Chitosan-Modified AgNPs Efficiently Inhibit Swine Coronavirus-Induced Host Cell Infections via Targeting the Spike Protein
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation and Characterization of Chi-AgNPs
2.3. Cell Culture and Viruses
2.4. Cell Viability Assay and Hemolysis Assay
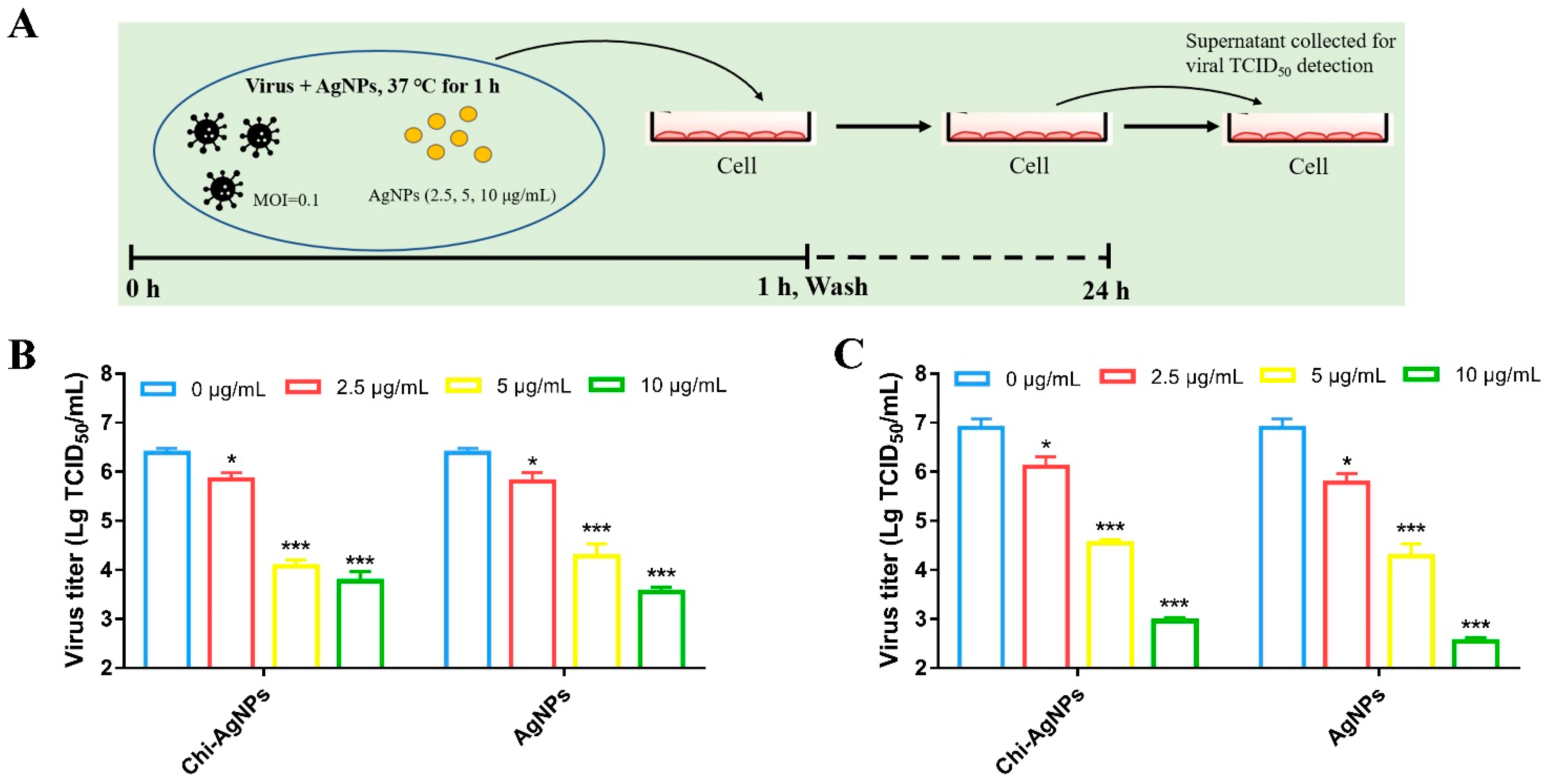
2.5. Measurement of Viral Titer
2.6. The Viricidal Effect of Chi-AgNPs on Virus
2.7. One-Step Growth Curves
2.8. Indirect Immunofluorescence Assay
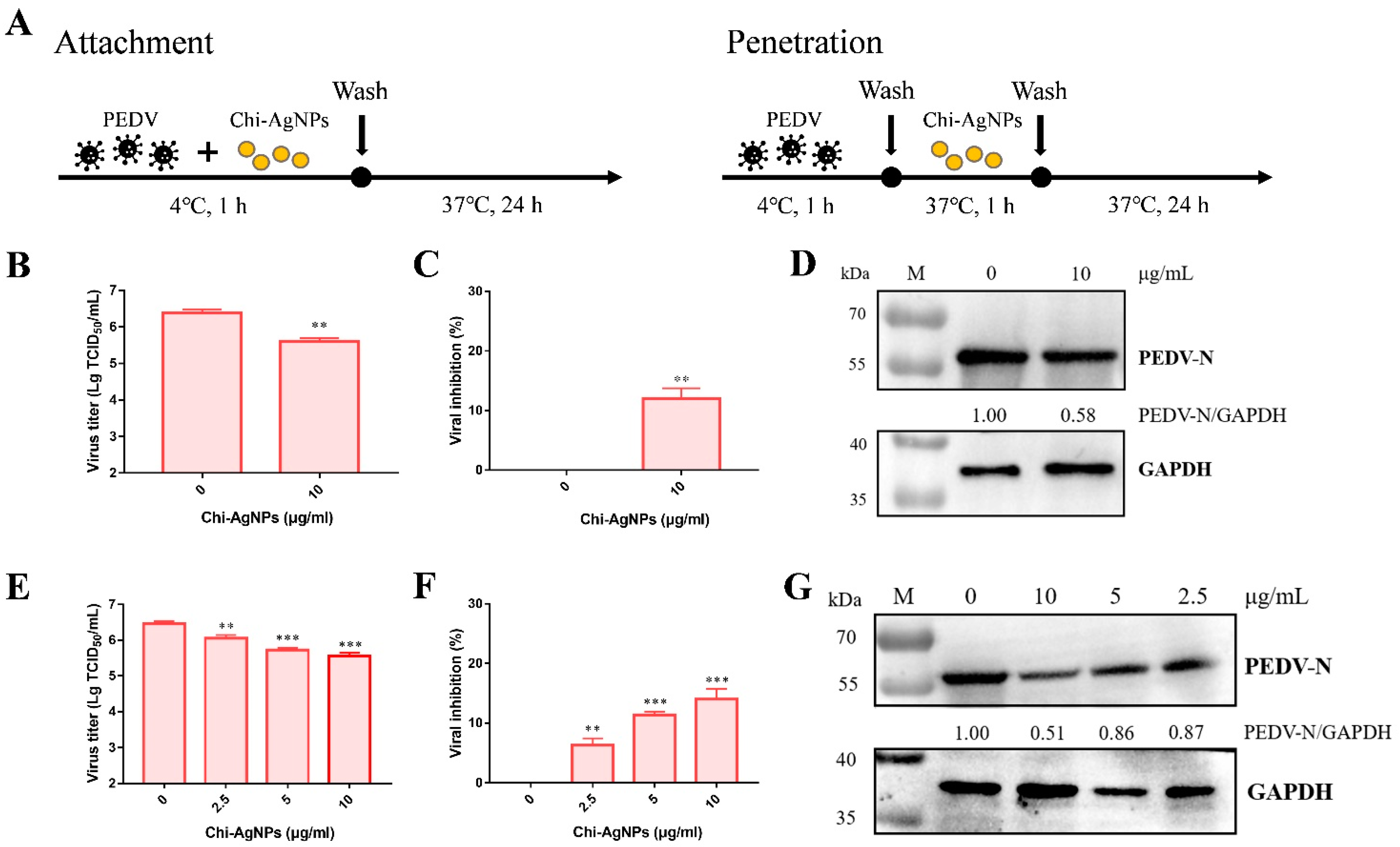
2.9. The Effect of Chi-AgNPs on Virus Attachment and Penetration
2.10. Western Blot
2.11. Determination of ROS Production
2.12. Detection of Mitochondrial Membrane Potential
2.13. Flow Cytometry Analysis of Cell Apoptosis
2.14. TEM Analysis of the Interaction between PEDV and Chi-AgNPs
2.15. Ellman’s Assay
2.16. Analysis of Secondary Structure by Circular Dichroism
2.17. Statistical Analysis
3. Results and Discussion
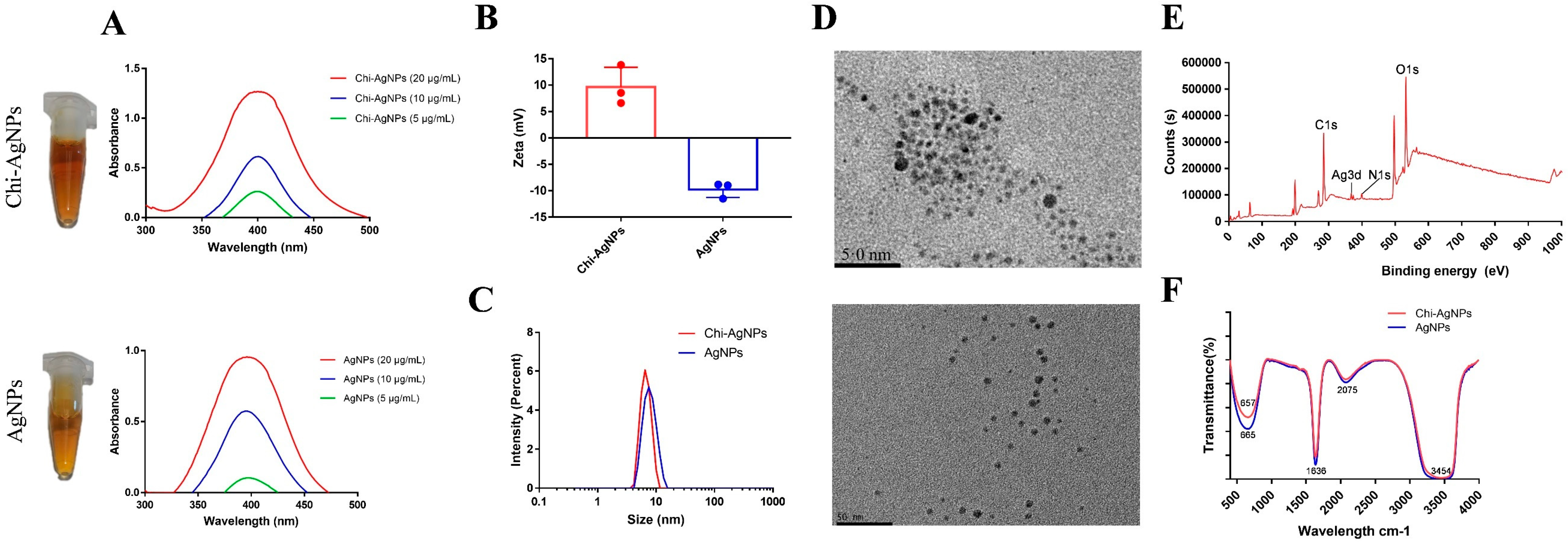
3.1. Characterization of Chi-AgNPs
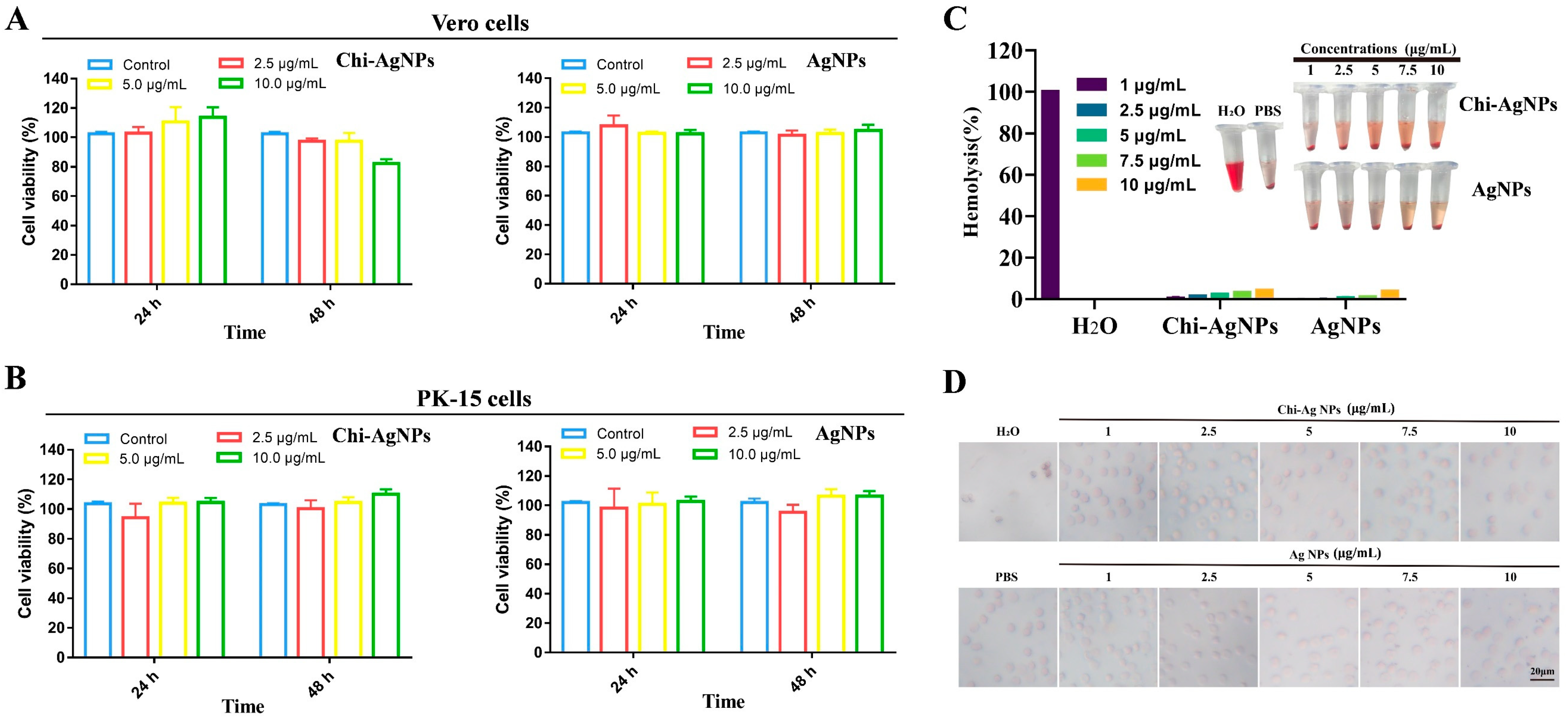
3.2. Biocompatibility Analysis of Chi-AgNPs
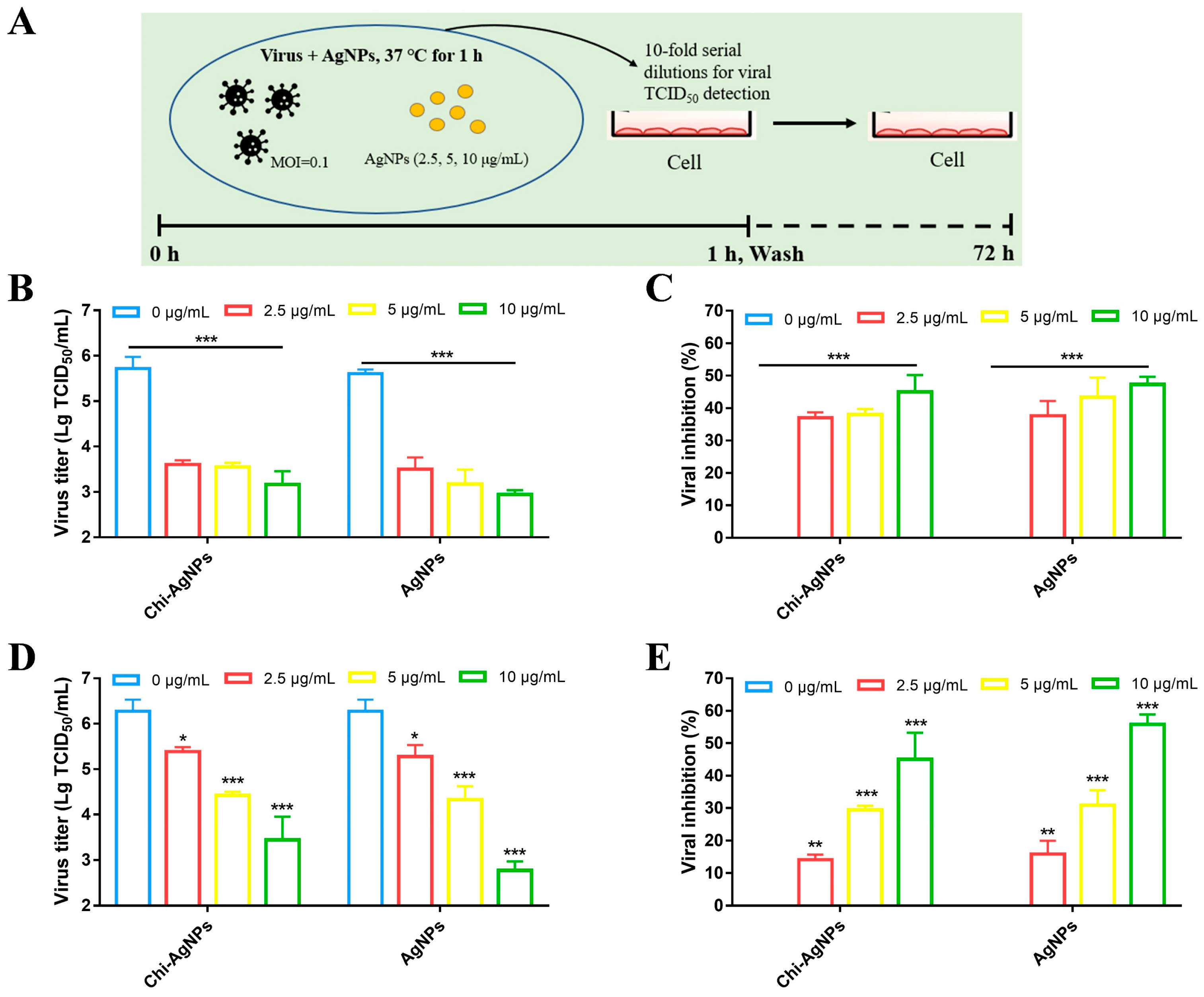
3.3. Chi-AgNPs Antiviral Effect by Directly Targeting Virions
3.4. The Inhibitory Effect of Chi-AgNPs on Virus Replication
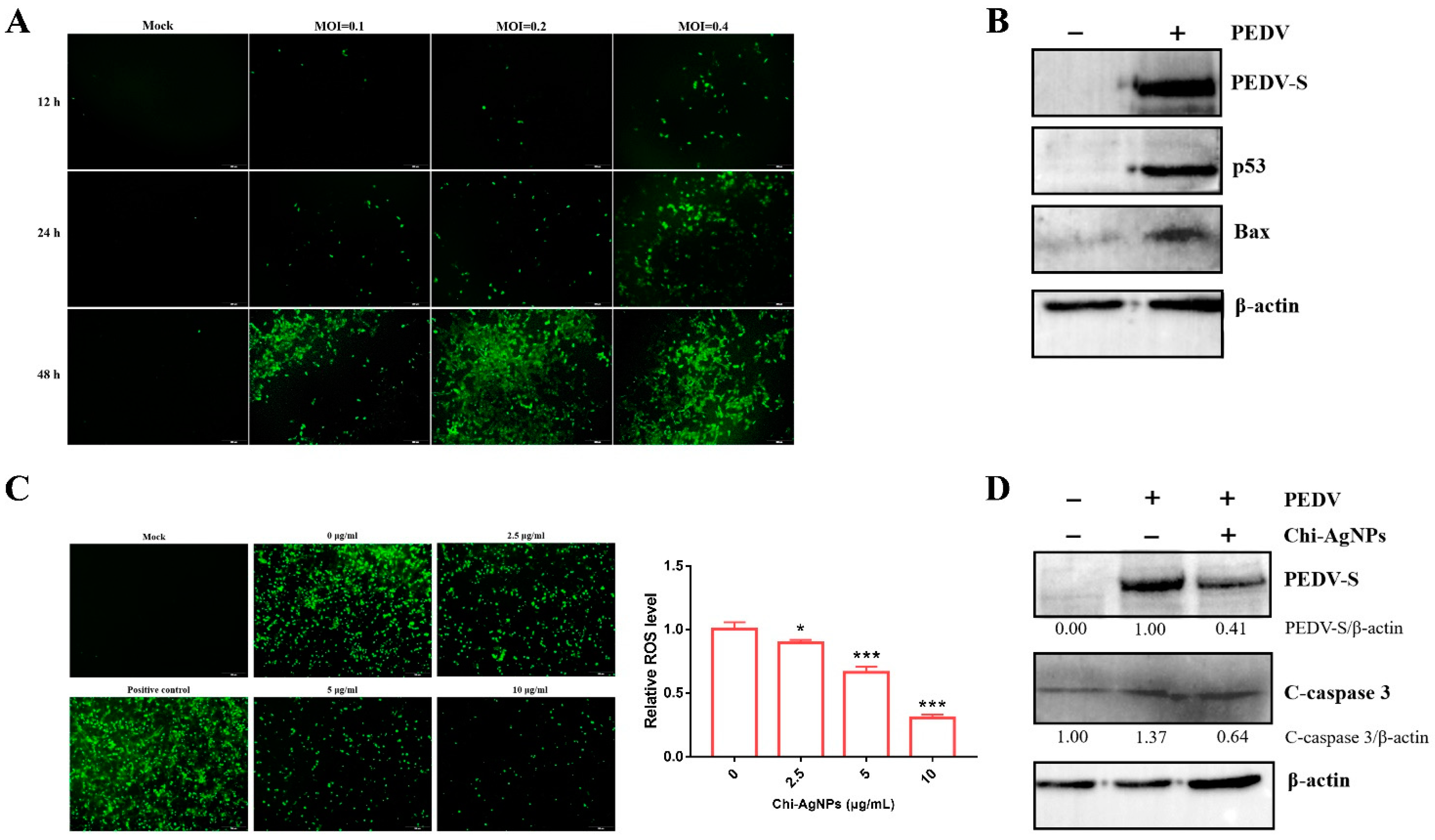
3.5. Chi-AgNPs Exhibit Antiviral Activity of PEDV
3.6. Chi-AgNPs Inhibit PEDV Infection at the Attachment and Penetration Step
