Inorganic Polyphosphate Modulates Chromosome Transmission Fidelity in the Fission Yeast Schizosaccharomyces pombe
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
2.2. Media and Growth Conditions
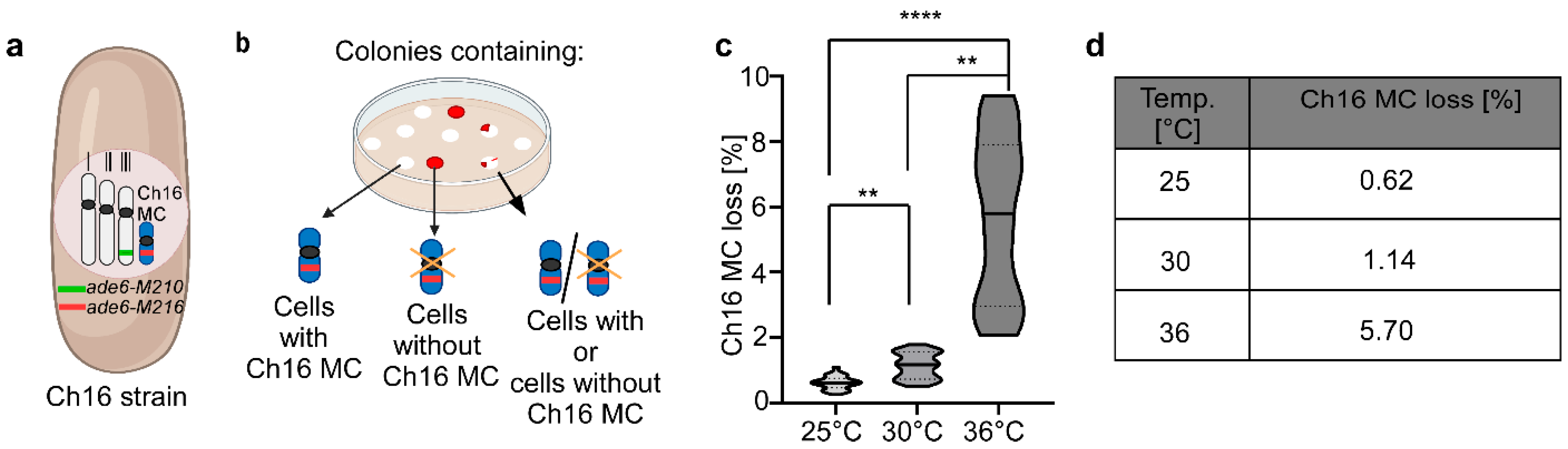
2.3. Mini-Chromosome Loss Assay
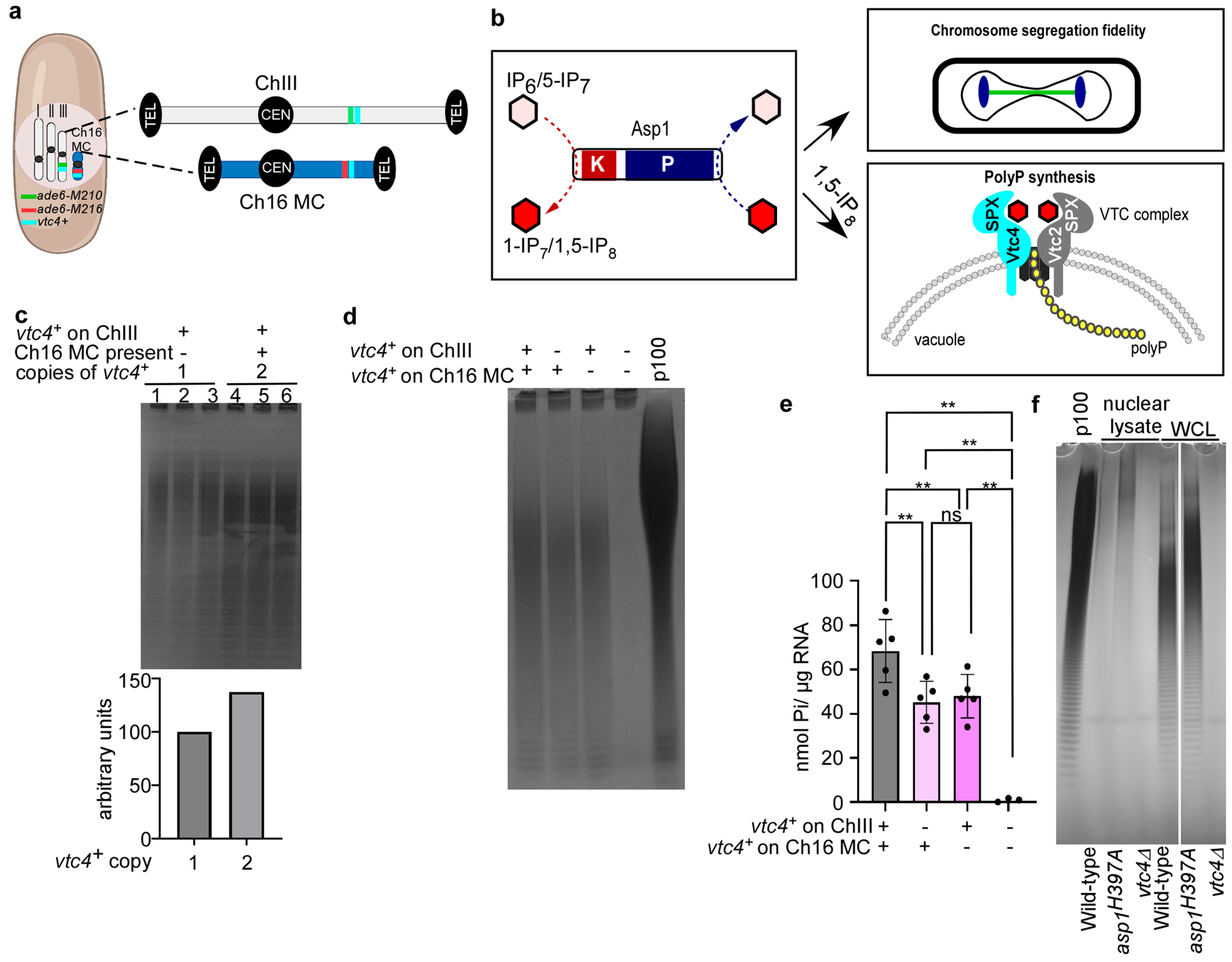
2.4. Isolation of Nuclei
2.5. Polyphosphate Extraction and Analysis
2.6. Western Blot Analysis
2.7. Microscopy
2.8. Programs and Statistical Analysis
3. Results
3.1. Chromosome Transmission Fidelity Is Temperature Dependent
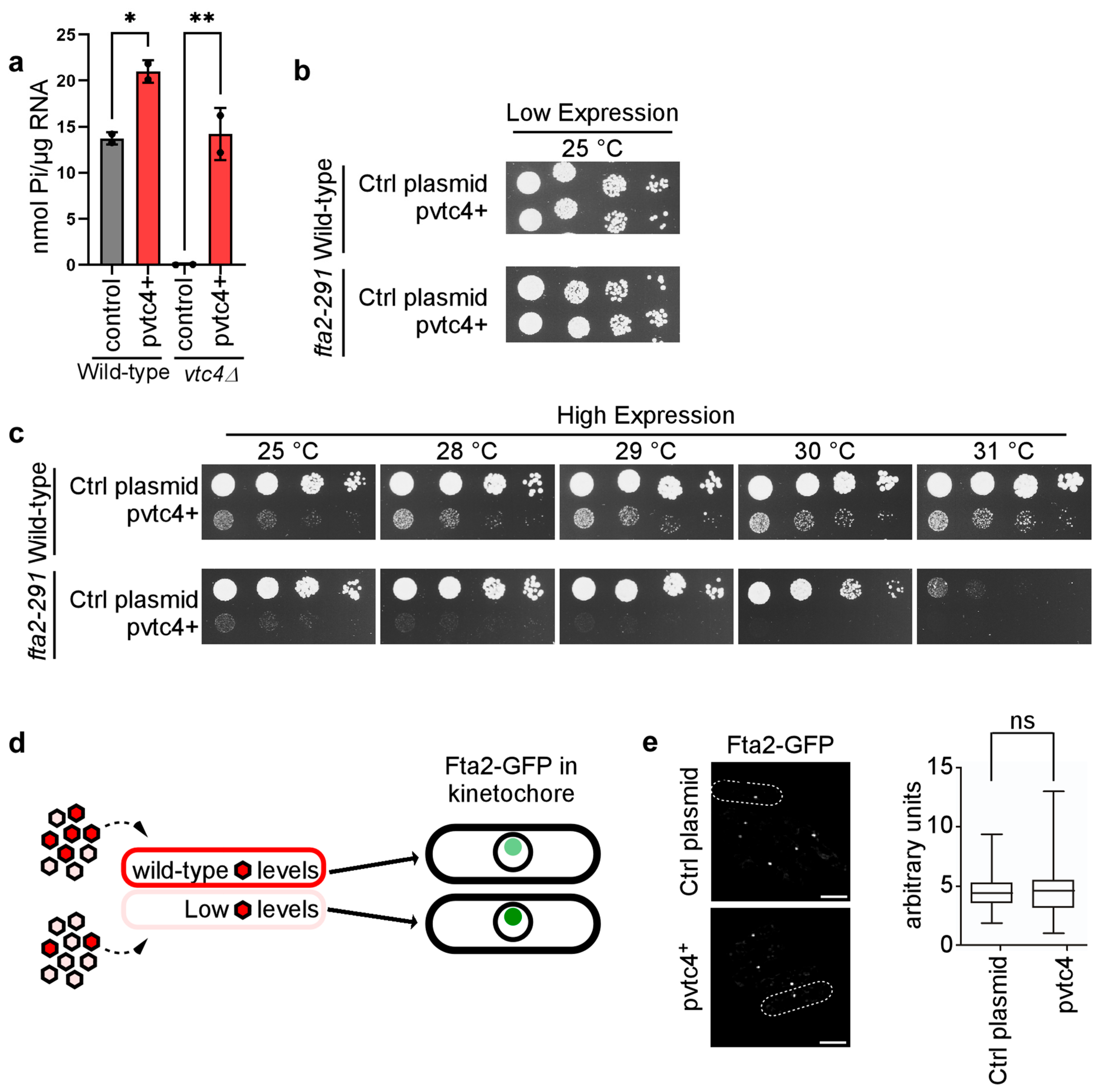
3.2. vtc4+ Gene Dosage Impacts polyP Levels
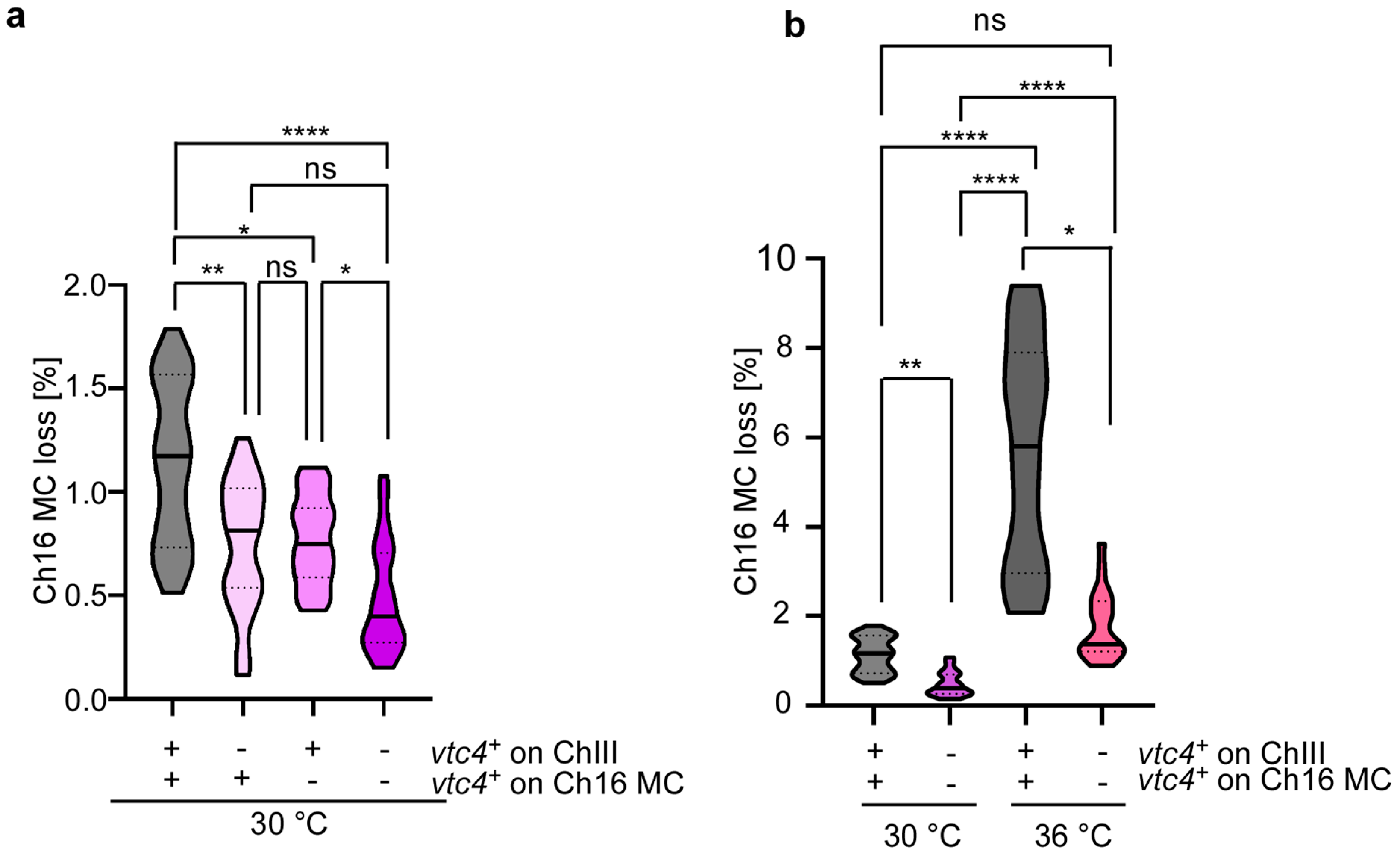
3.3. Cellular polyP Levels Impact Chromosome Transmission Fidelity in a Dose-Dependent Manner
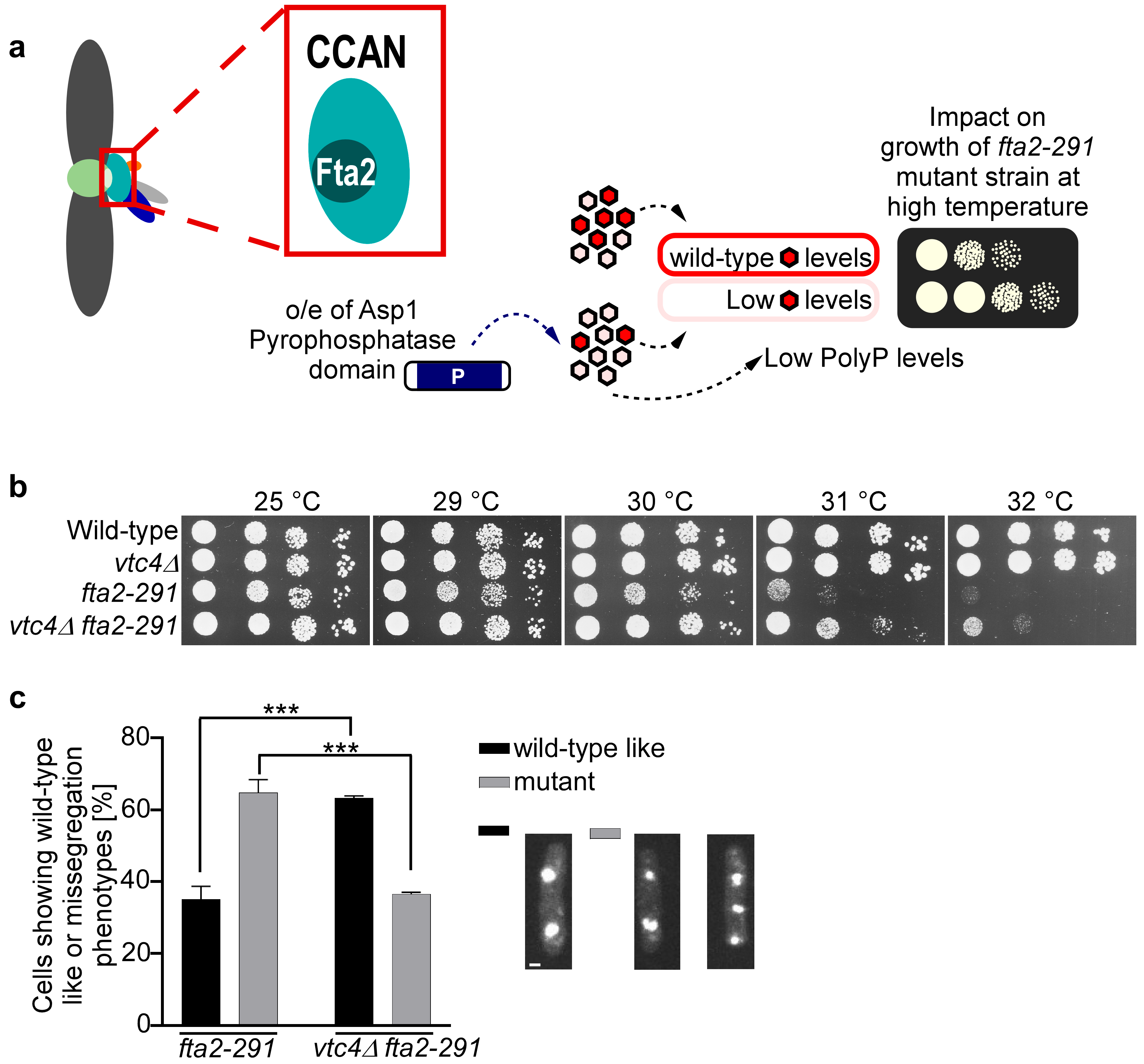
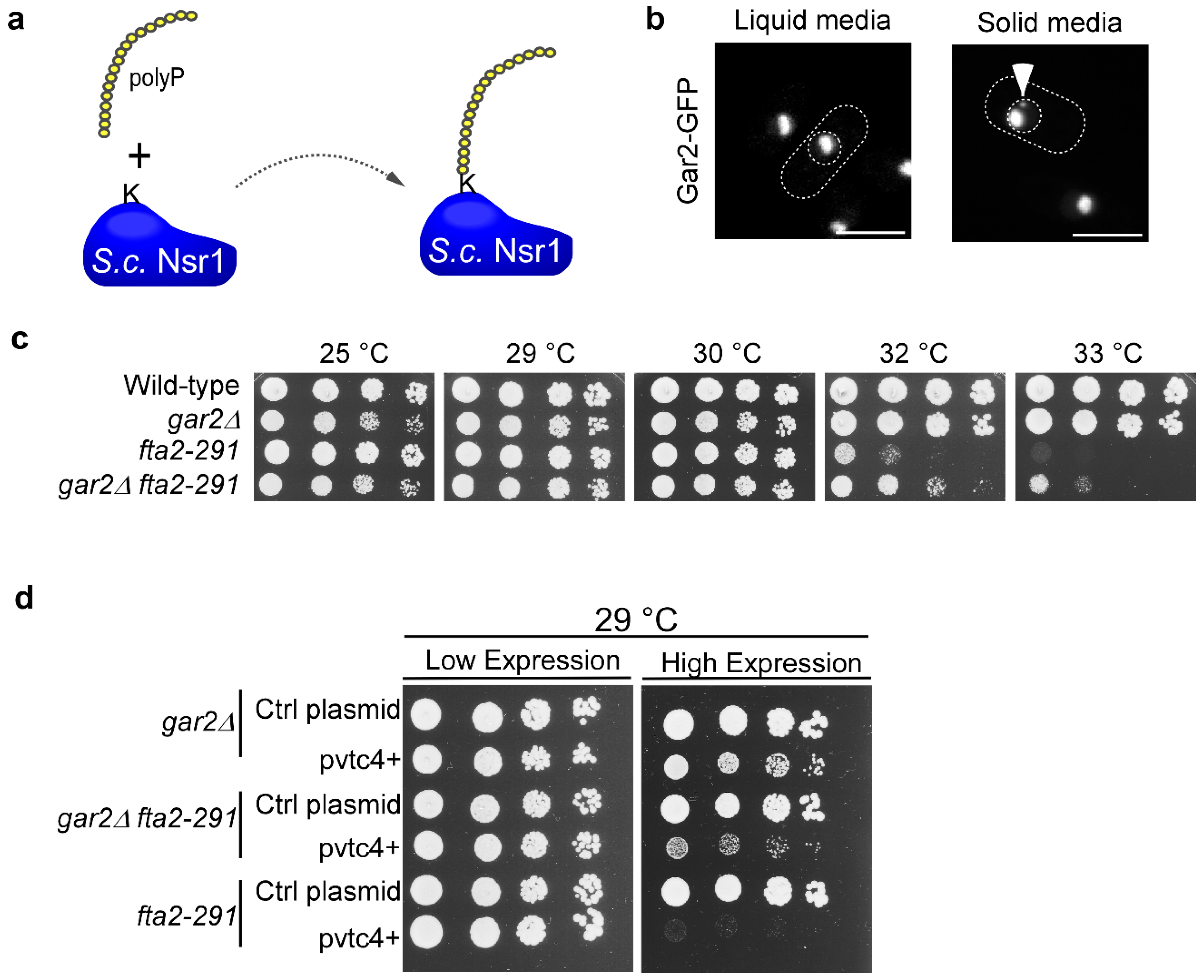
