Expression of Foxtail Millet bZIP Transcription Factor SibZIP67 Enhances Drought Tolerance in Arabidopsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Transcriptome Analysis of Differential Expression Genes
2.3. RNA Extraction and Expression Analysis
2.4. Vector Construction and Plant Transformation
2.5. Phylogenetic Analysis
2.6. Stress Treatments and the Response of the Seeds to Mannitol and ABA
2.7. Statistical Analysis
3. Results
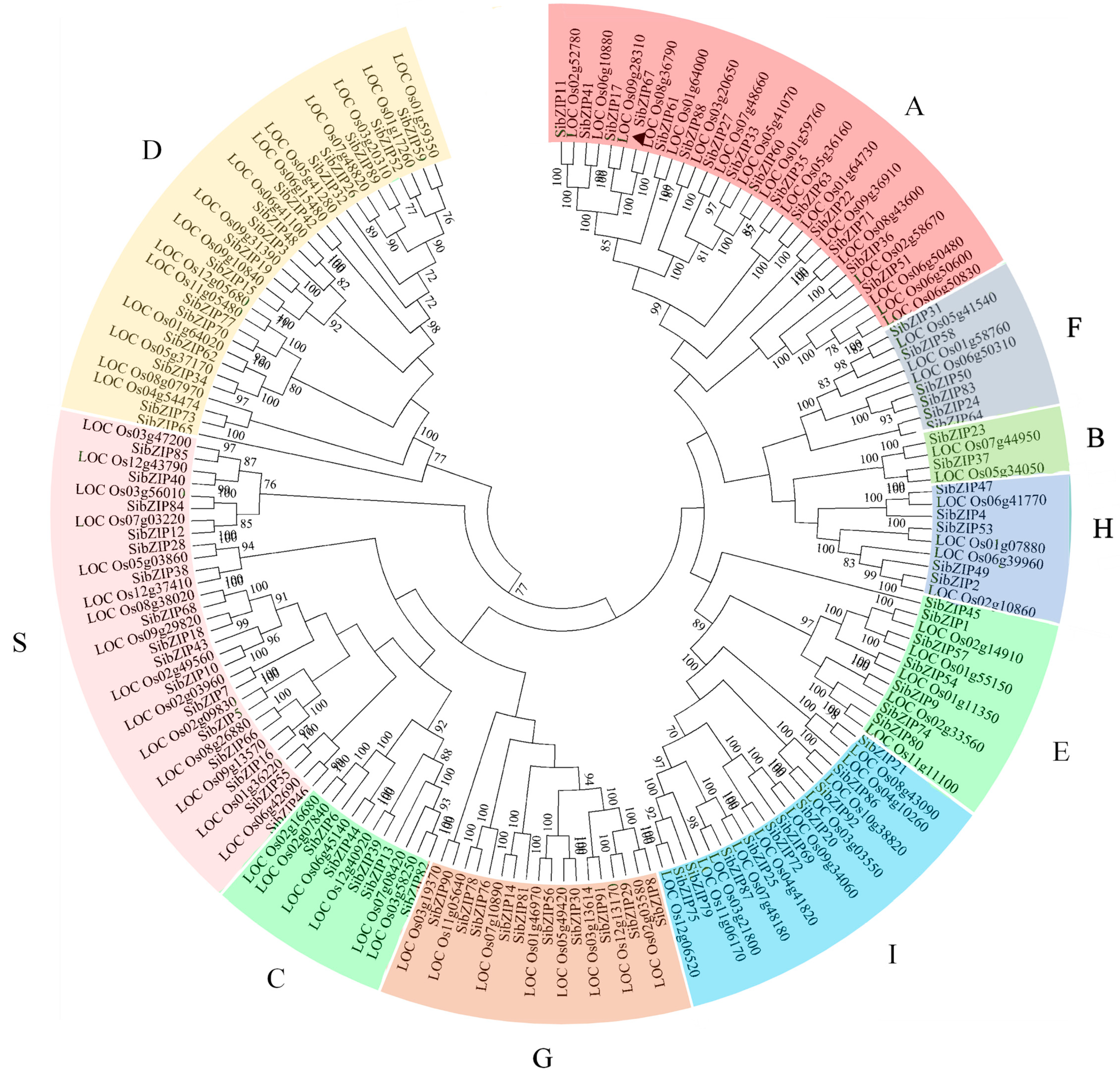
3.1. Isolation and Classification Analysis of SibZIP Genes
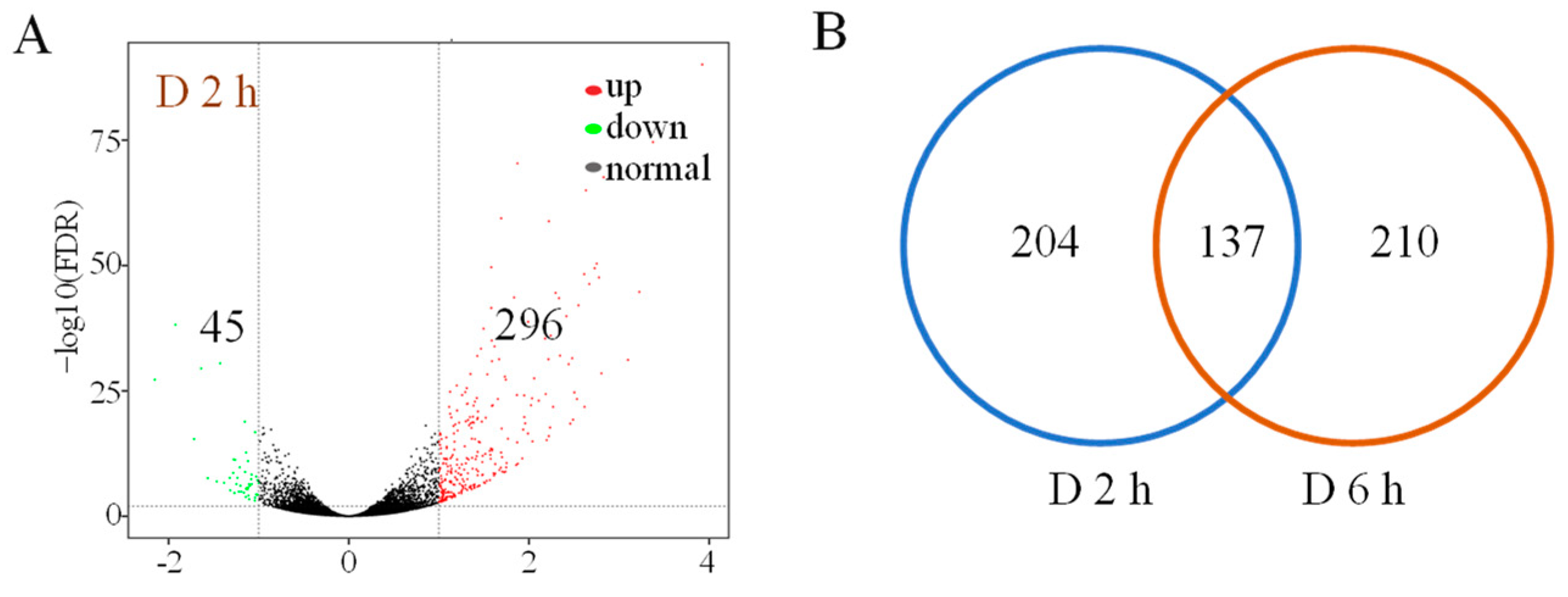
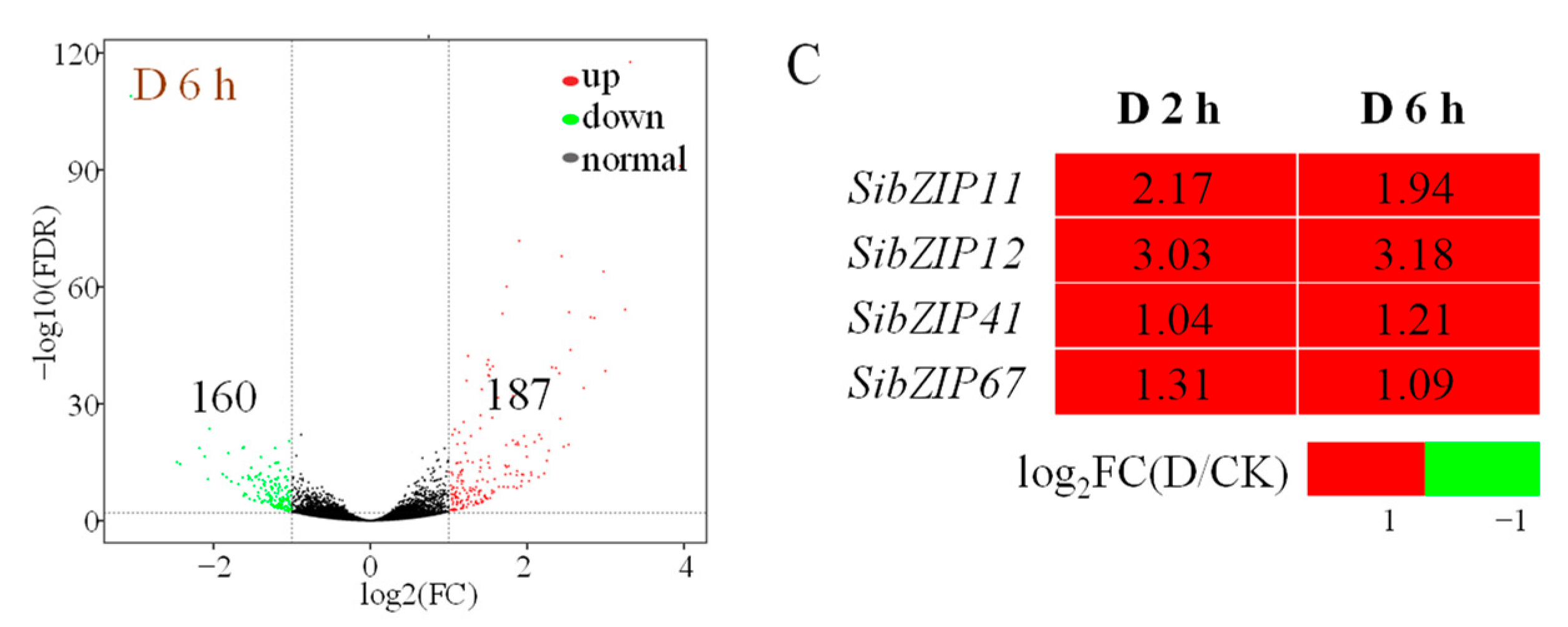
3.2. Differentially Expressed Genes after PEG Treatment in Foxtail Millet
3.3. Bioinformatic and Expression Analysis of the SibZIP67 Gene
3.4. Analysis of Seed Germination and Growth with Exogenous ABA and Osmotic Treatments
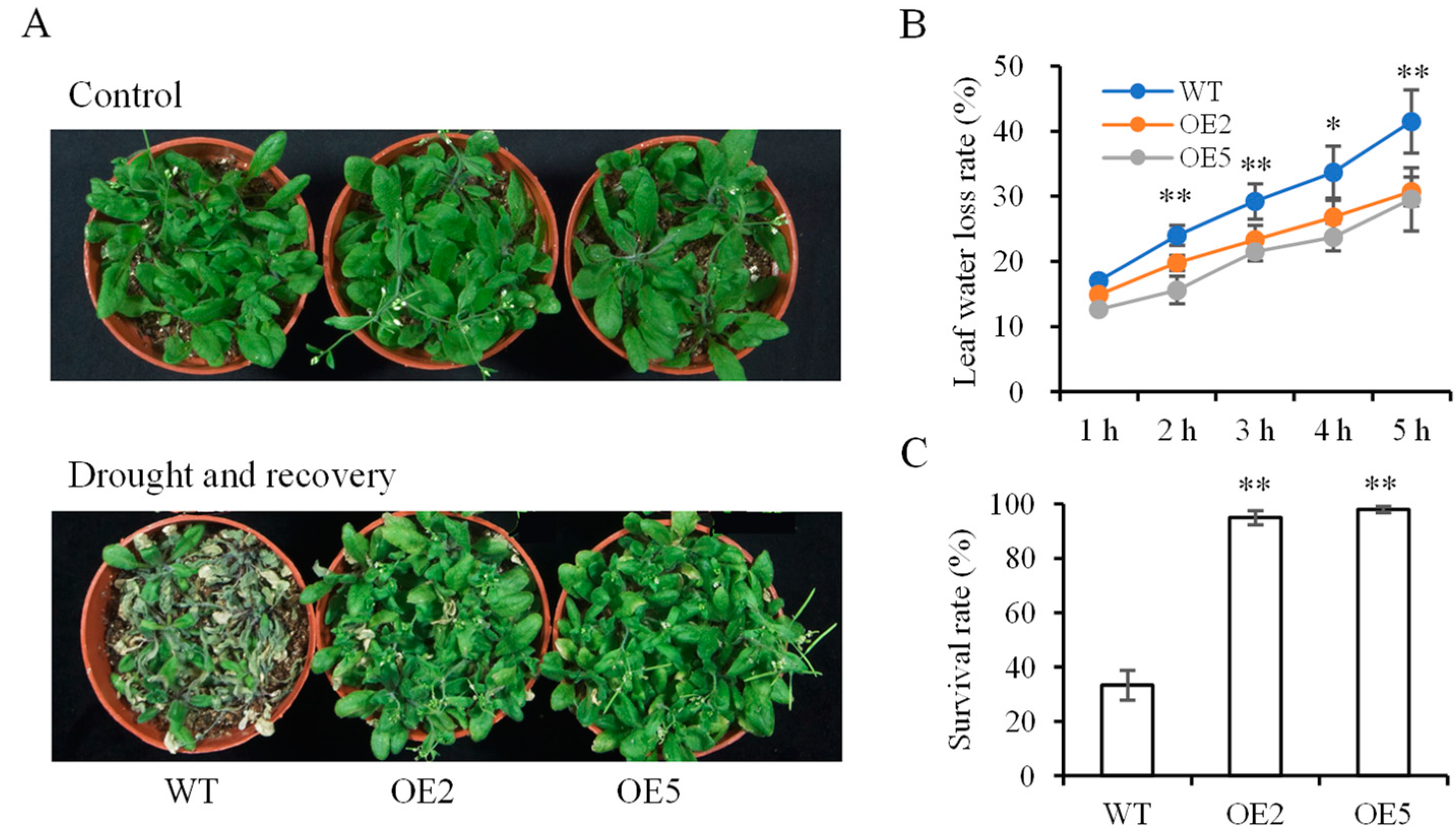
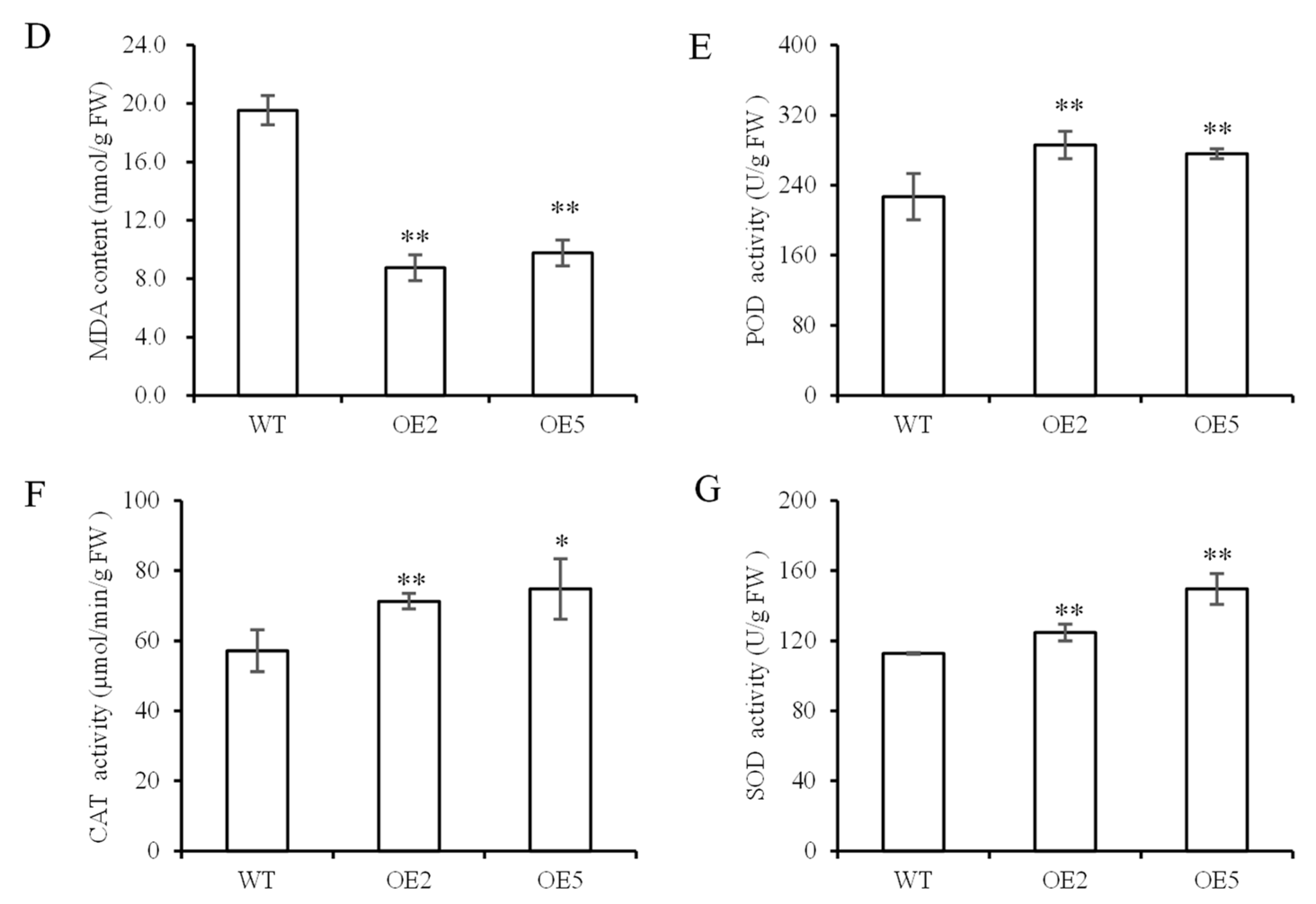
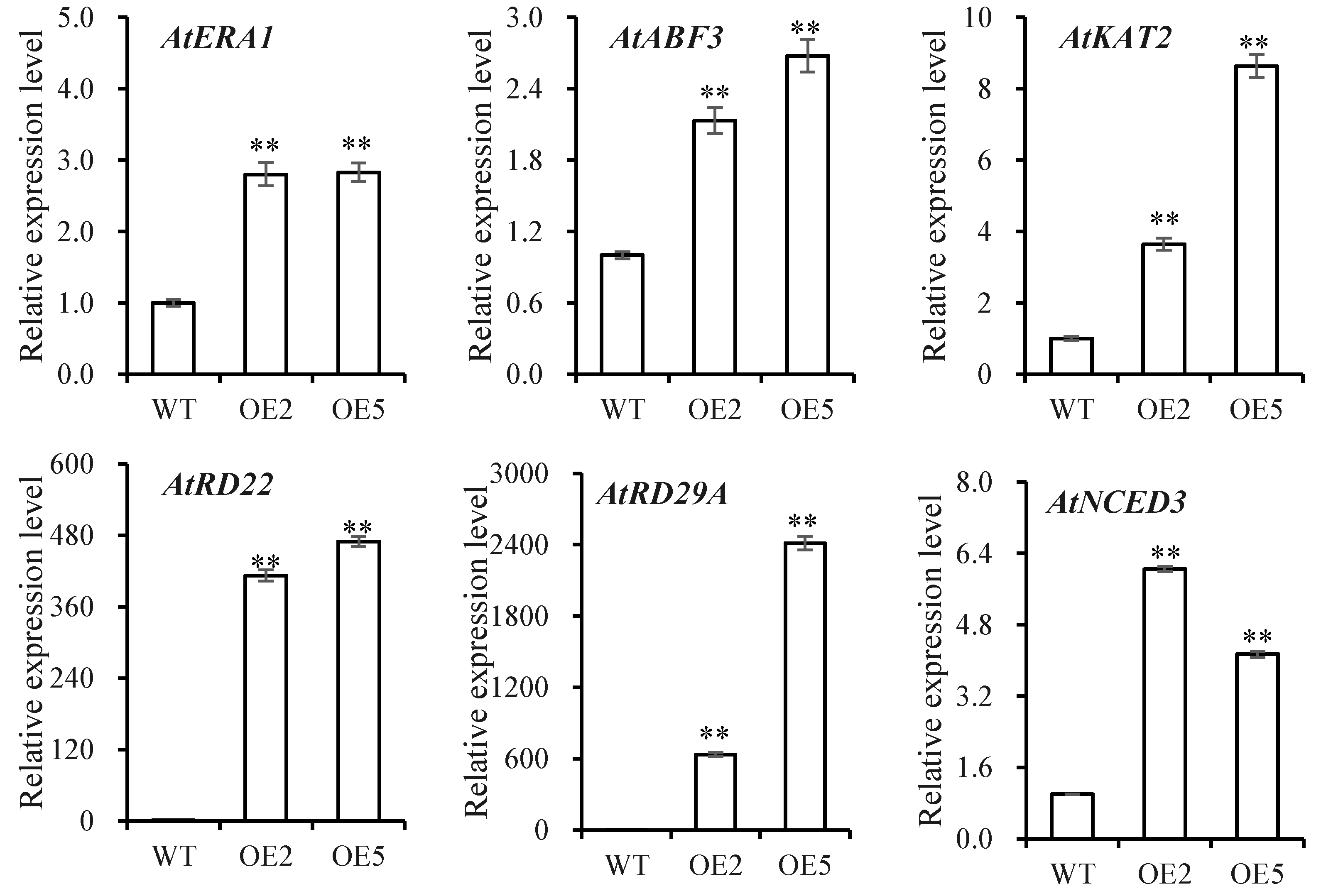
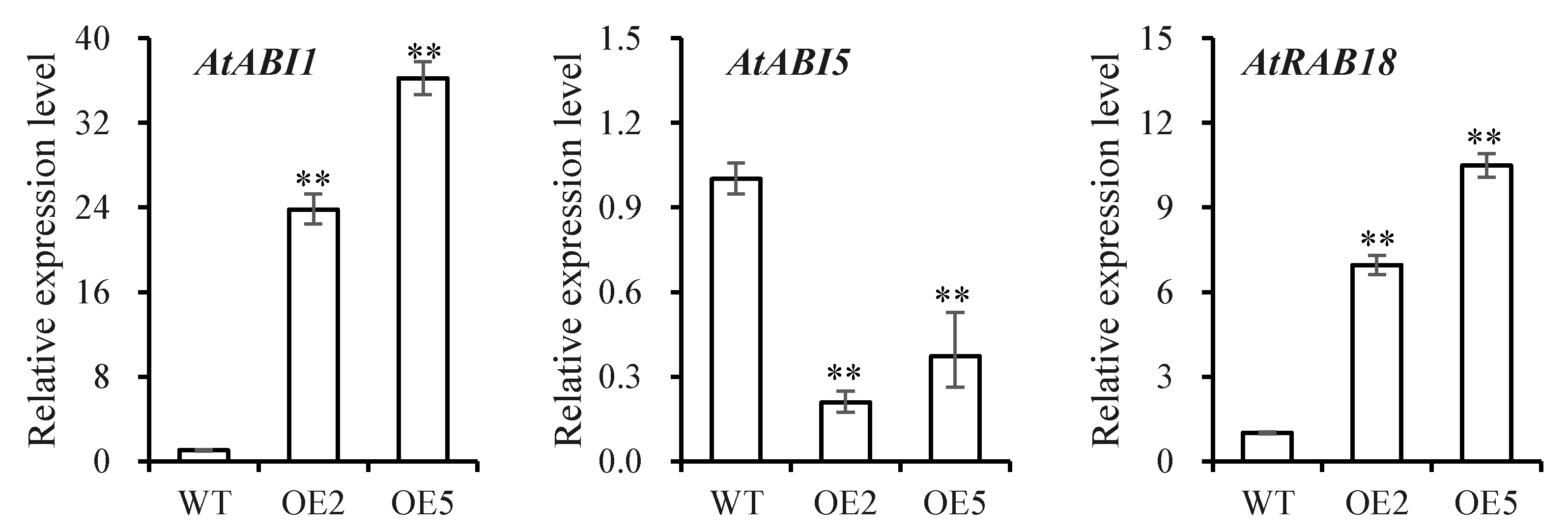
3.5. Analysis of Plants under Drought Stress
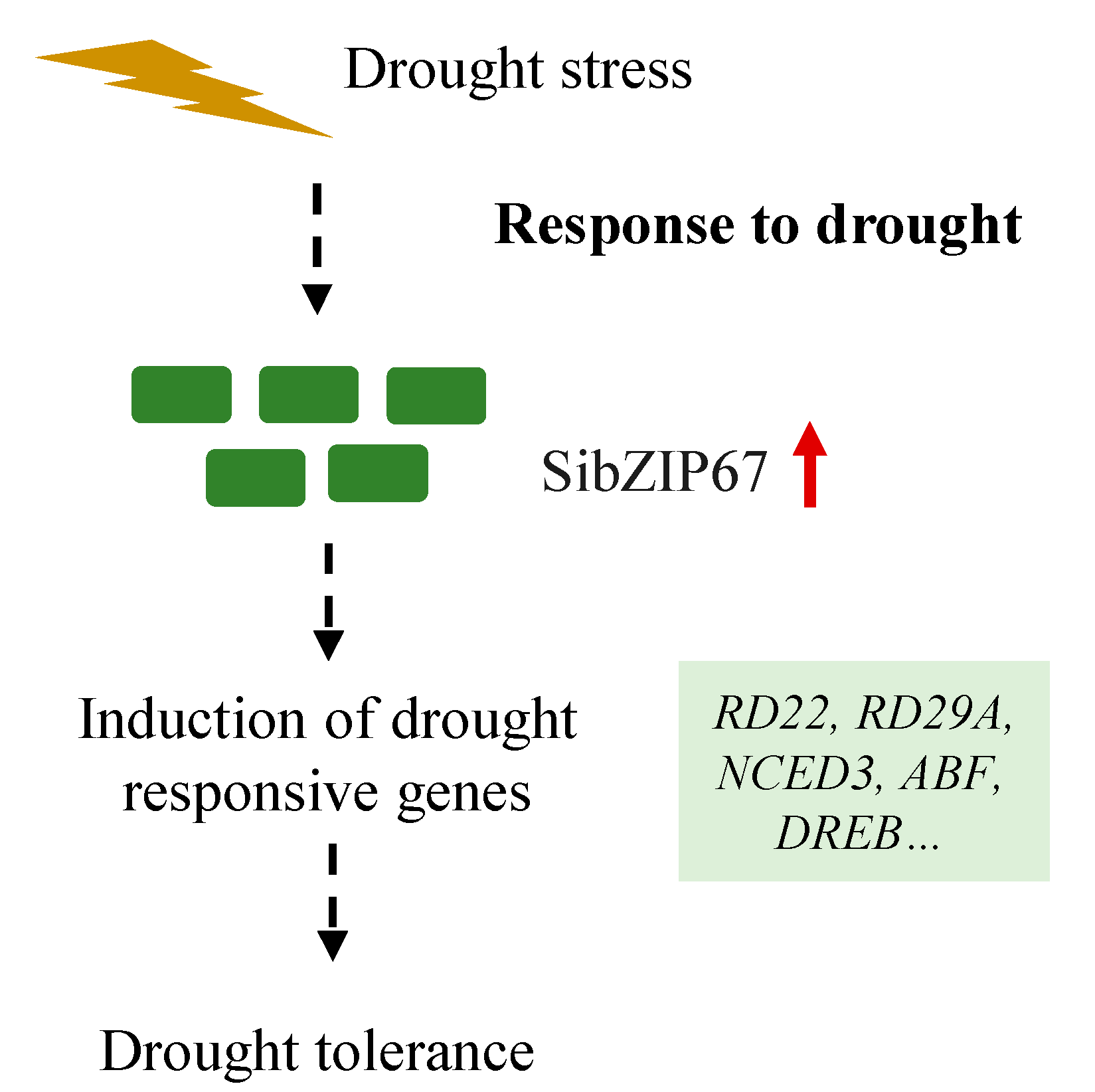
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.; Li, J.; Wang, P.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Nykiel, M.; Gietler, M.; Fidler, J.; Prabucka, B.; Labudda, M. Abiotic stress signaling and responses in plants. Plants 2023, 12, 3405. [Google Scholar] [CrossRef]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef]