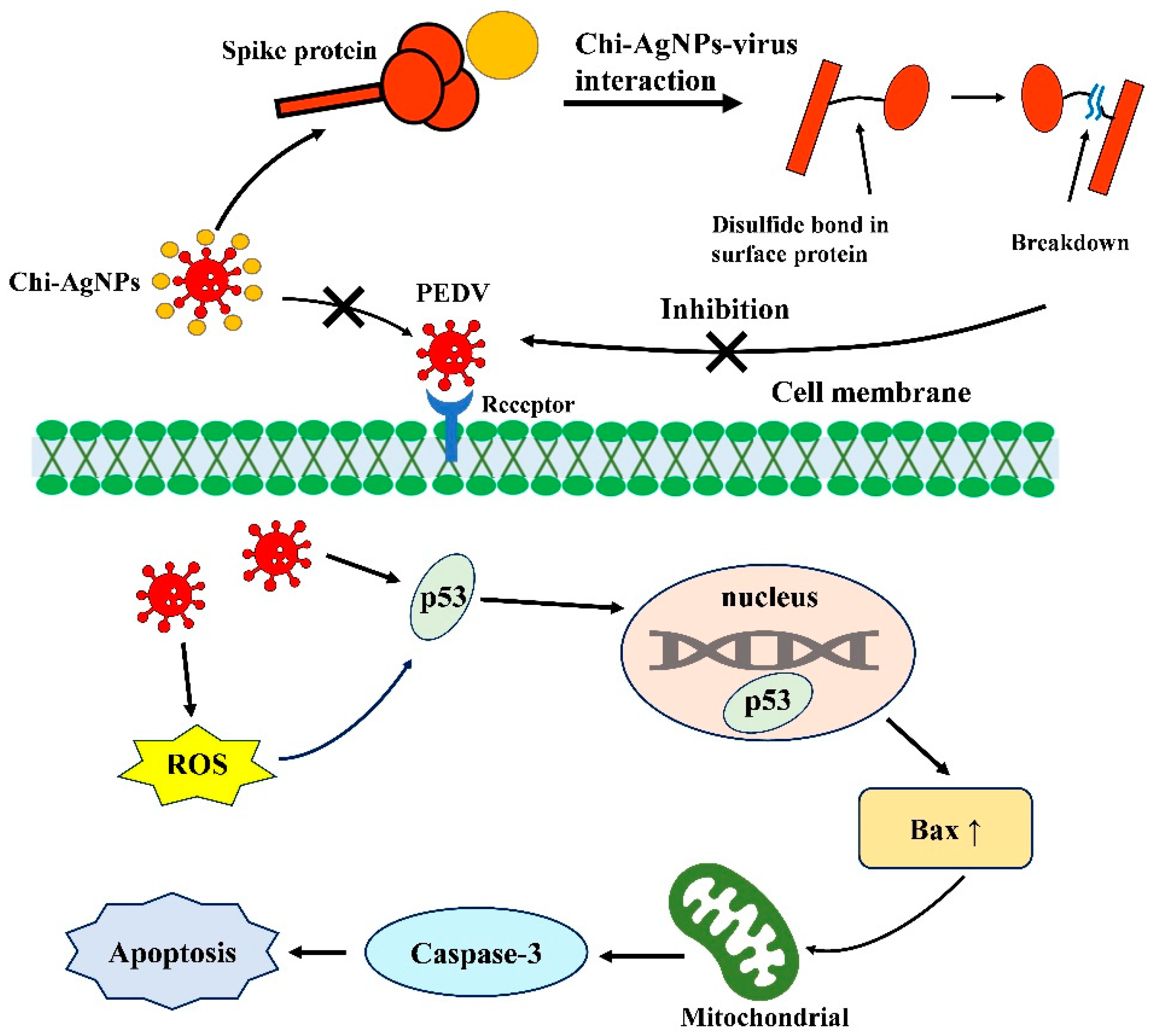
3.7. The Interaction Mechanism between Chi-AgNPs and PEDV
3.8. Chi-AgNPs Reduce PEDV-Induced Apoptosis by Inhibiting ROS Production
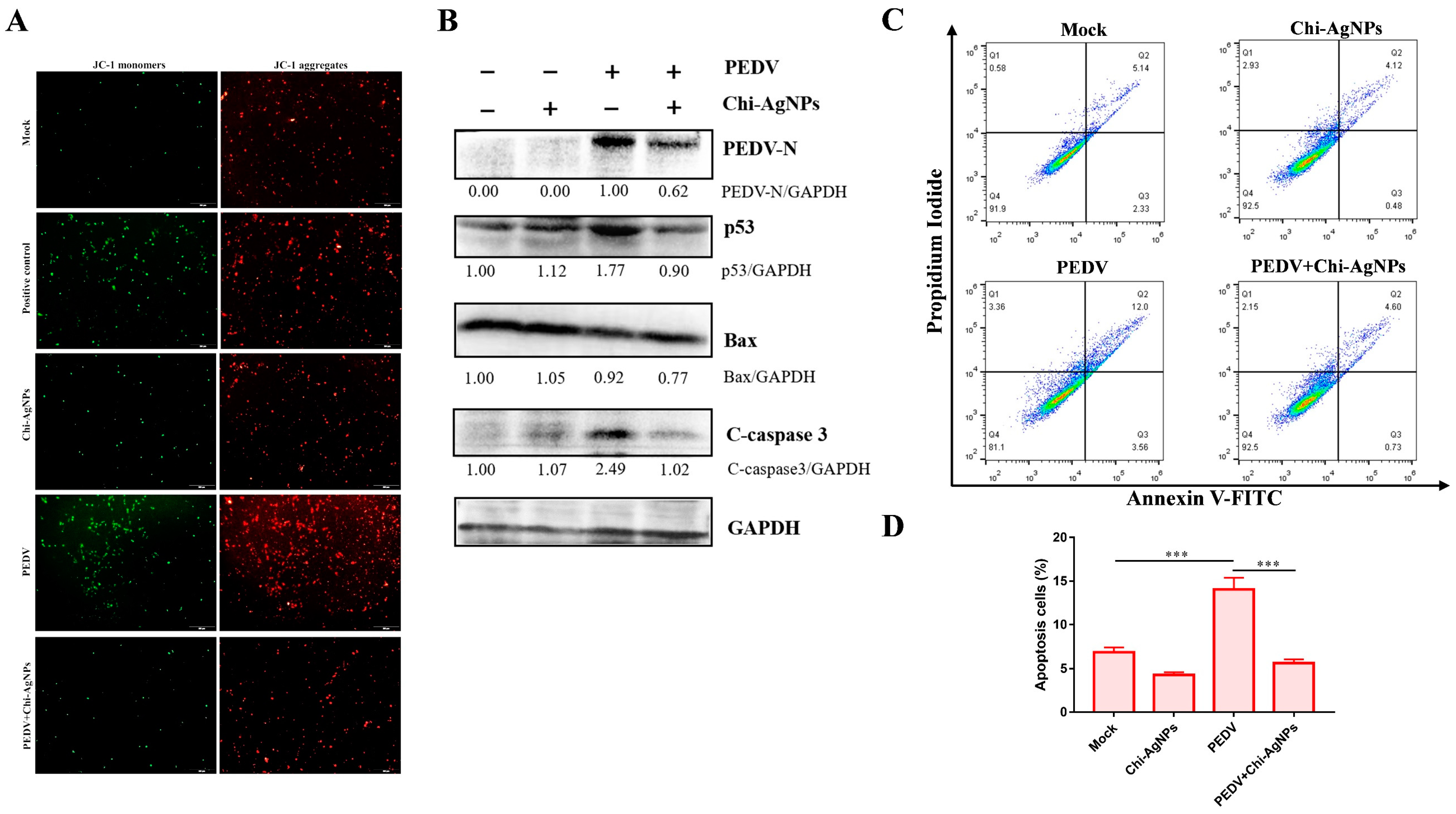
3.9. Chi-AgNPs Inhibit Cell Apoptosis during PEDV Infection via Regulating p53-Mediated Apoptotic Pathway
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Zhang, J.; Li, C.; Song, T.; Li, P.; Liu, J.; Du, X.; Wang, S. Gold/Silver hybrid Nanoparticles with enduring inhibition of coronavirus multiplication through multisite mechanisms. Bioconjugate Chem. 2020, 31, 2553–2563. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.B. Detection of respiratory viruses by molecular methods. Clin. Microbiol. Rev. 2008, 21, 716–747. [Google Scholar] [CrossRef]
- Wu, D.; Wu, T.; Liu, Q.; Yang, Z. The SARS-CoV-2 outbreak: What we know. Int. J. Infect. Dis. 2020, 94, 44–48. [Google Scholar] [CrossRef]
- Wu, J.T.; Leung, K.; Leung, G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. Lancet 2020, 395, 689–697. [Google Scholar] [CrossRef]
- Alphandéry, E. The potential of various nanotechnologies for coronavirus diagnosis/treatment highlighted through a literature analysis. Bioconjugate Chem. 2020, 31, 1873–1882. [Google Scholar] [CrossRef] [PubMed]
