Multiple Mechanisms to Regulate Actin Functions: “Fundamental” Versus Lineage-Specific Mechanisms and Hierarchical Relationships
Abstract
:1. Introduction
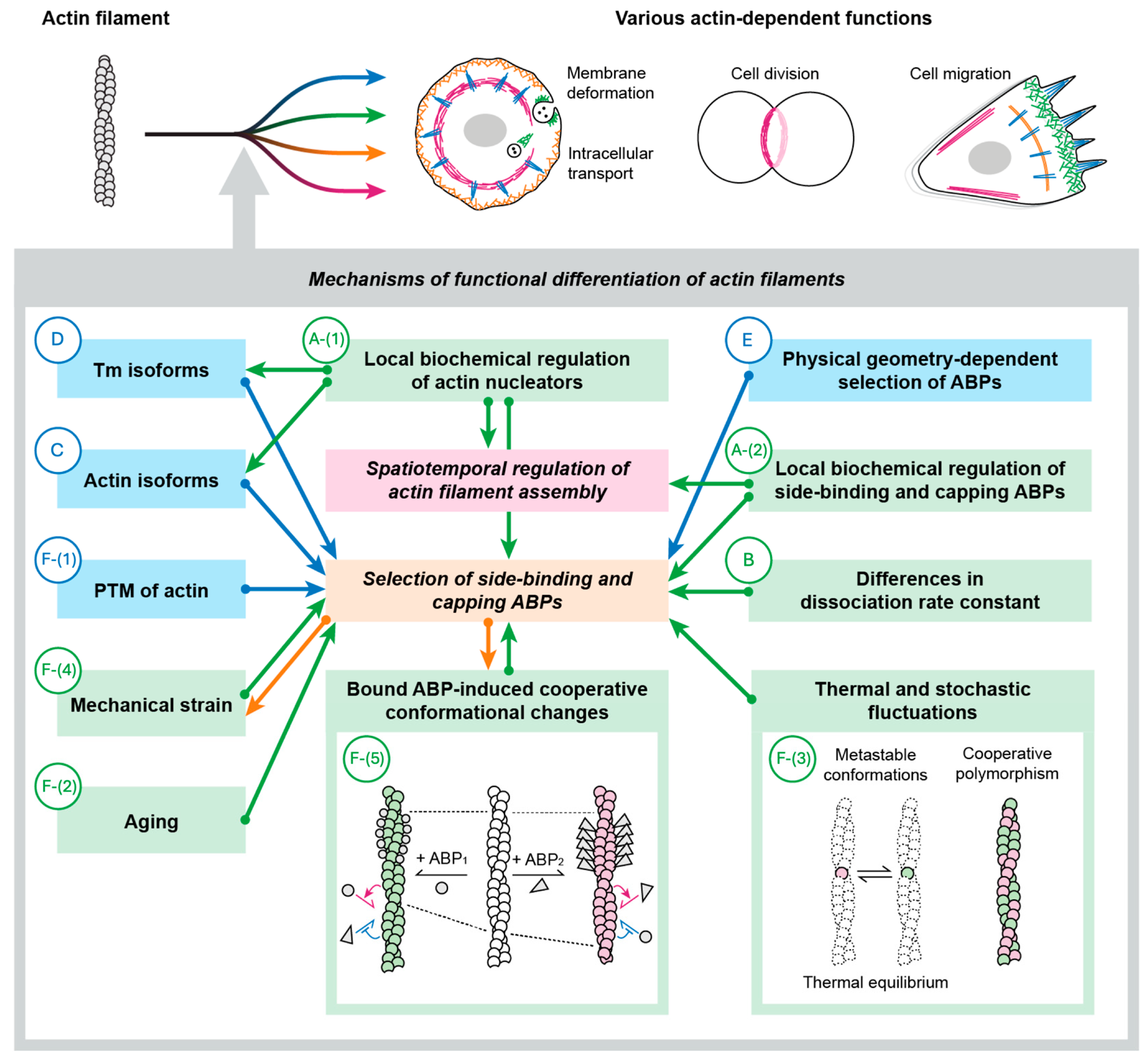
2. Mechanisms to Create Functionally Distinct Actin Filaments
- A:
- Local biochemical regulation of specific ABPs.
- A-(1): Local biochemical regulation of actin nucleators.
- A-(2): Local biochemical regulation of filament-side binding and capping ABPs.
- B:
- Differences in dissociation rate constants from actin among ABPs.
- C:
- Actin isoform-dependent selection of ABPs.
- D:
- Tropomyosin (Tm) isoform-dependent selection of ABPs.
- E:
- Physical geometry-dependent selection of ABPs.
- F:
- Filament conformation-dependent selection of ABPs.
- F-(1): Post-translational modification (PTM) of actin.
- F-(2): Aging.
- F-(3): Thermal and stochastic fluctuations.
- F-(4): Mechanical strain.
- F-(5): Bound ABP-induced cooperative conformational changes.
3. A: Local Biochemical Regulation of Specific ABPs
3.1. A-(1): Local Biochemical Regulation of Actin Nucleators
3.2. A-(2): Local Biochemical Regulation of Filament Side-Binding and Capping ABPs
4. B: Differences in Dissociation Rate Constants Among ABPs
5. C: Actin Isoform-Dependent Selection of ABPs
6. D: Tropomyosin (Tm) Isoform-Dependent Selection of ABPs
7. E: Physical Geometry-Dependent Selection of ABPs
8. F: Filament Conformation-Dependent Selection of ABPs
8.1. F-(1): Post-Translational Modification (PTM) of Actin
8.2. F-(2): Aging
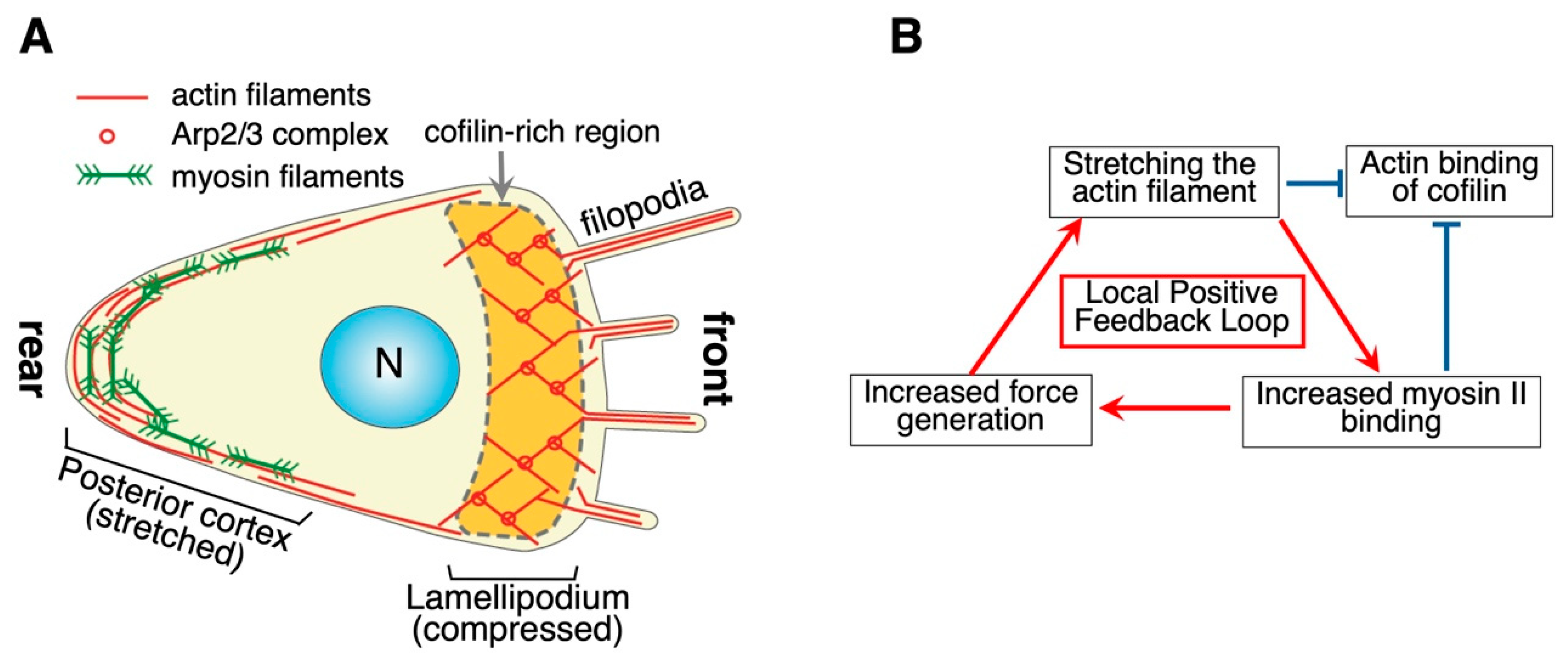
8.3. F-(3): Thermal and Stochastic Fluctuations
8.4. F-(4): Mechanical Strain
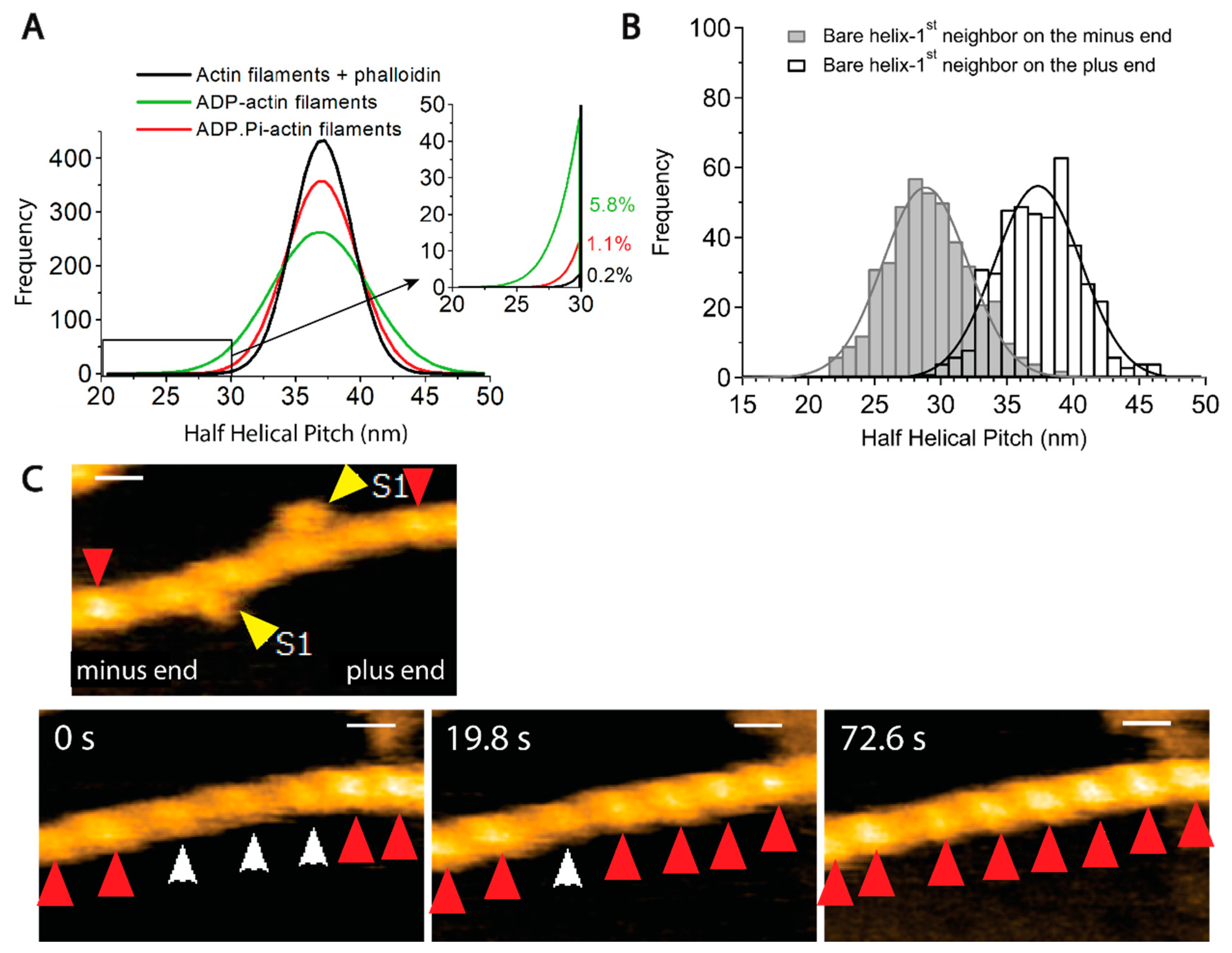
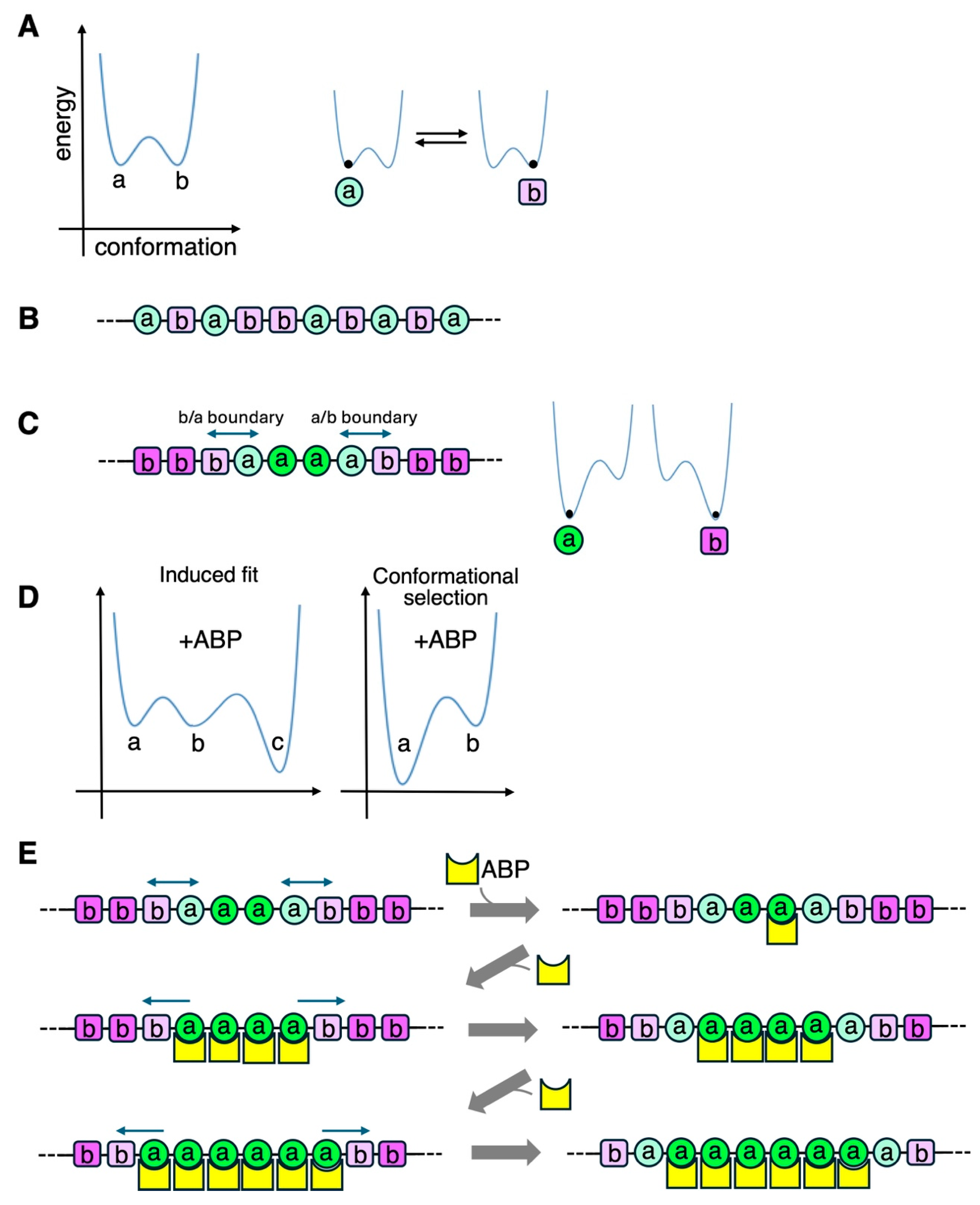
8.5. F-(5): Bound ABP-Induced Cooperative Conformational Changes
| ABP | Detected by | ||||
|---|---|---|---|---|---|
| Cryo-EM | AFM | Cooperative Binding (Cluster Formation) | Cooperative Binding (Binding Ratio) | Other Biophysical Methods | |
| α-catenin | [120] | ||||
| Cofilin (short range) | [11,155,161] | [159] | [159,161,162] | [161,225,226,227] | [228,229] |
| Cofilin (long range) | [201,202,203] | ||||
| Coronin 1-C | [118] | ||||
| Drebrin | [125,204] | ||||
| ABD of dystrophin | [119] | ||||
| ABD1 of fimbrin | [185] | ||||
| ABD2 of fimbrin | [186] | [124] | |||
| Formin | [211,212] | ||||
| Gelsolin | [230] | ||||
| Lifeact | [188] | ||||
| Myosin II motor (−ATP) | [121] | [190,191,192,193,194] | |||
| Myosin II motor (+ATP) | [122,123] | ||||
| Myosin V +ATP | [136] | ||||
| ABD of Rng2 | [189] | ||||
| Tm | [84] | ||||
| Jasplakinolide * | [14] | ||||
| Phalloidin * | [13,14] | ||||
9. Fundamental vs. Lineage-Specific Regulatory Mechanisms and the Hierarchy Among Them
10. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Pollard, T.D.; Cooper, J.A. Actin, a central player in cell shape and movement. Science 2009, 326, 1208–1212. [Google Scholar] [CrossRef]
- Fernandez, M.K.; Sinha, M.; Zidan, M.; Renz, M. Nuclear actin filaments—A historical perspective. Nucleus 2024, 15, 2320656. [Google Scholar] [CrossRef] [PubMed]
- Uyeda, T.Q.P.; Iwadate, Y.; Umeki, N.; Nagasaki, A.; Yumura, S. Stretching actin filaments within cells enhances their affinity for the myosin II motor domain. PLoS ONE 2011, 6, e26200. [Google Scholar] [CrossRef] [PubMed]
- Michelot, A.; Drubin, D.G. Building distinct actin filament networks in a common cytoplasm. Curr. Biol. 2011, 21, R560–R569. [Google Scholar] [CrossRef] [PubMed]
- Kovar, D.R.; Sirotkin, V.; Lord, M. Three’s company: The fission yeast actin cytoskeleton. Trends Cell Biol. 2011, 21, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Tokuraku, K.; Kuragano, M.; Uyeda, T.Q.P. Long-Range and Directional Allostery of Actin Filaments Plays Important Roles in Various Cellular Activities. Int. J. Mol. Sci. 2020, 21, 3209. [Google Scholar] [CrossRef]
- von der Ecken, J.; Muller, M.; Lehman, W.; Manstein, D.J.; Penczek, P.A.; Raunser, S. Structure of the F-actin-tropomyosin complex. Nature 2015, 519, 114–117. [Google Scholar] [CrossRef]
- Kabsch, W.; Mannherz, H.G.; Suck, D.; Pai, E.F.; Holmes, K.C. Atomic structure of the actin: DNase I complex. Nature 1990, 347, 37–44. [Google Scholar] [CrossRef]
- Murakami, K.; Yasunaga, T.; Noguchi, T.Q.P.; Gomibuchi, Y.; Ngo, K.X.; Uyeda, T.Q.P.; Wakabayashi, T. Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell 2010, 143, 275–287. [Google Scholar] [CrossRef]
- Oda, T.; Iwasa, M.; Aihara, T.; Maéda, Y.; Narita, A. The nature of the globular- to fibrous-actin transition. Nature 2009, 457, 441–445. [Google Scholar] [CrossRef]
- Tanaka, K.; Takeda, S.; Mitsuoka, K.; Oda, T.; Kimura-Sakiyama, C.; Maéda, Y.; Narita, A. Structural basis for cofilin binding and actin filament disassembly. Nat. Commun. 2018, 9, 1860. [Google Scholar] [CrossRef]
- Oda, T.; Takeda, S.; Narita, A.; Maeda, Y. Structural Polymorphism of Actin. J. Mol. Biol. 2019, 431, 3217–3228. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ge, P.; Oztug Durer, Z.A.; Grintsevich, E.E.; Zhou, Z.H.; Reisler, E. D-loop Dynamics and Near-Atomic-Resolution Cryo-EM Structure of Phalloidin-Bound F-Actin. Structure 2020, 28, 586–593.e3. [Google Scholar] [CrossRef] [PubMed]
- Pospich, S.; Merino, F.; Raunser, S. Structural Effects and Functional Implications of Phalloidin and Jasplakinolide Binding to Actin Filaments. Structure 2020, 28, 437–449.e5. [Google Scholar] [CrossRef]
- Otterbein, L.R.; Graceffa, P.; Dominguez, R. The crystal structure of uncomplexed actin in the ADP state. Science 2001, 293, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Spang, A.; Saw, J.H.; Jorgensen, S.L.; Zaremba-Niedzwiedzka, K.; Martijn, J.; Lind, A.E.; van Eijk, R.; Schleper, C.; Guy, L.; Ettema, T.J.G. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 2015, 521, 173–179. [Google Scholar] [CrossRef]
- Liu, Y.; Makarova, K.S.; Huang, W.C.; Wolf, Y.I.; Nikolskaya, A.N.; Zhang, X.; Cai, M.; Zhang, C.J.; Xu, W.; Luo, Z.; et al. Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature 2021, 593, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Akil, C.; Robinson, R.C. Genomes of Asgard archaea encode profilins that regulate actin. Nature 2018, 562, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Akil, C.; Tran, L.T.; Orhant-Prioux, M.; Baskaran, Y.; Manser, E.; Blanchoin, L.; Robinson, R.C. Insights into the evolution of regulated actin dynamics via characterization of primitive gelsolin/cofilin proteins from Asgard archaea. Proc. Natl. Acad. Sci. USA 2020, 117, 19904–19913. [Google Scholar] [CrossRef]
- Ebashi, S.; Endo, M.; Ohtsuki, I. Control of muscle contraction. Q. Rev. Biophys. 1969, 2, 351–384. [Google Scholar] [CrossRef]
- Tall, E.G.; Spector, I.; Pentyala, S.N.; Bitter, I.; Rebecchi, M.J. Dynamics of phosphatidylinositol 4,5-bisphosphate in actin-rich structures. Curr. Biol. 2000, 10, 743–746. [Google Scholar] [CrossRef] [PubMed]