- Huang, G.; Ma, S.; Bai, L.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef]
- Strader, L.; Weijers, D.; Wagner, D. Plant transcription factors—Being in the right place with the right company. Curr. Opin. Plant Biol. 2022, 65, 102136. [Google Scholar] [CrossRef]
- Xiong, W.; Xu, X.; Zhang, L.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Genome-wide analysis of the WRKY gene family in physic nut (Jatropha curcas L.). Gene 2013, 524, 124–132. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, 15–45. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- de Souza, C.R.B.; Serrao, C.P.; Barros, N.L.F.; Dos Reis, S.P.; Marques, D.N. Plant bZIP proteins: Potential use in agriculture—A review. Curr. Protein Pept. Sci. 2024, 25, 107–119. [Google Scholar] [CrossRef]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP transcription factors in plant salt stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef]
- Landschulz, W.H.; Johnson, P.F.; Mcknight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, J.; Zhang, B.; Vanitha, J.; Ramachandran, S.; Jiang, S. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J. Integr. Plant Biol. 2011, 53, 212–231. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef]
- Liang, Y.; Xia, J.; Jiang, Y.; Bao, Y.; Chen, H.; Wang, D.; Zhang, D.; Yu, J.; Cang, J. Genome-wide identification and analysis of bZIP gene family and resistance of TaABI5 (TabZIP96) under freezing stress in wheat (Triticum aestivum). Int. J. Mol. Sci. 2022, 23, 2351. [Google Scholar] [CrossRef]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef]