- Turlewicz-Podbielska, H.; Pomorska-Mól, M. Porcine coronaviruses: Overview of the state of the art. Virol. Sin. 2021, 36, 833–851. [Google Scholar] [CrossRef]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, H.Y. Porcine enteric coronaviruses: An updated overview of the pathogenesis, prevalence, and diagnosis. Vet. Res. Commun. 2021, 45, 75–86. [Google Scholar] [CrossRef]
- Gerdts, V.; Zakhartchouk, A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet. Microbiol. 2017, 206, 45–51. [Google Scholar] [CrossRef]
- Lin, C.M.; Saif, L.J.; Marthaler, D.; Wang, Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016, 226, 20–39. [Google Scholar] [CrossRef]
- He, W.T.; Bollen, N.; Xu, Y.; Zhao, J.; Dellicour, S.; Yan, Z.; Gong, W.; Zhang, C.; Zhang, L.; Lu, M.; et al. Phylogeography reveals association between swine trade and the spread of porcine epidemic diarrhea virus in China and across the world. Mol. Biol. Evol. 2022, 39, msab364. [Google Scholar] [CrossRef]
- Tang, Z.; Kong, N.; Zhang, X.; Liu, Y.; Hu, P.; Mou, S.; Liljeström, P.; Shi, J.; Tan, W.; Kim, J.S.; et al. A materials-science perspective on tackling COVID-19. Nat. Rev. Mater. 2020, 5, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Bull, R.A.; Adikari, T.N.; Ferguson, J.M.; Hammond, J.M.; Stevanovski, I.; Beukers, A.G.; Naing, Z.; Yeang, M.; Verich, A.; Gamaarachchi, H.; et al. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat. Commun. 2020, 11, 6272. [Google Scholar] [CrossRef]