- Merlot, S.; Firtel, R.A. Leading the way: Directional sensing through phosphatidylinositol 3-kinase and other signaling pathways. J. Cell Sci. 2003, 116, 3471–3478. [Google Scholar] [CrossRef]
- Oikawa, T.; Yamaguchi, H.; Itoh, T.; Kato, M.; Ijuin, T.; Yamazaki, D.; Suetsugu, S.; Takenawa, T. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat. Cell Biol. 2004, 6, 420–426. [Google Scholar] [CrossRef]
- Iijima, M.; Huang, Y.E.; Luo, H.R.; Vazquez, F.; Devreotes, P.N. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J. Biol. Chem. 2004, 279, 16606–16613. [Google Scholar] [CrossRef]
- Devreotes, P.; Horwitz, A.R. Signaling networks that regulate cell migration. Cold Spring Harb. Perspect. Biol. 2015, 7, a005959. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.M.; Veltman, D.; Kay, R.R. Chemotaxis of a model organism: Progress with Dictyostelium. Curr. Opin. Cell Biol. 2015, 36, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 451–477. [Google Scholar] [CrossRef]
- Rottner, K.; Hanisch, J.; Campellone, K.G. WASH, WHAMM and JMY: Regulation of Arp2/3 complex and beyond. Trends Cell Biol. 2010, 20, 650–661. [Google Scholar] [CrossRef]
- Lebensohn, A.M.; Kirschner, M.W. Activation of the WAVE complex by coincident signals controls actin assembly. Mol. Cell 2009, 36, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Higgs, H.N. Formin proteins: A domain-based approach. Trends Biochem. Sci. 2005, 30, 342–353. [Google Scholar] [CrossRef]
- Survery, S.; Hurtig, F.; Haq, S.R.; Eriksson, J.; Guy, L.; Rosengren, K.J.; Lindas, A.C.; Chi, C.N. Heimdallarchaea encodes profilin with eukaryotic-like actin regulation and polyproline binding. Commun. Biol. 2021, 4, 1024. [Google Scholar] [CrossRef]
- Pandey, D.K.; Chaudhary, B. Evolutionary expansion and structural functionalism of the ancient family of profilin proteins. Gene 2017, 626, 70–86. [Google Scholar] [CrossRef]
- Fritz-Laylin, L.K.; Titus, M.A. The evolution and diversity of actin-dependent cell migration. Mol. Biol. Cell 2023, 34, pe6. [Google Scholar] [CrossRef]
- Chalkia, D.; Nikolaidis, N.; Makalowski, W.; Klein, J.; Nei, M. Origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol. Biol. Evol. 2008, 25, 2717–2733. [Google Scholar] [CrossRef]
- Baum, J.; Tonkin, C.J.; Paul, A.S.; Rug, M.; Smith, B.J.; Gould, S.B.; Richard, D.; Pollard, T.D.; Cowman, A.F. A malaria parasite formin regulates actin polymerization and localizes to the parasite-erythrocyte moving junction during invasion. Cell Host Microbe 2008, 3, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Velle, K.B.; Swafford, A.J.M.; Garner, E.; Fritz-Laylin, L.K. Actin network evolution as a key driver of eukaryotic diversification. J. Cell Sci. 2024, 137, jcs261660. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, M.E.; Heuser, J.E.; Kerkhoff, E.; Mullins, R.D. Drosophila Spire is an actin nucleation factor. Nature 2005, 433, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, R.; Pinyol, R.; Reichenbach, N.; Custer, L.; Klingensmith, J.; Kessels, M.M.; Qualmann, B. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell 2007, 131, 337–350. [Google Scholar] [CrossRef]
- Sanger, J.W.; Wang, J.; Fan, Y.; White, J.; Mi-Mi, L.; Dube, D.K.; Sanger, J.M.; Pruyne, D. Assembly and Maintenance of Myofibrils in Striated Muscle. In The Actin Cytoskeleton; Handbook of Experimental Pharmacology; Springer: Cham, Switzerland, 2017; Volume 235, pp. 39–75. [Google Scholar] [CrossRef]
- Fowler, V.M.; Dominguez, R. Tropomodulins and Leiomodins: Actin Pointed End Caps and Nucleators in Muscles. Biophys. J. 2017, 112, 1742–1760. [Google Scholar] [CrossRef]
- Kong, S.G.; Yamazaki, Y.; Shimada, A.; Kijima, S.T.; Hirose, K.; Katoh, K.; Ahn, J.; Song, H.G.; Han, J.W.; Higa, T.; et al. CHLOROPLAST UNUSUAL POSITIONING 1 is a plant-specific actin polymerization factor regulating chloroplast movement. Plant Cell 2024, 36, 1159–1181. [Google Scholar] [CrossRef] [PubMed]
- Gunning, P.; O’Neill, G.; Hardeman, E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol. Rev. 2008, 88, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Insall, R.; Muller-Taubenberger, A.; Machesky, L.; Kohler, J.; Simmeth, E.; Atkinson, S.J.; Weber, I.; Gerisch, G. Dynamics of the Dictyostelium Arp2/3 complex in endocytosis, cytokinesis, and chemotaxis. Cell Motil. Cytoskelet. 2001, 50, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Aizawa, H.; Fukui, Y.; Yahara, I. Live dynamics of Dictyostelium cofilin suggests a role in remodeling actin latticework into bundles. J. Cell Sci. 1997, 110, 2333–2344. [Google Scholar] [CrossRef] [PubMed]
- Aizawa, H.; Sutoh, K.; Tsubuki, S.; Kawashima, S.; Ishii, A.; Yahara, I. Identification, characterization, and intracellular distribution of cofilin in Dictyostelium discoideum. J. Biol. Chem. 1995, 270, 10923–10932. [Google Scholar] [CrossRef]
- Yumura, S.; Mori, H.; Fukui, Y. Localization of actin and myosin for the study of ameboid movement in Dictyostelium using improved immunofluorescence. J. Cell Biol. 1984, 99, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Moores, S.L.; Sabry, J.H.; Spudich, J.A. Myosin dynamics in live Dictyostelium cells. Proc. Natl. Acad. Sci. USA 1996, 93, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Luck-Vielmetter, D.; Schleicher, M.; Grabatin, B.; Wippler, J.; Gerisch, G. Replacement of threonine residues by serine and alanine in a phosphorylatable heavy chain fragment of Dictyostelium myosin II. FEBS Lett. 1990, 269, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Steimle, P.A.; Yumura, S.; Côté, G.P.; Medley, Q.G.; Polyakov, M.V.; Leppert, B.; Egelhoff, T.T. Recruitment of a myosin heavy chain kinase to actin-rich protrusions in Dictyostelium. Curr. Biol. 2001, 11, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Yumura, S.; Yoshida, M.; Betapudi, V.; Licate, L.S.; Iwadate, Y.; Nagasaki, A.; Uyeda, T.Q.P.; Egelhoff, T.T. Multiple myosin II heavy chain kinases: Roles in filament assembly control and proper cytokinesis in Dictyostelium. Mol. Biol. Cell 2005, 16, 4256–4266. [Google Scholar] [CrossRef] [PubMed]
- Egelhoff, T.T.; Brown, S.S.; Spudich, J.A. Spatial and temporal control of nonmuscle myosin localization: Identification of a domain that is necessary for myosin filament disassembly in vivo. J. Cell Biol. 1991, 112, 677–688. [Google Scholar] [CrossRef]
- Bamburg, J.R. Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 1999, 15, 185–230. [Google Scholar] [CrossRef] [PubMed]