3.4. PolyP Impacts Kinetochore Function
3.5. Nucleolin Plays a Role in CCAN Kinetochore Complex Function
4. Discussion
4.1. Moderate Growth Temperature Changes and Chromosome Transmission Fidelity
4.2. PolyP Dosage and Chromosome Transmission Fidelity
4.3. PolyP and the Kinetochore
4.4. PolyP and Asp1-Modulated Chromosome Segregation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Knouse, K.A.; Davoli, T.; Elledge, S.J.; Amon, A. Aneuploidy in Cancer: Seq-Ing Answers to Old Questions. Annu. Rev. Cancer Biol. 2017, 1, 335–354. [Google Scholar] [CrossRef]
- Sdeor, E.; Okada, H.; Saad, R.; Ben-Yishay, T.; Ben-David, U. Aneuploidy as a Driver of Human Cancer. Nat. Genet. 2024, 56, 2014–2026. [Google Scholar] [CrossRef]
- Torres, E.M.; Sokolsky, T.; Tucker, C.M.; Chan, L.Y.; Boselli, M.; Dunham, M.J.; Amon, A. Effects of Aneuploidy on Cellular Physiology and Cell Division in Haploid Yeast. Science 2007, 317, 916–924. [Google Scholar] [CrossRef]
- Sheltzer, J.M.; Torres, E.M.; Dunham, M.J.; Amon, A. Transcriptional Consequences of Aneuploidy. Proc. Natl. Acad. Sci. USA 2012, 109, 12644–12649. [Google Scholar] [CrossRef]
- Zhu, J.; Tsai, H.-J.; Gordon, M.R.; Li, R. Cellular Stress Associated with Aneuploidy. Dev. Cell 2018, 44, 420–431. [Google Scholar] [CrossRef]
- Gilchrist, C.; Stelkens, R. Aneuploidy in Yeast: Segregation Error or Adaptation Mechanism? Yeast 2019, 36, 525–539. [Google Scholar] [CrossRef]
- Torres, E.M. Consequences of Gaining an Extra Chromosome. Chromosome Res. 2023, 31, 24. [Google Scholar] [CrossRef]
- Anders, K.R.; Kudrna, J.R.; Keller, K.E.; Kinghorn, B.; Miller, E.M.; Pauw, D.; Peck, A.T.; Shellooe, C.E.; Strong, I.J.T. A Strategy for Constructing Aneuploid Yeast Strains by Transient Nondisjunction of a Target Chromosome. BMC Genet. 