- Mushtaq, A.; Iqbal, M.Z.; Kong, X. Antiviral effects of coinage metal-based nanomaterials to combat COVID-19 and its variants. J. Mater. Chem. B 2022, 10, 5323–5343. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yeom, M.; Lee, T.; Kim, H.O.; Na, W.; Kang, A.; Lim, J.W.; Park, G.; Park, C.; Song, D.; et al. Porous gold nanoparticles for attenuating infectivity of influenza A virus. J. Nanobiotechnol. 2020, 18, 54. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Ono, T.; Miyahira, Y.; Nguyen, V.Q.; Matsui, T.; Ishihara, M. Antiviral activity of silver nanoparticle/chitosan composites against H1N1 influenza A virus. Nanoscale Res. Lett. 2013, 8, 93. [Google Scholar] [CrossRef]
- Yang, X.X.; Li, C.M.; Huang, C.Z. Curcumin modified silver nanoparticles for highly efficient inhibition of respiratory syncytial virus infection. Nanoscale 2016, 8, 3040–3048. [Google Scholar] [CrossRef] [PubMed]
- Lara, H.H.; Ayala-Nuñez, N.V.; Ixtepan-Turrent, L.; Rodriguez-Padilla, C. Mode of antiviral action of silver nanoparticles against HIV-1. J. Nanobiotechnol. 2010, 8, 1. [Google Scholar] [CrossRef]
- Lu, L.; Sun, R.W.; Chen, R.; Hui, C.K.; Ho, C.M.; Luk, J.M.; Lau, G.K.; Che, C.M. Silver nanoparticles inhibit hepatitis B virus replication. Antivir. Ther. 2008, 13, 253–262. [Google Scholar] [CrossRef]