- Ngo, K.X.; Umeki, N.; Kijima, S.T.; Kodera, N.; Ueno, H.; Furutani-Umezu, N.; Nakajima, J.; Noguchi, T.Q.P.; Nagasaki, A.; Tokuraku, K.; et al. Allosteric regulation by cooperative conformational changes of actin filaments drives mutually exclusive binding with cofilin and myosin. Sci. Rep. 2016, 6, 35449. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Tatsumi, H.; Sokabe, M. Actin filaments function as a tension sensor by tension dependent binding of cofilin to the filament. J. Cell Biol. 2011, 195, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, M.G.; Janzen, D.; Hwang, R.; Roldan, J.; Jarchum, I.; Knecht, D.A. Visualization of the actin cytoskeleton: Different F-actin-binding probes tell different stories. Cytoskeleton 2014, 71, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Fukui, Y.; Kitanishi-Yumura, T.; Yumura, S. Myosin II-independent F-actin flow contributes to cell locomotion in Dictyostelium. J. Cell Sci. 1999, 112 Pt 6, 877–886. [Google Scholar] [CrossRef]
- Ruprecht, V.; Wieser, S.; Callan-Jones, A.; Smutny, M.; Morita, H.; Sako, K.; Barone, V.; Ritsch-Marte, M.; Sixt, M.; Voituriez, R.; et al. Cortical contractility triggers a stochastic switch to fast amoeboid cell motility. Cell 2015, 160, 673–685. [Google Scholar] [CrossRef]
- Yumura, S.; Itoh, G.; Kikuta, Y.; Kikuchi, T.; Kitanishi-Yumura, T.; Tsujioka, M. Cell-scale dynamic recycling and cortical flow of the actin-myosin cytoskeleton for rapid cell migration. Biol. Open 2013, 2, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, S.; Taniguchi, D.; Tanaka, S.; Kiuchi, T.; Vavylonis, D.; Watanabe, N. Convection-Induced Biased Distribution of Actin Probes in Live Cells. Biophys. J. 2019, 116, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, S.; Rutkowski, D.M.; Lynch, K.A.; Liu, Y.; Vavylonis, D.; Watanabe, N. Force transmission by retrograde actin flow-induced dynamic molecular stretching of Talin. Nat. Commun. 2023, 14, 8468. [Google Scholar] [CrossRef]
- Ivanova, J.R.; Benk, A.S.; Schaefer, J.V.; Dreier, B.; Hermann, L.O.; Pluckthun, A.; Missirlis, D.; Spatz, J.P. Designed Ankyrin Repeat Proteins as Actin Labels of Distinct Cytoskeletal Structures in Living Cells. ACS Nano 2024, 18, 8919–8933. [Google Scholar] [CrossRef] [PubMed]
- Nagasaki, A.; Kijima, S.T.; Shinkai, Y.; Ohtsuka, Y.; Ochiishi, T.; Sasaki, Y.T.F.; Hata, S.; Hirano, K.; Kato, Y.; Doi, M.; et al. Actin Painting: A multicolor fluorescence staining method based on cell type-specific differences in the composition and distribution of filamentous actin structures. bioRxiv 2025. [Google Scholar] [CrossRef]
- Gallwitz, D.; Seidel, R. Molecular cloning of the actin gene from yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1980, 8, 1043–1059. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Gallwitz, D. A single intronless actin gene in the fission yeast Schizosaccharomyces pombe: Nucleotide sequence and transcripts formed in homologous and heterologous yeast. Nucleic Acids Res. 1987, 15, 7369–7379. [Google Scholar] [CrossRef] [PubMed]
- Sugase, Y.; Hirono, M.; Kindle, K.L.; Kamiya, R. Cloning and characterization of the actin-encoding gene of Chlamydomonas reinhardtii. Gene 1996, 168, 117–121. [Google Scholar] [CrossRef]
- Cupples, C.G.; Pearlman, R.E. Isolation and characterization of the actin gene from Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 1986, 83, 5160–5164. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.M.; Fey, P.; Ramalingam, N.; Liu, X.I.; Rohlfs, M.; Noegel, A.A.; Müller-Taubenberger, A.; Glöckner, G.; Schleicher, M. The actinome of Dictyostelium discoideum in comparison to actins and actin-related proteins from other organisms. PLoS ONE 2008, 3, e2654. [Google Scholar] [CrossRef] [PubMed]
- Dugina, V.B.; Shagieva, G.S.; Kopnin, P.B. Biological Role of Actin Isoforms in Mammalian Cells. Biochemistry 2019, 84, 583–592. [Google Scholar] [CrossRef]
- Shah, R.; Panagiotou, T.C.; Cole, G.B.; Moraes, T.F.; Lavoie, B.D.; McCulloch, C.A.; Wilde, A. The DIAPH3 linker specifies a β-actin network that maintains RhoA and Myosin-II at the cytokinetic furrow. Nat. Commun. 2024, 15, 5250. [Google Scholar] [CrossRef] [PubMed]
- Belyantseva, I.A.; Perrin, B.J.; Sonnemann, K.J.; Zhu, M.; Stepanyan, R.; McGee, J.; Frolenkov, G.I.; Walsh, E.J.; Friderici, K.H.; Friedman, T.B.; et al. γ-actin is required for cytoskeletal maintenance but not development. Proc. Natl. Acad. Sci. USA 2009, 106, 9703–9708. [Google Scholar] [CrossRef] [PubMed]
- Shawlot, W.; Deng, J.M.; Fohn, L.E.; Behringer, R.R. Restricted β-galactosidase expression of a hygromycin-lacZ gene targeted to the β-actin locus and embryonic lethality of β-actin mutant mice. Transgenic Res. 1998, 7, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Vedula, P.; Kurosaka, S.; Leu, N.A.; Wolf, Y.I.; Shabalina, S.A.; Wang, J.; Sterling, S.; Dong, D.W.; Kashina, A. Diverse functions of homologous actin isoforms are defined by their nucleotide, rather than their amino acid sequence. eLife 2017, 6, e31661. [Google Scholar] [CrossRef] [PubMed]
- McDowell, J.M.; Huang, S.; McKinney, E.C.; An, Y.Q.; Meagher, R.B. Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 1996, 142, 587–602. [Google Scholar] [CrossRef]
- Kijima, S.T.; Hirose, K.; Kong, S.G.; Wada, M.; Uyeda, T.Q.P. Distinct Biochemical Properties of Arabidopsis thaliana Actin Isoforms. Plant Cell Physiol. 2016, 57, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, M.K.; McKinney, E.C.; Meagher, R.B. A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant Cell 2009, 21, 701–718. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, M.K.; Burgos-Rivera, B.; McKinney, E.C.; Ruzicka, D.R.; Meagher, R.B. Class-specific interaction of profilin and ADF isovariants with actin in the regulation of plant development. Plant Cell 2007, 19, 3111–3126. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Arora, P.D.; McCulloch, C.A.; Wilde, A. Cytokinesis requires localized β-actin filament production by an actin isoform specific nucleator. Nat. Commun. 2017, 8, 1530. [Google Scholar] [CrossRef] [PubMed]
- Lubit, B.W.; Schwartz, J.H. An antiactin antibody that distinguishes between cytoplasmic and skeletal muscle actins. J. Cell Biol. 1980, 86, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Kijima, S.T.; Staiger, C.J.; Katoh, K.; Nagasaki, A.; Ito, K.; Uyeda, T.Q.P. Arabidopsis vegetative actin isoforms, AtACT2 and AtACT7, generate distinct filament arrays in living plant cells. Sci. Rep. 2018, 8, 4381. [Google Scholar] [CrossRef] [PubMed]
- Bailey, K. Tropomyosin: A new asymmetric protein component of muscle. Nature 1946, 157, 368. [Google Scholar] [CrossRef]
- Lehman, W.; Vibert, P.; Uman, P.; Craig, R. Steric-blocking by tropomyosin visualized in relaxed vertebrate muscle thin filaments. J. Mol. Biol. 1995, 251, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Lazarides, E. Tropomyosin antibody: The specific localization of tropomyosin in nonmuscle cells. J. Cell Biol. 1975, 65, 549–561. [Google Scholar] [CrossRef]
- Gunning, P.W.; Hardeman, E.C.; Lappalainen, P.; Mulvihill, D.P. Tropomyosin–master regulator of actin filament function in the cytoskeleton. J. Cell Sci. 2015, 128, 2965–2974. [Google Scholar] [CrossRef] [PubMed]
- Butters, C.A.; Willadsen, K.A.; Tobacman, L.S. Cooperative interactions between adjacent troponin-tropomyosin complexes may be transmitted through the actin filament. J. Biol. Chem. 1993, 268, 15565–15570. [Google Scholar] [CrossRef] [PubMed]