- Droge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Y.; Zhang, S.; Xie, K.; Zhang, C.; Xi, Y.; Sun, F. Genome-wide analysis of the abiotic stress-related bZIP family in switchgrass. Mol. Biol. Rep. 2020, 47, 4439–4454. [Google Scholar] [CrossRef]
- Tu, M.; Wang, X.; Feng, T.; Sun, X.; Wang, Y.; Huang, L.; Gao, M.; Wang, Y.; Wang, X. Expression of a grape (Vitis vinifera) bZIP transcription factor, VlbZIP36, in Arabidopsis thaliana confers tolerance of drought stress during seed germination and seedling establishment. Plant Sci. 2016, 252, 311–323. [Google Scholar] [CrossRef]
- Cao, L.; Lu, X.; Zhang, P.; Wang, G.; Wei, L.; Wang, T. Systematic analysis of differentially expressed maize ZmbZIP genes between drought and rewatering transcriptome reveals bZIP family members involved in abiotic stress responses. Int. J. Mol. Sci. 2019, 20, 4103. [Google Scholar] [CrossRef]
- Lu, G.; Gao, C.; Zheng, X.; Han, B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 2009, 229, 605–615. [Google Scholar] [CrossRef]
- Park, S.H.; Jeong, J.S.; Lee, K.H.; Kim, Y.S.; Choi, Y.D.; Kim, J.K. OsbZIP23 and OsbZIP45, members of the rice basic leucine zipper transcription factor family, are involved in drought tolerance. Plant Biotechnol. Rep. 2015, 9, 89–96. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008, 148, 1938–1952. [Google Scholar] [CrossRef]