- Orlowski, P.; Tomaszewska, E.; Gniadek, M.; Baska, P.; Nowakowska, J.; Sokolowska, J.; Nowak, Z.; Donten, M.; Celichowski, G.; Grobelny, J.; et al. Tannic acid modified silver nanoparticles show antiviral activity in herpes simplex virus type 2 infection. PLoS ONE 2014, 9, e104113. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Lu, J.; Liu, L.; Dong, N.; Fang, L.; Xiao, S.; Han, H. Antiviral Activity of graphene oxide-silver nanocomposites by preventing viral entry and activation of the antiviral innate immune response. ACS Appl. Bio Mater. 2018, 1, 1286–1293. [Google Scholar] [CrossRef]
- Milić, M.; Leitinger, G.; Pavičić, I.; Zebić Avdičević, M.; Dobrović, S.; Goessler, W.; Vinković Vrček, I. Cellular uptake and toxicity effects of silver nanoparticles in mammalian kidney cells. J. Appl. Toxicol. 2015, 35, 581–592. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Lu, J.; Liu, N.; Lu, W.; Li, Y.; Shang, C.; Li, X.; Hu, L.; Jiang, G. Antiviral properties of silver nanoparticles against SARS-CoV-2: Effects of surface coating and particle size. Nanomaterials 2022, 12, 990. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Liu, Y.; Luo, X.; Lei, W.; Xie, L. Antiviral and virucidal effects of curcumin on transmissible gastroenteritis virus in vitro. J. Gen. Virol. 2020, 101, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Yazdi, F.; Anbia, M.; Sepehrian, M. Recent advances in removal of inorganic anions from water by chitosan-based composites: A comprehensive review. Carbohydr. Polym. 2023, 320, 121230. [Google Scholar] [CrossRef]