- Tobacman, L.S. Cooperative binding of tropomyosin to actin. Adv. Exp. Med. Biol. 2008, 644, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Gateva, G.; Kremneva, E.; Reindl, T.; Kotila, T.; Kogan, K.; Gressin, L.; Gunning, P.W.; Manstein, D.J.; Michelot, A.; Lappalainen, P. Tropomyosin Isoforms Specify Functionally Distinct Actin Filament Populations In Vitro. Curr. Biol. 2017, 27, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Bryce, N.S.; Schevzov, G.; Ferguson, V.; Percival, J.M.; Lin, J.J.; Matsumura, F.; Bamburg, J.R.; Jeffrey, P.L.; Hardeman, E.C.; Gunning, P.; et al. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol. Biol. Cell 2003, 14, 1002–1016. [Google Scholar] [CrossRef]
- Creed, S.J.; Desouza, M.; Bamburg, J.R.; Gunning, P.; Stehn, J. Tropomyosin isoform 3 promotes the formation of filopodia by regulating the recruitment of actin-binding proteins to actin filaments. Exp. Cell Res. 2011, 317, 249–261. [Google Scholar] [CrossRef]
- Ostap, E.M. Tropomyosins as discriminators of myosin function. Adv. Exp. Med. Biol. 2008, 644, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Gunning, P.W.; Ghoshdastider, U.; Whitaker, S.; Popp, D.; Robinson, R.C. The evolution of compositionally and functionally distinct actin filaments. J. Cell Sci. 2015, 128, 2009–2019. [Google Scholar] [CrossRef]
- Eichinger, L.; Noegel, A.A. Crawling into a new era–the Dictyostelium genome project. EMBO J. 2003, 22, 1941–1946. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Le Berre, M.; Lautenschlaeger, F.; Maiuri, P.; Callan-Jones, A.; Heuze, M.; Takaki, T.; Voituriez, R.; Piel, M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell 2015, 160, 659–672. [Google Scholar] [CrossRef]
- Alexandrova, A.Y.; Chikina, A.S.; Svitkina, T.M. Actin cytoskeleton in mesenchymal-to-amoeboid transition of cancer cells. Int. Rev. Cell Mol. Biol. 2020, 356, 197–256. [Google Scholar] [CrossRef]
- Balasubramanian, M.K.; Helfman, D.M.; Hemmingsen, S.M. A new tropomyosin essential for cytokinesis in the fission yeast S. pombe. Nature 1992, 360, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Mabuchi, I. Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol. Biol. Cell 2006, 17, 1933–1945. [Google Scholar] [CrossRef]
- Liu, H.P.; Bretscher, A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell 1989, 57, 233–242. [Google Scholar] [CrossRef]
- Drees, B.; Brown, C.; Barrell, B.G.; Bretscher, A. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J. Cell Biol. 1995, 128, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Dhar, A.; Bagyashree, V.T.; Biswas, S.; Kumari, J.; Sridhara, A.; Jeevan Subodh, B.; Shekhar, S.; Palani, S. Functional redundancy and formin-independent localization of tropomyosin isoforms in Saccharomyces cerevisiae. bioRxiv 2024. [Google Scholar] [CrossRef]
- Okada, K.; Ravi, H.; Smith, E.M.; Goode, B.L. Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: A coordinated mechanism for severing and capping filaments. Mol. Biol. Cell 2006, 17, 2855–2868. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.W.; Bamburg, J.R. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF). Cell Motil. 1982, 2, 1–8. [Google Scholar] [CrossRef]
- Mabuchi, I. Effects of muscle proteins on the interaction between actin and an actin-depolymerizing protein from starfish oocytes. J. Biochem. 1982, 92, 1439–1447. [Google Scholar] [CrossRef] [PubMed]
- Nishida, E.; Muneyuki, E.; Maekawa, S.; Ohta, Y.; Sakai, H. An actin-depolymerizing protein (destrin) from porcine kidney. Its action on F-actin containing or lacking tropomyosin. Biochemistry 1985, 24, 6624–6630. [Google Scholar] [CrossRef] [PubMed]
- Ono, S.; Ono, K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol. 2002, 156, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.R.; Hocky, G.M.; Homa, K.E.; Morganthaler, A.N.; Hitchcock-DeGregori, S.E.; Voth, G.A.; Kovar, D.R. Competition between Tropomyosin, Fimbrin, and ADF/Cofilin drives their sorting to distinct actin filament networks. eLife 2017, 6, e23152. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; East, D.A.; Mulvihill, D.P. Formins determine the functional properties of actin filaments in yeast. Curr. Biol. 2014, 24, 1525–1530. [Google Scholar] [CrossRef] [PubMed]
- Hook, J.; Lemckert, F.; Schevzov, G.; Fath, T.; Gunning, P. Functional identity of the gamma tropomyosin gene: Implications for embryonic development, reproduction and cell viability. Bioarchitecture 2011, 1, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Kamasaki, T.; Arai, R.; Osumi, M.; Mabuchi, I. Directionality of F-actin cables changes during the fission yeast cell cycle. Nat. Cell Biol. 2005, 7, 916–917. [Google Scholar] [CrossRef]
- Kamasaki, T.; Osumi, M.; Mabuchi, I. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J. Cell Biol. 2007, 178, 765–771. [Google Scholar] [CrossRef]
- Reymann, A.C.; Boujemaa-Paterski, R.; Martiel, J.L.; Guerin, C.; Cao, W.; Chin, H.F.; De La Cruz, E.M.; Thery, M.; Blanchoin, L. Actin network architecture can determine myosin motor activity. Science 2012, 336, 1310–1314. [Google Scholar] [CrossRef]
- Lombardo, A.T.; Nelson, S.R.; Kennedy, G.G.; Trybus, K.M.; Walcott, S.; Warshaw, D.M. Myosin Va transport of liposomes in three-dimensional actin networks is modulated by actin filament density, position, and polarity. Proc. Natl. Acad. Sci. USA 2019, 116, 8326–8335. [Google Scholar] [CrossRef]
- Lo Presti, L.; Chang, F.; Martin, S.G. Myosin vs organize actin cables in fission yeast. Mol. Biol. Cell 2012, 23, 4579–4591. [Google Scholar] [CrossRef] [PubMed]
- Nagy, S.; Ricca, B.L.; Norstrom, M.F.; Courson, D.S.; Brawley, C.M.; Smithback, P.A.; Rock, R.S. A myosin motor that selects bundled actin for motility. Proc. Natl. Acad. Sci. USA 2008, 105, 9616–9620. [Google Scholar] [CrossRef] [PubMed]
- Truong Quang, B.A.; Peters, R.; Cassani, D.A.D.; Chugh, P.; Clark, A.G.; Agnew, M.; Charras, G.; Paluch, E.K. Extent of myosin penetration within the actin cortex regulates cell surface mechanics. Nat. Commun. 2021, 12, 6511. [Google Scholar] [CrossRef]
- Winkelman, J.D.; Suarez, C.; Hocky, G.M.; Harker, A.J.; Morganthaler, A.N.; Christensen, J.R.; Voth, G.A.; Bartles, J.R.; Kovar, D.R. Fascin- and α-Actinin-Bundled Networks Contain Intrinsic Structural Features that Drive Protein Sorting. Curr. Biol. 2016, 26, 2697–2706. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.R.; Homa, K.E.; Morganthaler, A.N.; Brown, R.R.; Suarez, C.; Harker, A.J.; O’Connell, M.E.; Kovar, D.R. Cooperation between tropomyosin and α-actinin inhibits fimbrin association with actin filament networks in fission yeast. eLife 2019, 8, e47279. [Google Scholar] [CrossRef] [PubMed]
- Carlier, M.F.; Laurent, V.; Santolini, J.; Melki, R.; Didry, D.; Xia, G.X.; Hong, Y.; Chua, N.H.; Pantaloni, D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: Implication in actin-based motility. J. Cell Biol. 1997, 136, 1307–1322. [Google Scholar] [CrossRef] [PubMed]
