Unraveling the Engagement of Kinases to CRBN Through a Shared Structural Motif to Optimize PROTACs Efficacy
Abstract
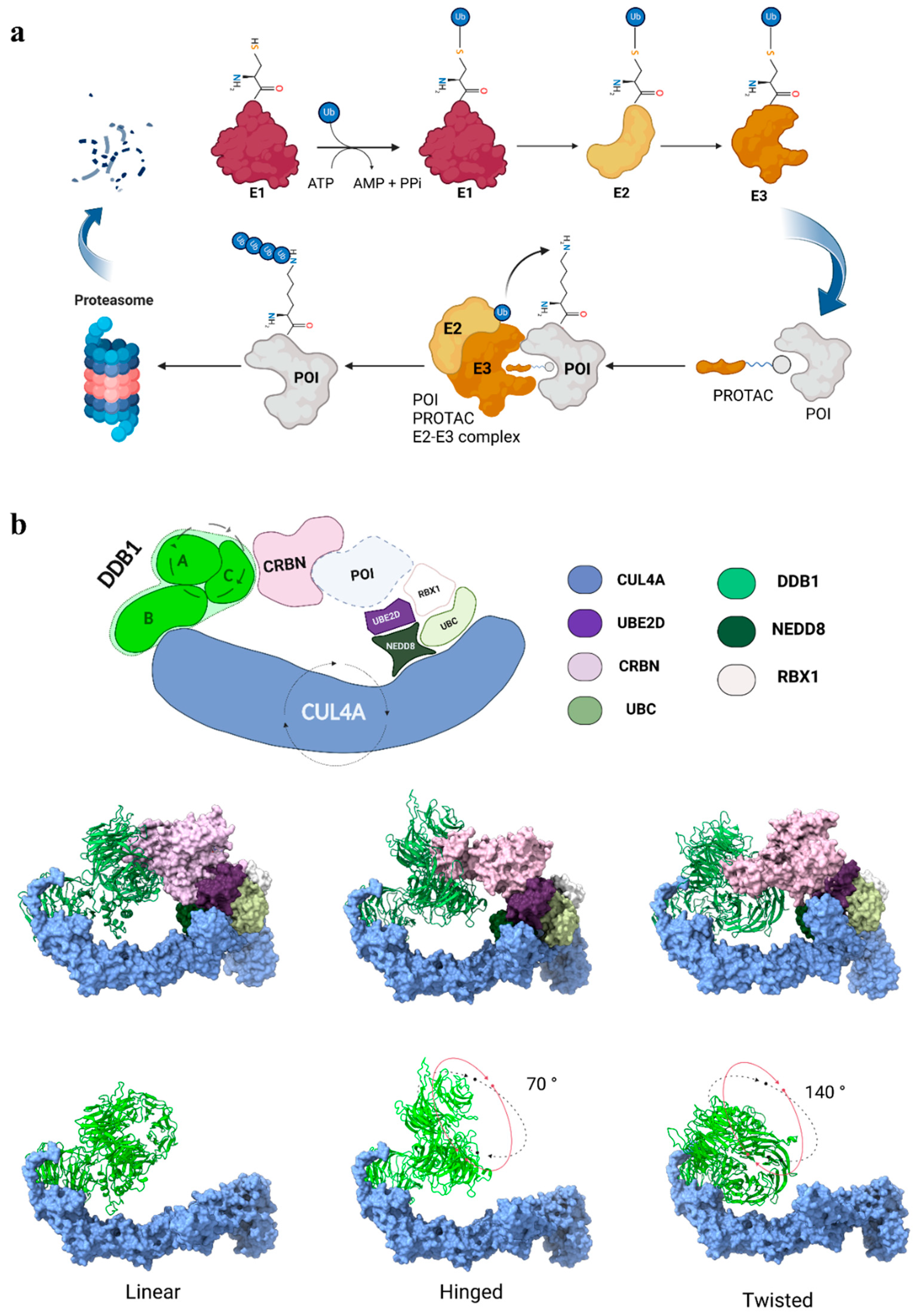
1. Introduction
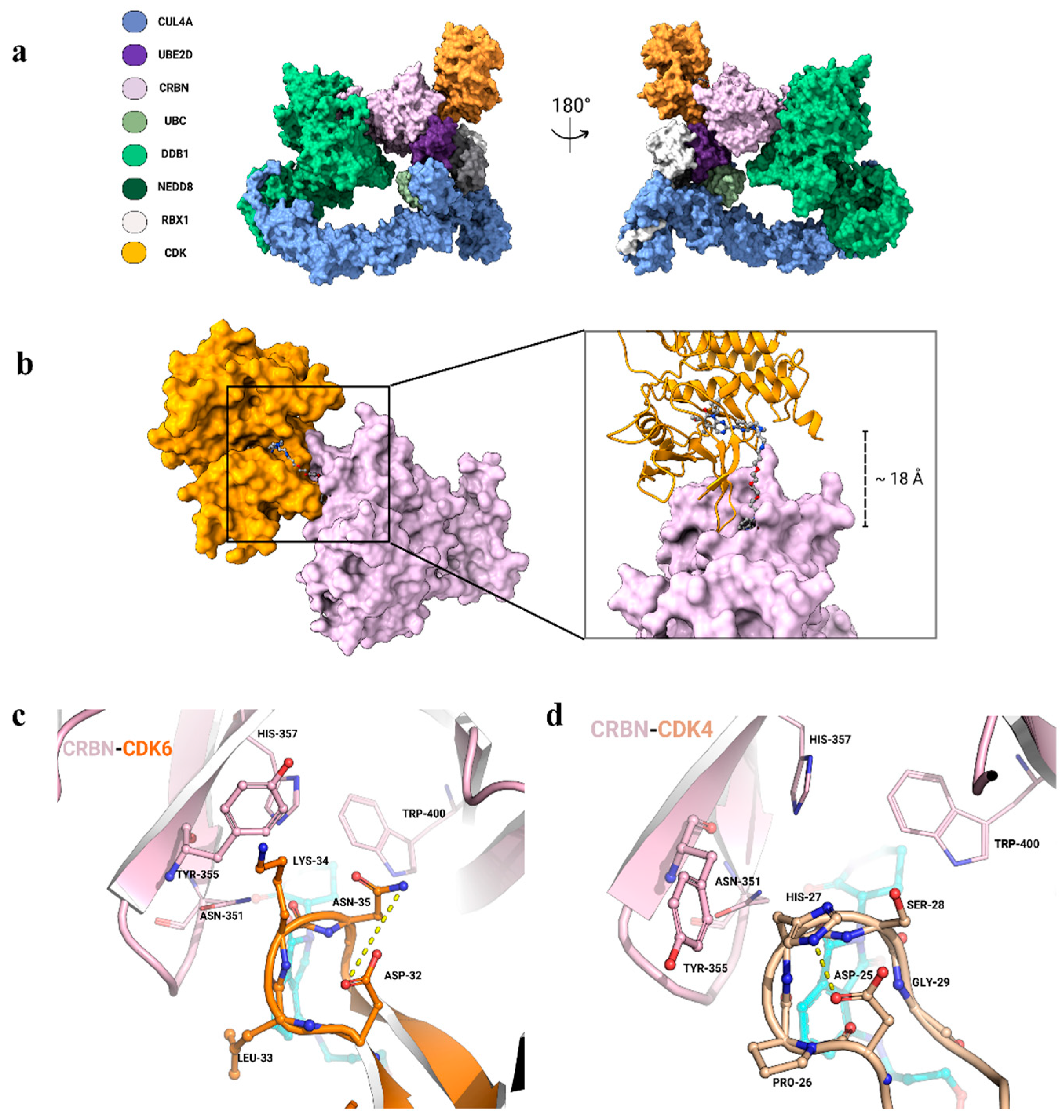
2. Rationalizing Selectivity of CDK Kinases-Targeting PROTACs
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Z.; Hu, M.; Yang, Y.; Du, C.; Zhou, H.; Liu, C.; Chen, Y.; Fan, L.; Ma, H.; Gong, Y.; et al. An overview of PROTACs: A promising drug discovery paradigm. Mol. Biomed. 2022, 3, 46. [Google Scholar] [CrossRef] [PubMed]
- Sincere, N.I.; Anand, K.; Ashique, S.; Yang, J.; You, C. PROTACs: Emerging Targeted Protein Degradation Approaches for Advanced Druggable Strategies. Molecules 2023, 28, 4014. [Google Scholar] [CrossRef] [PubMed]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef]
- Li, K.; Crews, C.M. PROTACs: Past, present and future. Chem. Soc. Rev. 2022, 51, 5214–5236. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liu, M.; Yang, Z.; Li, J.; Gao, Y.; Tan, R. Proteolysis-Targeting Chimeras (PROTACs) in Cancer Therapy: Present and Future. Molecules 2022, 27, 8828. [Google Scholar] [CrossRef]
- Neklesa, T.K.; Winkler, J.D.; Crews, C.M. Targeted protein degradation by PROTACs. Pharmacol. Ther. 2017, 174, 138–144. [Google Scholar] [CrossRef]
- Harper, J.W.; Schulman, B.A. Cullin-RING Ubiquitin Ligase Regulatory Circuits: A Quarter Century Beyond the F-Box Hypothesis. Annu. Rev. Biochem. 2021, 90, 403–429. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Schulman, B.A.; Song, L.; Miller, J.J.; Jeffrey, P.D.; Wang, P.; Chu, C.; Koepp, D.M.; Elledge, S.J.; Pagano, M.; et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 2002, 416, 703–709. [Google Scholar] [CrossRef]