2009, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.M.; Macedo, J.C.; Mattingly, A.J.; Wangsa, D.; Camps, J.; Lima, V.; Gomes, A.M.; Dória, S.; Ried, T.; Logarinho, E.; et al. Chromosome Mis-Segregation and Cytokinesis Failure in Trisomic Human Cells. eLife 2015, 4, e05068. [Google Scholar] [CrossRef]
- Loegler, V.; Friedrich, A.; Schacherer, J. Overview of the Saccharomyces cerevisiae Population Structure through the Lens of 3034 Genomes. G3 Genes Genomes Genet. 2024, 14, jkae245. [Google Scholar] [CrossRef] [PubMed]
- Sunshine, A.B.; Payen, C.; Ong, G.T.; Liachko, I.; Tan, K.M.; Dunham, M.J. The Fitness Consequences of Aneuploidy Are Driven by Condition-Dependent Gene Effects. PLoS Biol. 2015, 13, e1002155. [Google Scholar] [CrossRef]
- Berman, J. Ploidy Plasticity: A Rapid and Reversible Strategy for Adaptation to Stress. FEMS Yeast Res. 2016, 16, fow020. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Pinski, D.F.; Forsburg, S.L. Diploidy Confers Genomic Instability in Schizosaccharomyces pombe. Genetics 2025, 230, iyaf078. [Google Scholar] [CrossRef]
- Folco, H.D.; Chalamcharla, V.R.; Sugiyama, T.; Thillainadesan, G.; Zofall, M.; Balachandran, V.; Dhakshnamoorthy, J.; Mizuguchi, T.; Grewal, S.I.S. Untimely Expression of Gametogenic Genes in Vegetative Cells Causes Uniparental Disomy. Nature 2017, 543, 126–130. [Google Scholar] [CrossRef]
- Niwa, O.; Yanagida, M. Triploid Meiosis and Aneuploidy in Schizosaccharomyces pombe: An Unstable Aneuploid Disomic for Chromosome III. Curr. Genet. 1985, 9, 463–470. [Google Scholar] [CrossRef]