- Orlova, A.; Rybakova, I.N.; Prochniewicz, E.; Thomas, D.D.; Ervasti, J.M.; Egelman, E.H. Binding of dystrophin’s tandem calponin homology domain to F-actin is modulated by actin’s structure. Biophys. J. 2001, 80, 1926–1931. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.T.; Roadcap, D.W.; Holoweckyj, N.; Bear, J.E. Coronin 1C harbours a second actin-binding site that confers co-operative binding to F-actin. Biochem. J. 2012, 444, 89–96. [Google Scholar] [CrossRef]
- Way, M.; Pope, B.; Cross, R.A.; Kendrick-Jones, J.; Weeds, A.G. Expression of the N-terminal domain of dystrophin in E. coli and demonstration of binding to F-actin. FEBS Lett. 1992, 301, 243–245. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.D.; Kwiatkowski, A.V.; Ouyang, C.Y.; Liu, H.; Pokutta, S.; Watkins, S.C.; Volkmann, N.; Hanein, D.; Weis, W.I.; Mullins, R.D.; et al. αE-catenin actin-binding domain alters actin filament conformation and regulates binding of nucleation and disassembly factors. Mol. Biol. Cell 2013, 24, 3710–3720. [Google Scholar] [CrossRef]
- Orlova, A.; Egelman, E.H. Cooperative rigor binding of myosin to actin is a function of F-actin structure. J. Mol. Biol. 1997, 265, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Tokuraku, K.; Kurogi, R.; Toya, R.; Uyeda, T.Q.P. Novel mode of cooperative binding between myosin and Mg2+-actin filaments in the presence of low concentrations of ATP. J. Mol. Biol. 2009, 386, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, R.; Nishikawa, Y.; Uyeda, T.Q.P.; Tokuraku, K. Unidirectional growth of heavy meromyosin clusters along actin filaments revealed by real-time fluorescence microscopy. Cytoskeleton 2017, 74, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Kuragano, M.; Yoshino, A.; Shibata, K.; Uyeda, T.Q.P.; Tokuraku, K. Unidirectional cooperative binding of fimbrin actin-binding domain 2 to actin filament. Biochem. Biophys. Res. Commun. 2021, 552, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Grintsevich, E.E.; Hsueh, C.; Reisler, E.; Gimzewski, J.K. Molecular cooperativity of drebrin1-300 binding and structural remodeling of F-actin. Biophys. J. 2012, 103, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Ooi, T.; Mihashi, K.; Kobayashi, H. On the polymerization of tropomyosin. Arch. Biochem. Biophys. 1962, 98, 1–11. [Google Scholar] [CrossRef] [PubMed]
- McLachlan, A.D.; Stewart, M. Tropomyosin coiled-coil interactions: Evidence for an unstaggered structure. J. Mol. Biol. 1975, 98, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Reinke, P.Y.A.; Heiringhoff, R.S.; Reindl, T.; Baker, K.; Taft, M.H.; Meents, A.; Mulvihill, D.P.; Davies, O.R.; Fedorov, R.; Zahn, M.; et al. Crystal structures of cables formed by the acetylated and unacetylated forms of the Schizosaccharomyces pombe tropomyosin ortholog TpmCdc8. J. Biol. Chem. 2024, 300, 107925. [Google Scholar] [CrossRef]
- Obeidy, P.; Sobey, T.; Nicovich, P.R.; Coster, A.C.F.; Pandzic, E. Tropomyosin Isoforms Segregate into Distinct Clusters on Single Actin Filaments. Biomolecules 2024, 14, 1240. [Google Scholar] [CrossRef]
- Rayment, I.; Holden, H.M.; Whittaker, M.; Yohn, C.B.; Lorenz, M.; Holmes, K.C.; Milligan, R.A. Structure of the actin-myosin complex and its implications for muscle contraction. Science 1993, 261, 58–65. [Google Scholar] [CrossRef]
- Washington, R.W.; Knecht, D.A. Actin binding domains direct actin-binding proteins to different cytoskeletal locations. BMC Cell Biol. 2008, 9, 10. [Google Scholar] [CrossRef]
- Shibata, K.; Nagasaki, A.; Adachi, H.; Uyeda, T.Q.P. Actin binding domain of filamin distinguishes posterior from anterior actin filaments in migrating Dictyostelium cells. Biophys. Physicobiol. 2016, 13, 321–331. [Google Scholar] [CrossRef]
- Harris, A.R.; Jreij, P.; Belardi, B.; Joffe, A.M.; Bausch, A.R.; Fletcher, D.A. Biased localization of actin binding proteins by actin filament conformation. Nat. Commun. 2020, 11, 5973. [Google Scholar] [CrossRef] [PubMed]
- Tsujioka, M.; Uyeda, T.Q.P.; Iwadate, Y.; Patel, H.; Shibata, K.; Yumoto, T.; Yonemura, S. Actin-binding domains mediate the distinct distribution of two Dictyostelium Talins through different affinities to specific subsets of actin filaments during directed cell migration. PLoS ONE 2019, 14, e0214736. [Google Scholar] [CrossRef] [PubMed]
- Egelman, E.H.; Francis, N.; DeRosier, D.J. F-actin is a helix with a random variable twist. Nature 1982, 298, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Kozuka, J.; Yokota, H.; Arai, Y.; Ishii, Y.; Yanagida, T. Dynamic polymorphism of single actin molecules in the actin filament. Nat. Chem. Biol. 2006, 2, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, S.; Inoue, Y.; Hojo, M.; Sokabe, M.; Adachi, T. Effect of tensile force on the mechanical behavior of actin filaments. J. Biomech. 2011, 44, 1776–1781. [Google Scholar] [CrossRef] [PubMed]
- Ngo, K.X.; Vu, H.T.; Umeda, K.; Trinh, M.N.; Kodera, N.; Uyeda, T. Deciphering the actin structure-dependent preferential cooperative binding of cofilin. eLife 2024, 13, RP95257. [Google Scholar] [CrossRef]
- Fineberg, A.; Takagi, Y.; Thirumurugan, K.; Andrecka, J.; Billington, N.; Young, G.; Cole, D.; Burgess, S.A.; Curd, A.P.; Hammer, J.A.; et al. Myosin-5 varies its step length to carry cargo straight along the irregular F-actin track. Proc. Natl. Acad. Sci. USA 2024, 121, e2401625121. [Google Scholar] [CrossRef] [PubMed]
- Varland, S.; Vandekerckhove, J.; Drazic, A. Actin Post-translational Modifications: The Cinderella of Cytoskeletal Control. Trends Biochem. Sci. 2019, 44, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Drazic, A.; Aksnes, H.; Marie, M.; Boczkowska, M.; Varland, S.; Timmerman, E.; Foyn, H.; Glomnes, N.; Rebowski, G.; Impens, F.; et al. NAA80 is actin’s N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc. Natl. Acad. Sci. USA 2018, 115, 4399–4404. [Google Scholar] [CrossRef]
- Hung, R.J.; Pak, C.W.; Terman, J.R. Direct redox regulation of F-actin assembly and disassembly by Mical. Science 2011, 334, 1710–1713. [Google Scholar] [CrossRef] [PubMed]
- Grintsevich, E.E.; Ge, P.; Sawaya, M.R.; Yesilyurt, H.G.; Terman, J.R.; Zhou, Z.H.; Reisler, E. Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Nat. Commun. 2017, 8, 2183. [Google Scholar] [CrossRef] [PubMed]
- Rajan, S.; Terman, J.R.; Reisler, E. MICAL-mediated oxidation of actin and its effects on cytoskeletal and cellular dynamics. Front. Cell Dev. Biol. 2023, 11, 1124202. [Google Scholar] [CrossRef] [PubMed]
- Jungbluth, A.; Eckerskorn, C.; Gerisch, G.; Lottspeich, F.; Stocker, S.; Schweiger, A. Stress-induced tyrosine phosphorylation of actin in Dictyostelium cells and localization of the phosphorylation site to tyrosine-53 adjacent to the DNase I binding loop. FEBS Lett. 1995, 375, 87–90. [Google Scholar] [CrossRef]