- Eltaweil, A.S.; Omer, A.M.; El-Aqapa, H.G.; Gaber, N.M.; Attia, N.F.; El-Subruiti, G.M.; Mohy-Eldin, M.S.; Abd El-Monaem, E.M. Chitosan based adsorbents for the removal of phosphate and nitrate: A critical review. Carbohydr. Polym. 2021, 274, 118671. [Google Scholar] [CrossRef]
- Li, D.; Gao, X.; Huang, X.; Liu, P.; Xiong, W.; Wu, S.; Hao, F.; Luo, H. Preparation of organic-inorganic chitosan@silver/sepiolite composites with high synergistic antibacterial activity and stability. Carbohydr. Polym. 2020, 249, 116858. [Google Scholar] [CrossRef]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef]
- Rai, M.; Deshmukh, S.D.; Ingle, A.P.; Gupta, I.R.; Galdiero, M.; Galdiero, S. Metal nanoparticles: The protective nanoshield against virus infection. Crit. Rev. Microbiol. 2016, 42, 46–56. [Google Scholar] [CrossRef]
- Wrapp, D.; McLellan, J.S. The 3.1-Angstrom cryo-electron microscopy structure of the porcine epidemic diarrhea virus spike protein in the prefusion conformation. J. Virol. 2019, 93, e00919–e00923. [Google Scholar] [CrossRef]
- Li, W.; van Kuppeveld, F.J.M.; He, Q.; Rottier, P.J.M.; Bosch, B.J. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016, 226, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Grishin, A.M.; Dolgova, N.V.; Landreth, S.; Fisette, O.; Pickering, I.J.; George, G.N.; Falzarano, D.; Cygler, M. Disulfide bonds play a critical role in the structure and function of the receptor-binding domain of the SARS-CoV-2 spike antigen. J. Mol. Biol. 2022, 434, 167357. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xu, Y.; Zhang, Q.; Yang, F.; Yin, Z.; Wang, L.; Li, Q. Porcine epidemic diarrhea virus infections induce apoptosis in Vero cells via a reactive oxygen species (ROS)/p53, but not p38 MAPK and SAPK/JNK signalling pathways. Vet. Microbiol. 2019, 232, 1–12. [Google Scholar] [CrossRef]
- Chen, Y.M.; Burrough, E. The Effects of swine coronaviruses on ER stress, autophagy, apoptosis, and alterations in cell morphology. Pathogens 2022, 11, 940. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin, Z.; Guo, M.; Zhao, M.; Xia, Y.; Wang, C.; Xu, T.; Zhu, B. Inhibition of H1N1 influenza virus-induced apoptosis by functionalized selenium nanoparticles with amantadine through ROS-mediated AKT signaling pathways. Int. J. Nanomed. 2018, 13, 2005–2016. [Google Scholar] [CrossRef]
- Yang, L.; Wang, C.; Shu, J.; Feng, H.; He, Y.; Chen, J.; Shu, J. Porcine epidemic diarrhea virus induces Vero cell apoptosis via the p53-PUMA signaling pathway. Viruses 2021, 13, 1218. [Google Scholar] [CrossRef]
- Jiang, X.; Foldbjerg, R.; Miclaus, T.; Wang, L.; Singh, R.; Hayashi, Y.; Sutherland, D.; Chen, C.; Autrup, H.; Beer, C. Multi-platform genotoxicity analysis of silver nanoparticles in the model cell line CHO-K1. Toxicol. Lett. 2013, 222, 55–63. [Google Scholar] [CrossRef]