- Hannah, J.; Zhou, P. Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene 2015, 573, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Lian, Q.; Gao, Y.; Li, Q.; He, X.; Jiang, X.; Pu, Z.; Xu, G. Cereblon Promotes the Ubiquitination and Proteasomal Degradation of Interleukin Enhancer-Binding Factor 2. Protein J. 2020, 39, 411–421. [Google Scholar] [CrossRef]
- Duda, D.M.; Borg, L.A.; Scott, D.C.; Hunt, H.W.; Hammel, M.; Schulman, B.A. Structural insights into NEDD8 activation of cullin-RING ligases: Conformational control of conjugation. Cell 2008, 134, 995–1006. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.; Scott, D.C.; Schulman, B.A. NEDD8 and ubiquitin ligation by cullin-RING E3 ligases. Curr. Opin. Struct. Biol. 2021, 67, 101–109. [Google Scholar] [CrossRef]
- Furukawa, M.; Zhang, Y.; McCarville, J.; Ohta, T.; Xiong, Y. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell Biol. 2000, 20, 8185–8197. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, D.T.; Ayrault, O.; Hunt, H.W.; Taherbhoy, A.M.; Duda, D.M.; Scott, D.C.; Borg, L.A.; Neale, G.; Murray, P.J.; Roussel, M.F.; et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell 2009, 33, 483–495. [Google Scholar] [CrossRef]
- Fischer, E.S.; Scrima, A.; Böhm, K.; Matsumoto, S.; Lingaraju, G.M.; Faty, M.; Yasuda, T.; Cavadini, S.; Wakasugi, M.; Hanaoka, F.; et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 2011, 147, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Watson, E.R.; Novick, S.; Matyskiela, M.E.; Chamberlain, P.P.; de la Peña, A.H.; Zhu, J.; Tran, E.; Griffin, P.R.; Wertz, I.E.; Lander, G.C. Molecular glue CELMoD compounds are regulators of cereblon conformation. Science 2022, 378, 549–553. [Google Scholar] [CrossRef]
- Ito, T.; Yamaguchi, Y.; Handa, H. Exploiting ubiquitin ligase cereblon as a target for small-molecule compounds in medicine and chemical biology. Cell Chem. Biol. 2021, 28, 987–999. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, J.; Ito, T.; Yamaguchi, Y.; Handa, H. Discovery of CRBN as a target of thalidomide: A breakthrough for progress in the development of protein degraders. Chem. Soc. Rev. 2022, 51, 6234–6250. [Google Scholar] [CrossRef]