- Liu, X.; Shu, S.; Hong, M.S.; Levine, R.L.; Korn, E.D. Phosphorylation of actin Tyr-53 inhibits filament nucleation and elongation and destabilizes filaments. Proc. Natl. Acad. Sci. USA 2006, 103, 13694–13699. [Google Scholar] [CrossRef]
- Bertling, E.; Englund, J.; Minkeviciene, R.; Koskinen, M.; Segerstrale, M.; Castren, E.; Taira, T.; Hotulainen, P. Actin Tyrosine-53-Phosphorylation in Neuronal Maturation and Synaptic Plasticity. J. Neurosci. 2016, 36, 5299–5313. [Google Scholar] [CrossRef] [PubMed]
- Blanchoin, L.; Pollard, T.D. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry 2002, 41, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Melki, R.; Fievez, S.; Carlier, M.F. Continuous monitoring of Pi release following nucleotide hydrolysis in actin or tubulin assembly using 2-amino-6-mercapto-7-methylpurine ribonucleoside and purine-nucleoside phosphorylase as an enzyme-linked assay. Biochemistry 1996, 35, 12038–12045. [Google Scholar] [CrossRef]
- Zimmermann, D.; Santos, A.; Kovar, D.R.; Rock, R.S. Actin age orchestrates myosin-5 and myosin-6 run lengths. Curr. Biol. 2015, 25, 2057–2062. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.J.; Hachicho, C.; Carl, A.G.; Gong, R.; Alushin, G.M. Bending forces and nucleotide state jointly regulate F-actin structure. Nature 2022, 611, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Oosterheert, W.; Klink, B.U.; Belyy, A.; Pospich, S.; Raunser, S. Structural basis of actin filament assembly and aging. Nature 2022, 611, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Grabarek, Z.; Wang, C.L. Differential effects of caldesmon on the intermediate conformational states of polymerizing actin. J. Biol. Chem. 2010, 285, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Galkin, V.E.; Orlova, A.; Vos, M.R.; Schroder, G.F.; Egelman, E.H. Near-atomic resolution for one state of F-actin. Structure 2015, 23, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Galkin, V.E.; Orlova, A.; Kudryashov, D.S.; Solodukhin, A.; Reisler, E.; Schroder, G.F.; Egelman, E.H. Remodeling of actin filaments by ADF/cofilin proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 20568–20572. [Google Scholar] [CrossRef]
- Orlova, A.; Shvetsov, A.; Galkin, V.E.; Kudryashov, D.S.; Rubenstein, P.A.; Egelman, E.H.; Reisler, E. Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc. Natl. Acad. Sci. USA 2004, 101, 17664–17668. [Google Scholar] [CrossRef] [PubMed]
- Theriot, J.A.; Mitchison, T.J. Actin microfilament dynamics in locomoting cells. Nature 1991, 352, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Ponti, A.; Machacek, M.; Gupton, S.L.; Waterman-Storer, C.M.; Danuser, G. Two distinct actin networks drive the protrusion of migrating cells. Science 2004, 305, 1782–1786. [Google Scholar] [CrossRef] [PubMed]
- Ngo, K.X.; Kodera, N.; Katayama, E.; Ando, T.; Uyeda, T.Q.P. Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. eLife 2015, 4, e04806. [Google Scholar] [CrossRef] [PubMed]
- Isambert, H.; Venier, P.; Maggs, A.C.; Fattoum, A.; Kassab, R.; Pantaloni, D.; Carlier, M.F. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 1995, 270, 11437–11444. [Google Scholar] [CrossRef] [PubMed]
- McGough, A.; Pope, B.; Chiu, W.; Weeds, A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J. Cell Biol. 1997, 138, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Suarez, C.; Roland, J.; Boujemaa-Paterski, R.; Kang, H.; McCullough, B.R.; Reymann, A.C.; Guerin, C.; Martiel, J.L.; De La Cruz, E.M.; Blanchoin, L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr. Biol. 2011, 21, 862–868. [Google Scholar] [CrossRef] [PubMed]
- Galkin, V.E.; Orlova, A.; Lukoyanova, N.; Wriggers, W.; Egelman, E.H. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J. Cell Biol. 2001, 153, 75–86. [Google Scholar] [CrossRef]
- Huehn, A.R.; Bibeau, J.P.; Schramm, A.C.; Cao, W.; De La Cruz, E.M.; Sindelar, C.V. Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments. Proc. Natl. Acad. Sci. USA 2020, 117, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, T.Q.P.; Morimatsu, M.; Iwane, A.H.; Yanagida, T.; Uyeda, T.Q.P. The role of structural dynamics of actin in class-specific myosin motility. PLoS ONE 2015, 10, e0126262. [Google Scholar] [CrossRef]
- Galkin, V.E.; Orlova, A.; Schroder, G.F.; Egelman, E.H. Structural polymorphism in F-actin. Nat. Struct. Mol. Biol. 2010, 17, 1318–1323. [Google Scholar] [CrossRef]
- Changeux, J.P.; Edelstein, S. Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol. Rep. 2011, 3, 19. [Google Scholar] [CrossRef]
- Kojima, H.; Ishijima, A.; Yanagida, T. Direct measurement of stiffness of single actin filaments with and without tropomyosin by in vitro nanomanipulation. Proc. Natl. Acad. Sci. USA 1994, 91, 12962–12966. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, K.; Sugimoto, Y.; Tanaka, H.; Ueno, Y.; Takezawa, Y.; Amemiya, Y. X-ray diffraction evidence for the extensibility of actin and myosin filaments during muscle contraction. Biophys. J. 1994, 67, 2422–2435. [Google Scholar] [CrossRef]
- Okura, K.; Matsumoto, T.; Narita, A.; Tatsumi, H. Mechanical Stress Decreases the Amplitude of Twisting and Bending Fluctuations of Actin Filaments. J. Mol. Biol. 2023, 435, 168295. [Google Scholar] [CrossRef] [PubMed]
- Wioland, H.; Jégou, A.; Romet-Lemonne, G. Torsional stress generated by ADF/cofilin on cross-linked actin filaments boosts their severing. Proc. Natl. Acad. Sci. USA 2019, 116, 2595–2602. [Google Scholar] [CrossRef] [PubMed]
- Fukui, Y.; Lynch, T.J.; Brzeska, H.; Korn, E.D. Myosin I is located at the leading edges of locomoting Dictyostelium amoebae. Nature 1989, 341, 328–331. [Google Scholar] [CrossRef]
- Mei, L.; Espinosa de Los Reyes, S.; Reynolds, M.J.; Leicher, R.; Liu, S.; Alushin, G.M. Molecular mechanism for direct actin force-sensing by α-catenin. eLife 2020, 9, e62514. [Google Scholar] [CrossRef]
- Sun, X.; Phua, D.Y.Z.; Axiotakis, L., Jr.; Smith, M.A.; Blankman, E.; Gong, R.; Cail, R.C.; Espinosa de Los Reyes, S.; Beckerle, M.C.; Waterman, C.M.; et al. Mechanosensing through Direct Binding of Tensed F-Actin by LIM Domains. Dev. Cell 2020, 55, 468–482.e7. [Google Scholar] [CrossRef] [PubMed]
- Winkelman, J.D.; Anderson, C.A.; Suarez, C.; Kovar, D.R.; Gardel, M.L. Evolutionarily diverse LIM domain-containing proteins bind stressed actin filaments through a conserved mechanism. Proc. Natl. Acad. Sci. USA 2020, 117, 25532–25542. [Google Scholar] [CrossRef]
- Zsolnay, V.; Gardel, M.L.; Kovar, D.R.; Voth, G.A. Cracked actin filaments as mechanosensitive receptors. Biophys. J. 2024, 123, 3283–3294. [Google Scholar] [CrossRef] [PubMed]
- Tojkander, S.; Gateva, G.; Lappalainen, P. Actin stress fibers–assembly, dynamics and biological roles. J. Cell Sci. 2012, 125, 1855–1864. [Google Scholar] [CrossRef] [PubMed]