- Chen, Y.; Shen, J.; Zhang, L.; Qi, H.; Yang, L.; Wang, H.; Wang, J.; Wang, Y.; Du, H.; Tao, Z.; et al. Nuclear translocation of OsMFT1 that is impeded by OsFTIP1 promotes drought tolerance in rice. Mol. Plant 2021, 14, 1297–1311. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Xia, C.; Zhao, G.; Liu, J.; Jia, J.; Kong, X. A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiol. Plant 2015, 153, 538–554. [Google Scholar] [CrossRef]
- Joo, H.; Baek, W.; Lim, C.W.; Lee, S.C. Post-translational modifications of bZIP transcription factors in abscisic acid signaling and drought responses. Curr. Genomics 2021, 22, 4–15. [Google Scholar] [CrossRef]
- Tang, S.; Li, L.; Wang, Y.; Chen, Q.; Zhang, W.; Jia, G.; Zhi, H.; Zhao, B.; Diao, X. Genotype-specific physiological and transcriptomic responses to drought stress in Setaria italica (an emerging model for Panicoideae grasses). Sci. Rep. 2017, 7, 10009. [Google Scholar] [CrossRef]
- Wang, C.; Chen, J.; Zhi, H.; Yang, L.; Li, W.; Wang, Y.; Li, H.; Zhao, B.; Chen, M.; Diao, X. Population genetics of foxtail millet and its wild ancestor. BMC Genet. 2010, 11, 90. [Google Scholar] [CrossRef]
- Dai, K.; Wang, X.; Liu, H.; Qiao, P.; Wang, J.; Shi, W.; Guo, J.; Diao, X. Efficient identification of QTL for agronomic traits in foxtail millet (Setaria italica) using RTM- and MLM-GWAS. Theor. Appl. Genet. 2024, 137, 18. [Google Scholar] [CrossRef]