- Klein, V.G.; Bond, A.G.; Craigon, C.; Lokey, R.S.; Ciulli, A. Amide-to-Ester Substitution as a Strategy for Optimizing PROTAC Permeability and Cellular Activity. J. Med. Chem. 2021, 64, 18082–18101. [Google Scholar] [CrossRef]
- Dragovich, P.S.; Pillow, T.H.; Blake, R.A.; Sadowsky, J.D.; Adaligil, E.; Adhikari, P.; Bhakta, S.; Blaquiere, N.; Chen, J.; Cruz-Chuh, J.D.; et al. Antibody-Mediated Delivery of Chimeric BRD4 Degraders. Part 1: Exploration of Antibody Linker, Payload Loading, and Payload Molecular Properties. J. Med. Chem. 2021, 64, 2534–2575. [Google Scholar] [CrossRef]
- Dragovich, P.S.; Pillow, T.H.; Blake, R.A.; Sadowsky, J.D.; Adaligil, E.; Adhikari, P.; Chen, J.; Corr, N.; Cruz-Chuh, J.D.; Del Rosario, G.; et al. Antibody-Mediated Delivery of Chimeric BRD4 Degraders. Part 2: Improvement of In Vitro Antiproliferation Activity and In Vivo Antitumor Efficacy. J. Med. Chem. 2021, 64, 2576–2607. [Google Scholar] [CrossRef]
- Gadd, M.S.; Testa, A.; Lucas, X.; Chan, K.H.; Chen, W.Z.; Lamont, D.J.; Zengerle, M.; Ciulli, A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 2017, 13, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Farnaby, W.; Koegl, M.; Roy, M.J.; Whitworth, C.; Diers, E.; Trainor, N.; Zollman, D.; Steurer, S.; Karolyi-Oezguer, J.; Riedmueller, C.; et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 2019, 15, 672–680. [Google Scholar] [CrossRef]
- Haubrich, K.; Spiteri, V.A.; Farnaby, W.; Sobott, F.; Ciulli, A. Breaking free from the crystal lattice: Structural biology in solution to study protein degraders. Curr. Opin. Struct. Biol. 2023, 79, 102534. [Google Scholar] [CrossRef]