- Wolf, W.A.; Chew, T.L.; Chisholm, R.L. Regulation of cytokinesis. Cell. Mol. Life Sci. 1999, 55, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Takemasa, T.; Sugimoto, K.; Yamashita, K. Amplitude-dependent stress fiber reorientation in early response to cyclic strain. Exp. Cell Res. 1997, 230, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Verkhovsky, A.B.; Svitkina, T.M.; Borisy, G.G. Self-polarization and directional motility of cytoplasm. Curr. Biol. 1999, 9, 11–20. [Google Scholar] [CrossRef]
- Risca, V.I.; Wang, E.B.; Chaudhuri, O.; Chia, J.J.; Geissler, P.L.; Fletcher, D.A. Actin filament curvature biases branching direction. Proc. Natl. Acad. Sci. USA 2012, 109, 2913–2918. [Google Scholar] [CrossRef]
- Mizuno, H.; Higashida, C.; Yuan, Y.; Ishizaki, T.; Narumiya, S.; Watanabe, N. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science 2011, 331, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Tanaka, K.; Yamashiro, S.; Narita, A.; Watanabe, N. Helical rotation of the diaphanous-related formin mDia1 generates actin filaments resistant to cofilin. Proc. Natl. Acad. Sci. USA 2018, 115, E5000–E5007. [Google Scholar] [CrossRef] [PubMed]
- Norstrom, M.F.; Smithback, P.A.; Rock, R.S. Unconventional processive mechanics of non-muscle myosin IIB. J. Biol. Chem. 2010, 285, 26326–26334. [Google Scholar] [CrossRef] [PubMed]
- Hanein, D.; Matsudaira, P.; DeRosier, D.J. Evidence for a conformational change in actin induced by fimbrin (N375) binding. J. Cell Biol. 1997, 139, 387–396. [Google Scholar] [CrossRef]
- Galkin, V.E.; Orlova, A.; Cherepanova, O.; Lebart, M.C.; Egelman, E.H. High-resolution cryo-EM structure of the F-actin-fimbrin/plastin ABD2 complex. Proc. Natl. Acad. Sci. USA 2008, 105, 1494–1498. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.F.; Sherman, M.B.; Matsudaira, P.; Chiu, W. Structure of the acrosomal bundle. Nature 2004, 431, 104–107. [Google Scholar] [CrossRef]
- Belyy, A.; Merino, F.; Sitsel, O.; Raunser, S. Structure of the Lifeact-F-actin complex. PLoS Biol. 2020, 18, e3000925. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, Y.; Takaine, M.; Ngo, K.X.; Imai, T.; Yamada, M.D.; Behjat, A.B.; Umeda, K.; Hirose, K.; Yurtsever, A.; Kodera, N.; et al. Actin-binding domain of Rng2 sparsely bound on F-actin strongly inhibits actin movement on myosin II. Life Sci. Alliance 2023, 6, e202201469. [Google Scholar] [CrossRef]
- Oosawa, F.; Fujime, S.; Ishiwata, S.; Mihashi, K. Dynamic property of F-actin and thin filament. Cold Spring Harb. Symp. Quant. Biol. 1973, 37, 277–285. [Google Scholar] [CrossRef]
- Loscalzo, J.; Reed, G.H.; Weber, A. Conformational change and cooperativity in actin filaments free of tropomyosin. Proc. Natl. Acad. Sci. USA 1975, 72, 3412–3415. [Google Scholar] [CrossRef]
- Thomas, D.D.; Seidel, J.C.; Gergely, J. Rotational dynamics of spin-labeled F-actin in the sub-millisecond time range. J. Mol. Biol. 1979, 132, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Miki, M.; Wahl, P.; Auchet, J.C. Fluorescence anisotropy of labeled F-actin: Influence of divalent cations on the interaction between F-actin and myosin heads. Biochemistry 1982, 21, 3661–3665. [Google Scholar] [CrossRef] [PubMed]
- Prochniewicz, E.; Thomas, D.D. Perturbations of functional interactions with myosin induce long-range allosteric and cooperative structural changes in actin. Biochemistry 1997, 36, 12845–12853. [Google Scholar] [CrossRef] [PubMed]
- Prochniewicz, E.; Chin, H.F.; Henn, A.; Hannemann, D.E.; Olivares, A.O.; Thomas, D.D.; De La Cruz, E.M. Myosin isoform determines the conformational dynamics and cooperativity of actin filaments in the strongly bound actomyosin complex. J. Mol. Biol. 2010, 396, 501–509. [Google Scholar] [CrossRef]
- Fujii, T.; Namba, K. Structure of actomyosin rigour complex at 5.2 Å resolution and insights into the ATPase cycle mechanism. Nat. Commun. 2017, 8, 13969. [Google Scholar] [CrossRef] [PubMed]
- Behrmann, E.; Muller, M.; Penczek, P.A.; Mannherz, H.G.; Manstein, D.J.; Raunser, S. Structure of the rigor actin-tropomyosin-myosin complex. Cell 2012, 150, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Umeki, N.; Hirose, K.; Uyeda, T.Q.P. Cofilin-induced cooperative conformational changes of actin subunits revealed using cofilin-actin fusion protein. Sci. Rep. 2016, 6, 20406. [Google Scholar] [CrossRef] [PubMed]
- Wioland, H.; Guichard, B.; Senju, Y.; Myram, S.; Lappalainen, P.; Jégou, A.; Romet-Lemonne, G. ADF/Cofilin Accelerates Actin Dynamics by Severing Filaments and Promoting Their Depolymerization at Both Ends. Curr. Biol. 2017, 27, 1956–1967.e7. [Google Scholar] [CrossRef] [PubMed]
- Gressin, L.; Guillotin, A.; Guerin, C.; Blanchoin, L.; Michelot, A. Architecture dependence of actin filament network disassembly. Curr. Biol. 2015, 25, 1437–1447. [Google Scholar] [CrossRef]
- Prochniewicz, E.; Janson, N.; Thomas, D.D.; De La Cruz, E.M. Cofilin increases the torsional flexibility and dynamics of actin filaments. J. Mol. Biol. 2005, 353, 990–1000. [Google Scholar] [CrossRef]
- Dedova, I.V.; Nikolaeva, O.P.; Mikhailova, V.V.; dos Remedios, C.G.; Levitsky, D.I. Two opposite effects of cofilin on the thermal unfolding of F-actin: A differential scanning calorimetric study. Biophys. Chem. 2004, 110, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Bobkov, A.A.; Muhlrad, A.; Pavlov, D.A.; Kokabi, K.; Yilmaz, A.; Reisler, E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J. Mol. Biol. 2006, 356, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Grintsevich, E.E.; Phillips, M.L.; Reisler, E.; Gimzewski, J.K. Atomic force microscopy reveals drebrin induced remodeling of f-actin with subnanometer resolution. Nano Lett. 2011, 11, 825–827. [Google Scholar] [CrossRef]
- Yanagida, T.; Nakase, M.; Nishiyama, K.; Oosawa, F. Direct observation of motion of single F-actin filaments in the presence of myosin. Nature 1984, 307, 58–60. [Google Scholar] [CrossRef] [PubMed]
- Moos, C.; Eisenberg, E. Effect of myosin on actin-bound nucleotide exchange in the presence and absence of ATP. Biochim. Biophys. Acta 1970, 223, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Nishida, E.; Maekawa, S.; Sakai, H. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry 1984, 23, 5307–5313. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Ohshima, S.; Obinata, T. A cofilin-like protein is involved in the regulation of actin assembly in developing skeletal muscle. J. Biochem. 1989, 106, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Fujime, S.; Ishiwata, S. Dynamic study of F-actin by quasielastic scattering of laser light. J. Mol. Biol. 1971, 62, 251–265. [Google Scholar] [CrossRef]
- Skau, C.T.; Courson, D.S.; Bestul, A.J.; Winkelman, J.D.; Rock, R.S.; Sirotkin, V.; Kovar, D.R. Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J. Biol. Chem. 2011, 286, 26964–26977. [Google Scholar] [CrossRef]
- Papp, G.; Bugyi, B.; Ujfalusi, Z.; Barko, S.; Hild, G.; Somogyi, B.; Nyitrai, M. Conformational changes in actin filaments induced by formin binding to the barbed end. Biophys. J. 2006, 91, 2564–2572. [Google Scholar] [CrossRef]