| Treatment | α Helix | β-Sheet | Beta-Turn | Rndm. Coil |
|---|---|---|---|---|
| S | 11.8% | 38.0% | 19.0% | 31.2% |
| S + Chi-AgNPs | 7.8% | 48.5% | 17.2% | 26.5% |
| S + TCEP | 6.4% | 53.4% | 13.9% | 26.3% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Yin, C.; Bai, Y.; Zhou, M.; Wang, N.; Tong, C.; Yang, Y.; Liu, B. Chitosan-Modified AgNPs Efficiently Inhibit Swine Coronavirus-Induced Host Cell Infections via Targeting the Spike Protein. Biomolecules 2024, 14, 1152. https://doi.org/10.3390/biom14091152
Wang D, Yin C, Bai Y, Zhou M, Wang N, Tong C, Yang Y, Liu B. Chitosan-Modified AgNPs Efficiently Inhibit Swine Coronavirus-Induced Host Cell Infections via Targeting the Spike Protein. Biomolecules. 2024; 14(9):1152. https://doi.org/10.3390/biom14091152
Chicago/Turabian StyleWang, Dongliang, Caiyun Yin, Yihan Bai, Mingxia Zhou, Naidong Wang, Chunyi Tong, Yi Yang, and Bin Liu. 2024. "Chitosan-Modified AgNPs Efficiently Inhibit Swine Coronavirus-Induced Host Cell Infections via Targeting the Spike Protein" Biomolecules 14, no. 9: 1152. https://doi.org/10.3390/biom14091152
APA StyleWang, D., Yin, C., Bai, Y., Zhou, M., Wang, N., Tong, C., Yang, Y., & Liu, B. (2024). Chitosan-Modified AgNPs Efficiently Inhibit Swine Coronavirus-Induced Host Cell Infections via Targeting the Spike Protein. Biomolecules, 14(9), 1152. https://doi.org/10.3390/biom14091152