- Bai, N.; Riching, K.M.; Makaju, A.; Wu, H.; Acker, T.M.; Ou, S.-C.; Zhang, Y.; Shen, X.; Bulloch, D.N.; Rui, H.; et al. Modeling the CRL4A ligase complex to predict target protein ubiquitination induced by cereblon-recruiting PROTACs. J. Biol. Chem. 2022, 298, 101653. [Google Scholar] [CrossRef]
- Oleinikovas, V.; Gainza, P.; Ryckmans, T.; Fasching, B.; Thomä, N.H. From Thalidomide to Rational Molecular Glue Design for Targeted Protein Degradation. Annu. Rev. Pharmacol. Toxicol. 2024, 64, 291–312. [Google Scholar] [CrossRef]
- Petzold, G.; Gainza, P.; Annunziato, S.; Lamberto, I.; Trenh, P.; McAllister, L.A.; DeMarco, B.; Schwander, L.; Bunker, R.D.; Zlotosch, M.; et al. Mining the CRBN Target Space Redefines Rules for Molecular Glue-induced Neosubstrate Recognition. bioRxiv 2024. [Google Scholar] [CrossRef]
- Petzold, G.; Fischer, E.S.; Thomä, N.H. Structural basis of lenalidomide-induced CK1α degradation by the CRL4(CRBN) ubiquitin ligase. Nature 2016, 532, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Matyskiela, M.E.; Lu, G.; Ito, T.; Pagarigan, B.; Lu, C.-C.; Miller, K.; Fang, W.; Wang, N.-Y.; Nguyen, D.; Houston, J.; et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature 2016, 535, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Sievers, Q.L.; Petzold, G.; Bunker, R.D.; Renneville, A.; Słabicki, M.; Liddicoat, B.J.; Abdulrahman, W.; Mikkelsen, T.; Ebert, B.L.; Thomä, N.H. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 2018, 362, eaat0572. [Google Scholar] [CrossRef] [PubMed]
- Matyskiela, M.E.; Clayton, T.; Zheng, X.; Mayne, C.; Tran, E.; Carpenter, A.; Pagarigan, B.; McDonald, J.; Rolfe, M.; Hamann, L.G.; et al. Crystal structure of the SALL4-pomalidomide-cereblon-DDB1 complex. Nat. Struct. Mol. Biol. 2020, 27, 319–322. [Google Scholar] [CrossRef]