- Yi, F.; Huo, M.; Li, J.; Yu, J. Time-series transcriptomics reveals a drought-responsive temporal network and crosstalk between drought stress and the circadian clock in foxtail millet. Plant J. 2022, 110, 1213–1228. [Google Scholar] [CrossRef]
- Cui, X.; Wang, B.; Chen, Z.; Guo, J.; Zhang, T.; Zhang, W.; Shi, L. Comprehensive physiological, transcriptomic, and metabolomic analysis of the key metabolic pathways in millet seedling adaptation to drought stress. Physiol. Plant 2023, 175, e14122. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Khan, Y.; Prasad, M. Dehydration-responsive miRNAs in foxtail millet: Genome-wide identification, characterization and expression profiling. Planta 2016, 243, 749–766. [Google Scholar] [CrossRef]
- Wang, J.; Sun, Z.; Wang, X.; Tang, Y.; Li, X.; Ren, C.; Ren, J.; Wang, X.; Jiang, C.; Zhong, C.; et al. Transcriptome-based analysis of key pathways relating to yield formation stage of foxtail millet under different drought stress conditions. Front. Plant Sci. 2022, 13, 1110910. [Google Scholar] [CrossRef]
- Zhang, R.; Zhi, H.; Li, Y.; Guo, E.; Feng, G.; Tang, S.; Guo, W.; Zhang, L.; Jia, G.; Diao, X. Response of multiple tissues to drought revealed by a weighted gene co-expression network analysis in foxtail millet [Setaria italica (L.) P. Beauv.]. Front. Plant Sci. 2021, 12, 746166. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.; Zhao, J.; Wang, Z.; Cheng, K.; Zhang, P.; Tian, G.; Liu, X.; Guo, E.; Du, Y.; Wang, Y. Transcriptome and metabolite analysis reveal the drought tolerance of foxtail millet significantly correlated with phenylpropanoids-related pathways during germination process under PEG stress. BMC Plant Biol. 2020, 20, 274. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, M.; Chen, Y.; Wu, P.; Wu, G.; Jiang, H. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J. Plant Physiol. 2011, 168, 1952–1959. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, Z.; Zhang, Q.; Wang, R.; Xiao, L.; Ma, H.; Chong, K.; Xu, Y. SKP1 is involved in abscisic acid signalling to regulate seed germination, stomatal opening and root growth in Arabidopsis thaliana. Plant Cell Environ. 2012, 35, 952–965. [Google Scholar] [CrossRef]
- Shi, J.; Yu, L.; Song, B. Proteomics analysis of Xiangcaoliusuobingmi-treated Capsicum annuum L. infected with Cucumber mosaic virus. Pestic. Biochem. Physiol. 2018, 149, 113–122. [Google Scholar] [CrossRef]
- Qin, L.; Chen, E.; Li, F.; Yu, X.; Liu, Z.; Yang, Y.; Wang, R.; Zhang, H.; Wang, H.; Liu, B.; et al. Genome-wide gene expression profiles analysis reveal novel insights into drought stress in foxtail millet (Setaria italica L.). Int. J. Mol. Sci. 2020, 21, 8520. [Google Scholar] [CrossRef]
- Amoah, J.N.; Adu-Gyamfi, M.O.; Kwarteng, A.O. Effect of drought acclimation on antioxidant system and polyphenolic content of foxtail millet (Setaria italica L.). Physiol. Mol. Biol. Plants 2023, 29, 1577–1589. [Google Scholar] [CrossRef]