- Bugyi, B.; Papp, G.; Hild, G.; Lorinczy, D.; Nevalainen, E.M.; Lappalainen, P.; Somogyi, B.; Nyitrai, M. Formins regulate actin filament flexibility through long range allosteric interactions. J. Biol. Chem. 2006, 281, 10727–10736. [Google Scholar] [CrossRef]
- Michelot, A.; Costanzo, M.; Sarkeshik, A.; Boone, C.; Yates, J.R., 3rd; Drubin, D.G. Reconstitution and protein composition analysis of endocytic actin patches. Curr. Biol. 2010, 20, 1890–1899. [Google Scholar] [CrossRef] [PubMed]
- Homa, K.E.; Hocky, G.M.; Suarez, C.; Kovar, D.R. Arp2/3 complex- and formin-mediated actin cytoskeleton networks facilitate actin binding protein sorting in fission yeast. Eur. J. Cell Biol. 2024, 103, 151404. [Google Scholar] [CrossRef] [PubMed]
- Kron, S.J.; Spudich, J.A. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. USA 1986, 83, 6272–6276. [Google Scholar] [CrossRef] [PubMed]
- Tokuraku, K.; Uyeda, T.Q. Phalloidin affects the myosin-dependent sliding velocities of actin filaments in a bound-divalent cation dependent manner. J. Muscle Res. Cell Motil. 2001, 22, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Rula, S.; Suwa, T.; Kijima, S.T.; Haraguchi, T.; Wakatsuki, S.; Sato, N.; Duan, Z.; Tominaga, M.; Uyeda, T.Q.P.; Ito, K. Measurement of enzymatic and motile activities of Arabidopsis myosins by using Arabidopsis actins. Biochem. Biophys. Res. Commun. 2018, 495, 2145–2151. [Google Scholar] [CrossRef]
- Patterson, B.; Ruppel, K.M.; Wu, Y.; Spudich, J.A. Cold-sensitive mutants G680V and G691C of Dictyostelium myosin II confer dramatically different biochemical defects. J. Biol. Chem. 1997, 272, 27612–27617. [Google Scholar] [CrossRef]
- Iwase, K.; Tanaka, M.; Hirose, K.; Uyeda, T.Q.P.; Honda, H. Acceleration of the sliding movement of actin filaments with the use of a non-motile mutant myosin in in vitro motility assays driven by skeletal muscle heavy meromyosin. PLoS ONE 2017, 12, e0181171. [Google Scholar] [CrossRef] [PubMed]
- Kubota, H.; Mikhailenko, S.V.; Okabe, H.; Taguchi, H.; Ishiwata, S. D-loop of actin differently regulates the motor function of myosins II and V. J. Biol. Chem. 2009, 284, 35251–35258. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, T.Q.P.; Komori, T.; Umeki, N.; Demizu, N.; Ito, K.; Iwane, A.H.; Tokuraku, K.; Yanagida, T.; Uyeda, T.Q.P. G146V mutation at the hinge region of actin reveals a myosin class-specific requirement of actin conformations for motility. J. Biol. Chem. 2012, 287, 24339–24345. [Google Scholar] [CrossRef] [PubMed]
- Umeki, N.; Shibata, K.; Noguchi, T.Q.P.; Hirose, K.; Sako, Y.; Uyeda, T.Q.P. K336I mutant actin alters the structure of neighbouring protomers in filaments and reduces affinity for actin-binding proteins. Sci. Rep. 2019, 9, 5353. [Google Scholar] [CrossRef] [PubMed]
- Prochniewicz, E.; Yanagida, T. Inhibition of sliding movement of F-actin by crosslinking emphasizes the role of actin structure in the mechanism of motility. J. Mol. Biol. 1990, 216, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Prochniewicz, E.; Katayama, E.; Yanagida, T.; Thomas, D.D. Cooperativity in F-actin: Chemical modifications of actin monomers affect the functional interactions of myosin with unmodified monomers in the same actin filament. Biophys. J. 1993, 65, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, M.; Pope, B.; Maciver, S.K.; Weeds, A.G. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry 1993, 32, 9985–9993. [Google Scholar] [CrossRef] [PubMed]
- Hayden, S.M.; Miller, P.S.; Brauweiler, A.; Bamburg, J.R. Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry 1993, 32, 9994–10004. [Google Scholar] [CrossRef]
- De La Cruz, E.M. Cofilin binding to muscle and non-muscle actin filaments: Isoform-dependent cooperative interactions. J. Mol. Biol. 2005, 346, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Bobkov, A.A.; Muhlrad, A.; Shvetsov, A.; Benchaar, S.; Scoville, D.; Almo, S.C.; Reisler, E. Cofilin (ADF) affects lateral contacts in F-actin. J. Mol. Biol. 2004, 337, 93–104. [Google Scholar] [CrossRef]
- Muhlrad, A.; Kudryashov, D.; Michael Peyser, Y.; Bobkov, A.A.; Almo, S.C.; Reisler, E. Cofilin induced conformational changes in F-actin expose subdomain 2 to proteolysis. J. Mol. Biol. 2004, 342, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Prochniewicz, E.; Zhang, Q.; Janmey, P.A.; Thomas, D.D. Cooperativity in F-actin: Binding of gelsolin at the barbed end affects structure and dynamics of the whole filament. J. Mol. Biol. 1996, 260, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J. Horizontal gene transfer in eukaryotes: Aligning theory with data. Nat. Rev. Genet. 2024, 25, 416–430. [Google Scholar] [CrossRef] [PubMed]
- Galkin, V.E.; Orlova, A.; Egelman, E.H. Actin filaments as tension sensors. Curr. Biol. 2012, 22, R96–R101. [Google Scholar] [CrossRef] [PubMed]

| Mechanism | Hierarchy | Fundamental vs. Lineage-Specific * |
|---|---|---|
| Local biochemical regulation of actin nucleators | High | Fundamental ** |
| Local biochemical regulation of side-binding and capping ABPs | High | Mixed? |
| Differences in dissociation rates | Low | Fundamental? |
| Actin isoform-dependent selection of binding ABPs | Low | Lineage-specific |
| Tm isoform-dependent selection of binding ABPs | Low | Lineage-specific |
| Physical geometry-dependent selection of ABPs | Low | Lineage-specific |
| Post-translational modification of actin | Low | Lineage-specific |
| Aging | High | Fundamental |
| Thermal and stochastic fluctuations | High *** | Fundamental |
| Mechanical strain | High | Fundamental |
| Bound ABP-induced cooperative conformational changes of actin filaments | Low | Fundamental **** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uyeda, T.Q.P.; Yamazaki, Y.; Kijima, S.T.; Noguchi, T.Q.P.; Ngo, K.X. Multiple Mechanisms to Regulate Actin Functions: “Fundamental” Versus Lineage-Specific Mechanisms and Hierarchical Relationships. Biomolecules 2025, 15, 279. https://doi.org/10.3390/biom15020279
Uyeda TQP, Yamazaki Y, Kijima ST, Noguchi TQP, Ngo KX. Multiple Mechanisms to Regulate Actin Functions: “Fundamental” Versus Lineage-Specific Mechanisms and Hierarchical Relationships. Biomolecules. 2025; 15(2):279. https://doi.org/10.3390/biom15020279
Chicago/Turabian StyleUyeda, Taro Q. P., Yosuke Yamazaki, Saku T. Kijima, Taro Q. P. Noguchi, and Kien Xuan Ngo. 2025. "Multiple Mechanisms to Regulate Actin Functions: “Fundamental” Versus Lineage-Specific Mechanisms and Hierarchical Relationships" Biomolecules 15, no. 2: 279. https://doi.org/10.3390/biom15020279
APA StyleUyeda, T. Q. P., Yamazaki, Y., Kijima, S. T., Noguchi, T. Q. P., & Ngo, K. X. (2025). Multiple Mechanisms to Regulate Actin Functions: “Fundamental” Versus Lineage-Specific Mechanisms and Hierarchical Relationships. Biomolecules, 15(2), 279. https://doi.org/10.3390/biom15020279