- Wang, E.S.; Verano, A.L.; Nowak, R.P.; Yuan, J.C.; Donovan, K.A.; Eleuteri, N.A.; Yue, H.; Ngo, K.H.; Lizotte, P.H.; Gokhale, P.C.; et al. Acute pharmacological degradation of Helios destabilizes regulatory T cells. Nat. Chem. Biol. 2021, 17, 711–717. [Google Scholar] [CrossRef]
- Rovers, E.; Schapira, M. Benchmarking Methods for PROTAC Ternary Complex Structure Prediction. J. Chem. Inf. Model. 2024, 64, 6162–6173. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, N.; Singh, A.; Kumar, P.; Kaushik, M. Protein kinases: Role of their dysregulation in carcinogenesis, identification and inhibition. Drug Res. 2023, 73, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Green, J.R.; Mahalingaiah, P.K.S.; Gopalakrishnan, S.M.; Liguori, M.J.; Mittelstadt, S.W.; Blomme, E.A.; Van Vleet, T.R. Off-target pharmacological activity at various kinases: Potential functional and pathological side effects. J. Pharmacol. Toxicol. Methods 2023, 123, 107468. [Google Scholar] [CrossRef] [PubMed]
- Naso, F.D.; Boi, D.; Ascanelli, C.; Pamfil, G.; Lindon, C.; Paiardini, A.; Guarguaglini, G. Nuclear localisation of Aurora-A: Its regulation and significance for Aurora-A functions in cancer. Oncogene 2021, 40, 3917–3928. [Google Scholar] [CrossRef] [PubMed]
- Abdeldayem, A.; Raouf, Y.S.; Constantinescu, S.N.; Moriggl, R.; Gunning, P.T. Advances in covalent kinase inhibitors. Chem. Soc. Rev. 2020, 49, 2617–2687. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Cai, M.; Shao, L.; Zhang, J. Targeting Protein Kinases Degradation by PROTACs. Front. Chem. 2021, 9, 679120. [Google Scholar] [CrossRef]
- Smith, B.E.; Wang, S.L.; Jaime-Figueroa, S.; Harbin, A.; Wang, J.; Hamman, B.D.; Crews, C.M. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat. Commun. 2019, 10, 131. [Google Scholar] [CrossRef]