- Shi, W.; Cheng, J.; Wen, X.; Wang, J.; Shi, G.; Yao, J.; Hou, L.; Sun, Q.; Xiang, P.; Yuan, X.; et al. Transcriptomic studies reveal a key metabolic pathway contributing to a well-maintained photosynthetic system under drought stress in foxtail millet (Setaria italica L.). Peer J. 2018, 6, e4752. [Google Scholar] [CrossRef]
- Zhong, X.; Feng, X.; Li, Y.; Guzmán, C.; Lin, N.; Xu, Q.; Zhang, Y.; Tang, H.; Qi, P.; Deng, M.; et al. Genome-wide identification of bZIP transcription factor genes related to starch synthesis in barley (Hordeum vulgare L.). Genome 2021, 64, 1067–1080. [Google Scholar] [CrossRef]
- Friedrichsen, D.M.; Nemhauser, J.; Muramitsu, T.; Maloof, J.N.; Alonso, J.; Ecker, J.R.; Furuya, M.; Chory, J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genomics 2002, 162, 1445–1456. [Google Scholar] [CrossRef]
- Tang, N.; Zhang, H.; Li, X.; Xiao, J.; Xiong, L. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 2012, 158, 1755–1768. [Google Scholar] [CrossRef]
- Yoon, S.; Lee, D.K.; Yu, I.J.; Kim, Y.S.; Choi, Y.D.; Kim, J.K. Overexpression of the OsbZIP66 transcription factor enhances drought tolerance of rice plants. Plant Biotechnol. Rep. 2017, 11, 53–62. [Google Scholar] [CrossRef]
- Liu, H.; Song, S.; Zhang, H.; Li, Y.; Niu, L.; Zhang, J.; Wang, W. Signaling transduction of ABA, ROS, and Ca2+ in plant stomatal closure in response to drought. Int. J. Mol. Sci. 2022, 23, 14824. [Google Scholar] [CrossRef]
