase. Appl. Environ. Microbiol. 2005, 71, 5692–5701. [Google Scholar] [CrossRef] [PubMed]
- Tumlirsch, T.; Jendrossek, D. Proteins with CHADs (Conserved Histidine α-Helical Domains) Are Attached to Polyphosphate Granules In Vivo and Constitute a Novel Family of Polyphosphate-Associated Proteins (Phosins). Appl. Environ. Microbiol. 2017, 83, e03399-16. [Google Scholar] [CrossRef] [PubMed]
- Serafim, L.S.; Lemos, P.C.; Levantesi, C.; Tandoi, V.; Santos, H.; Reis, M.A. Methods for detection and visualisation of intracellular polymers stored by polyphosphate-accumulating microorganisms. J. Microbiol. Methods 2002, 51, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Gunther, S.; Trutnau, M.; Kleinsteuber, S.; Hause, G.; Bley, T.; Roske, I.; Harms, H.; Muller, S. Dynamics of polyphosphate-accumulating bacteria in wastewater treatment plant microbial communities detected via DAPI (4’,6’-diamidino-2-phenylindole) and tetracycline labeling. Appl. Environ. Microbiol. 2009, 75, 2111–2121. [Google Scholar] [CrossRef] [PubMed]
- Gunther, S.; Hubschmann, T.; Rudolf, M.; Eschenhagen, M.; Roske, I.; Harms, H.; Muller, S. Fixation procedures for flow cytometric analysis of environmental bacteria. J. Microbiol. Methods 2008, 75, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Lee, H.J.; Kim, S.Y.; Song, J.J.; Park, W.; Jeon, C.O. Analysis of the Fine-Scale Population Structure of “Accumulibacter phosphatis” in Enhanced Biological Phosphorus Removal Sludge, Using Fluorescence Hybridization and Flow Cytometric Sorting. Appl. Environ. Microb. 2010, 76, 3825–3835. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, V.; Nautiyal, C.S. A high throughput method and culture medium for rapid screening of phosphate accumulating microorganisms. Bioresour. Technol. 2011, 102, 8057–8062. [Google Scholar] [CrossRef]
- Jimenez-Nunez, M.D.; Moreno-Sanchez, D.; Hernandez-Ruiz, L.; Benitez-Rondan, A.; Ramos-Amaya, A.; Rodriguez-Bayona, B.; Medina, F.; Brieva, J.A.; Ruiz, F.A. Myeloma cells contain high levels of inorganic polyphosphate which is associated with nucleolar transcription. Haematologica 2012, 97, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Cosmidis, J.; Benzerara, K.; Menguy, N.; Arning, E. Microscopy evidence of bacterial microfossils in phosphorite crusts of the Peruvian shelf: Implications for phosphogenesis mechanisms. Chem. Geol. 2013, 359, 10–22. [Google Scholar] [CrossRef]
- Ge, H.Q.; Batstone, D.J.; Keller, J. Biological phosphorus removal from abattoir wastewater at very short sludge ages mediated by novel PAO clade. Water Res. 2015, 69, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Rivadeneyra, A.; Gonzalez-Martinez, A.; Gonzalez-Lopez, J.; Martin-Ramos, D.; Martinez-Toledo, M.V.; Rivadeneyra, M.A. Precipitation of Phosphate Minerals by Microorganisms Isolated from a Fixed-Biofilm Reactor Used for the Treatment of Domestic Wastewater. Int. J. Environ. Res. Public Health 2014, 11, 3689–3704. [Google Scholar] [CrossRef] [PubMed]
- Majed, N.; Li, Y.Y.; Gu, A.Z. Advances in techniques for phosphorus analysis in biological sources. Curr. Opin. Biotech. 2012, 23, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Schuster, K.C.; Urlaub, E.; Gapes, J.R. Single-cell analysis of bacteria by Raman microscopy: Spectral information on the chemical composition of cells and on the heterogeneity in a culture. J. Microbiol. Methods 2000, 42, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Majed, N.; Matthaus, C.; Diem, M.; Gu, A.Z. Evaluation of intracellular polyphosphate dynamics in enhanced biological phosphorus removal process using Raman microscopy. Environ. Sci. Technol. 2009, 43, 5436–5442. [Google Scholar] [CrossRef] [PubMed]
- Ohtomo, R.; Sekiguchi, Y.; Kojima, T.; Saito, M. Different chain length specificity among three polyphosphate quantification methods. Anal. Biochem. 2008, 383, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Hupfer, M.; Glöss, S.; Schmieder, P.; Grossart, H.P. Methods for detection and quantification of polyphosphate and polyphosphate accumulating microorganisms in aquatic sediments. Int. Rev. Hydrobiol. 2008, 93, 1–30. [Google Scholar] [CrossRef]
- Comolli, L.R.; Kundmann, M.; Downing, K.H. Characterization of intact subcellular bodies in whole bacteria by cryo-electron tomography and spectroscopic imaging. J. Microsc. 2006, 223, 40–52. [Google Scholar] [CrossRef]
- Forbes, C.M.; O’Leary, N.D.; Dobson, A.D.; Marchesi, J.R. The contribution of ‘omic’-based approaches to the study of enhanced biological phosphorus removal microbiology. FEMS Microbiol. Ecol. 2009, 69, 1–15. [Google Scholar] [CrossRef]
- McMahon, K.D.; Dojka, M.A.; Pace, N.R.; Jenkins, D.; Keasling, J.D. Polyphosphate kinase from activated sludge performing enhanced biological phosphorus removal. Appl. Environ. Microb. 2002, 68, 4971–4978. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, P.; Wexler, M.; Bond, P.L. Metaproteomics Provides Functional Insight into Activated Sludge Wastewater Treatment. PLoS ONE 2008, 3, e1778. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).