- Kumarasamy, V.; Gao, Z.; Zhao, B.; Jiang, B.; Rubin, S.M.; Burgess, K.; Witkiewicz, A.K.; Knudsen, E.S. PROTAC-mediated CDK degradation differentially impacts cancer cell cycles due to heterogeneity in kinase dependencies. Br. J. Cancer 2023, 129, 1238–1250. [Google Scholar] [CrossRef]
- Riching, K.M.; Schwinn, M.K.; Vasta, J.D.; Robers, M.B.; Machleidt, T.; Urh, M.; Daniels, D.L. CDK Family PROTAC Profiling Reveals Distinct Kinetic Responses and Cell Cycle-Dependent Degradation of CDK2. SLAS Discov. 2021, 26, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Casement, R.; Bond, A.; Craigon, C.; Ciulli, A. Mechanistic and Structural Features of PROTAC Ternary Complexes. Methods Mol. Biol. 2021, 2365, 79–113. [Google Scholar] [CrossRef] [PubMed]
- Casement, R.; Bond, A.; Craigon, C.; Ciulli, A. Selective targeting of non-centrosomal AURKA functions through use of a targeted protein degradation tool. Commun. Biol. 2021, 4, 640. [Google Scholar] [CrossRef]
- Rana, S.; Bendjennat, M.; Kour, S.; King, H.M.; Kizhake, S.; Zahid, M.; Natarajan, A. Selective degradation of CDK6 by a palbociclib based PROTAC. Bioorg. Med. Chem. Lett. 2019, 29, 1375–1379. [Google Scholar] [CrossRef] [PubMed]
- Braal, C.L.; Jongbloed, E.M.; Wilting, S.M.; Mathijssen, R.H.J.; Koolen, S.L.W.; Jager, A. Inhibiting CDK4/6 in Breast Cancer with Palbociclib, Ribociclib, and Abemaciclib: Similarities and Differences. Drugs 2021, 81, 317–331. [Google Scholar] [CrossRef]
- Kokic, G.; Yakoub, G.; Heuvel, D.v.D.; Wondergem, A.P.; van der Meer, P.J.; van der Weegen, Y.; Chernev, A.; Fianu, I.; Fokkens, T.J.; Lorenz, S.; et al. Structural basis for RNA polymerase II ubiquitylation and inactivation in transcription-coupled repair. Nat. Struct. Mol. Biol. 2024, 31, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Rosignoli, S.; Pacelli, M.; Manganiello, F.; Paiardini, A. An outlook on structural biology after AlphaFold: Tools, limits and perspectives. FEBS Open Bio 2024. [Google Scholar] [CrossRef] [PubMed]