- Hao, G.; Zhang, X.; Wang, Y.; Wu, Z.; Huang, C. Nucleotide variation in the NCED3 region of Arabidopsis thaliana and its association study with abscisic acid content under drought stress. J. Integr. Plant. Biol. 2009, 51, 175–183. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol. Gen. Genet. 1993, 238, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Huang, Y.; Tang, N.; Xiong, L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 2007, 115, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, X.; Gao, H.; Zhang, L.; Tang, W.; Wei, G.; Sun, J.; Xiong, W. Expression of Foxtail Millet bZIP Transcription Factor SibZIP67 Enhances Drought Tolerance in Arabidopsis. Biomolecules 2024, 14, 958. https://doi.org/10.3390/biom14080958
Jia X, Gao H, Zhang L, Tang W, Wei G, Sun J, Xiong W. Expression of Foxtail Millet bZIP Transcription Factor SibZIP67 Enhances Drought Tolerance in Arabidopsis. Biomolecules. 2024; 14(8):958. https://doi.org/10.3390/biom14080958
Chicago/Turabian StyleJia, Xinfeng, Hanchi Gao, Lingxin Zhang, Wei Tang, Guo Wei, Juan Sun, and Wangdan Xiong. 2024. "Expression of Foxtail Millet bZIP Transcription Factor SibZIP67 Enhances Drought Tolerance in Arabidopsis" Biomolecules 14, no. 8: 958. https://doi.org/10.3390/biom14080958
APA StyleJia, X., Gao, H., Zhang, L., Tang, W., Wei, G., Sun, J., & Xiong, W. (2024). Expression of Foxtail Millet bZIP Transcription Factor SibZIP67 Enhances Drought Tolerance in Arabidopsis. Biomolecules, 14(8), 958. https://doi.org/10.3390/biom14080958