- Janson, G.; Paiardini, A. PyMod 3: A complete suite for structural bioinformatics in PyMOL. Bioinformatics 2021, 37, 1471–1472. [Google Scholar] [CrossRef] [PubMed]
- Rosignoli, S.; Paiardini, A. DockingPie: A consensus docking plugin for PyMOL. Bioinformatics 2022, 38, 4233–4234. [Google Scholar] [CrossRef]
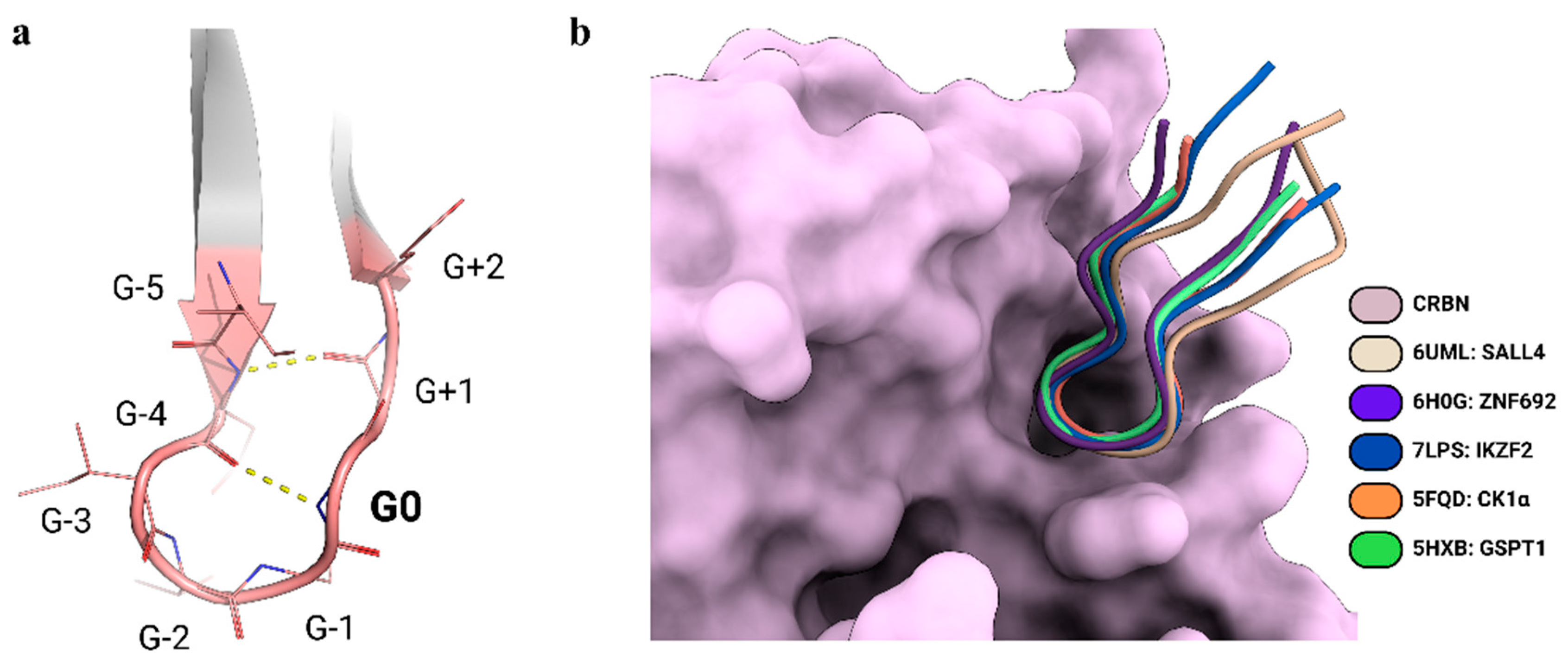
| Targeted Protein | PDB 1 | Compound | Ref. |
|---|---|---|---|
| Casein kinase 1 (CK1α) | 5FQD | Lenalidomide | [29] |
| GTP-binding subunit ERF3A (GSPT1) | 5HXB | CC-885 | [30] |
| Zinc finger protein 692 (ZNF692) | 6H0G | Pomalidomide | [31] |
| Sal-like protein 4 (SALL4) | 6UML | Pomalidomide | [32] |
| Zinc finger protein Helios (IKZF2) | 7LPS | ALV1 | [33] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosignoli, S.; Giordani, S.; Pacelli, M.; Guarguaglini, G.; Paiardini, A. Unraveling the Engagement of Kinases to CRBN Through a Shared Structural Motif to Optimize PROTACs Efficacy. Biomolecules 2025, 15, 206. https://doi.org/10.3390/biom15020206
Rosignoli S, Giordani S, Pacelli M, Guarguaglini G, Paiardini A. Unraveling the Engagement of Kinases to CRBN Through a Shared Structural Motif to Optimize PROTACs Efficacy. Biomolecules. 2025; 15(2):206. https://doi.org/10.3390/biom15020206
Chicago/Turabian StyleRosignoli, Serena, Sara Giordani, Maddalena Pacelli, Giulia Guarguaglini, and Alessandro Paiardini. 2025. "Unraveling the Engagement of Kinases to CRBN Through a Shared Structural Motif to Optimize PROTACs Efficacy" Biomolecules 15, no. 2: 206. https://doi.org/10.3390/biom15020206
APA StyleRosignoli, S., Giordani, S., Pacelli, M., Guarguaglini, G., & Paiardini, A. (2025). Unraveling the Engagement of Kinases to CRBN Through a Shared Structural Motif to Optimize PROTACs Efficacy. Biomolecules, 15(2), 206. https://doi.org/10.3390/biom15020206