- Niwa, O.; Matsumoto, T.; Yanagida, M. Construction of a Mini-Chromosome by Deletion and Its Mitotic and Meiotic Behaviour in Fission Yeast. Mol. Gen. Genet. MGG 1986, 203, 397–405. [Google Scholar] [CrossRef]
- Niwa, O.; Matsumoto, T.; Chikashige, Y.; Yanagida, M. Characterization of Schizosaccharomyces pombe Minichromosome Deletion Derivatives and a Functional Allocation of Their Centromere. EMBO J. 1989, 8, 3045–3052. [Google Scholar] [CrossRef]
- Chikashige, Y.; Tsutsumi, C.; Okamasa, K.; Yamane, M.; Nakayama, J.; Niwa, O.; Haraguchi, T.; Hiraoka, Y. Gene Expression and Distribution of Swi6 in Partial Aneuploids of the Fission Yeast Schizosaccharomyces pombe. Cell Struct. Funct. 2007, 32, 149–161. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pascual-Ortiz, M.; Walla, E.; Fleig, U.; Saiardi, A. The PPIP5K Family Member Asp1 Controls Inorganic Polyphosphate Metabolism in S. Pombe. J. Fungi 2021, 7, 626. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamada, H.; Yanagida, M. Fission Yeast Minichromosome Loss Mutants Mis Cause Lethal Aneuploidy and Replication Abnormality. Mol. Biol. Cell 1994, 5, 1145–1158. [Google Scholar] [CrossRef]
- Fleig, U.; Sen-Gupta, M.; Hegemann, J.H. Fission Yeast mal2+ Is Required for Chromosome Segregation. Mol. Cell. Biol. 1996, 16, 6169–6177. [Google Scholar] [CrossRef]
- Javerzat, J.-P.; Cranston, G.; Allshire, R.C. Fission Yeast Genes Which Disrupt Mitotic Chromosome Segregation When Overexpressed. Nucleic Acids Res. 1996, 24, 4676–4683. [Google Scholar] [CrossRef]
- Beinhauer, J.D.; Hagan, I.M.; Hegemann, J.H.; Fleig, U. Mal3, the Fission Yeast Homologue of the Human APC-Interacting Protein EB-1 Is Required for Microtubule Integrity and the Maintenance of Cell Form. J. Cell Biol. 1997, 139, 717–728. [Google Scholar] [CrossRef]
- Takahashi, K.; Chen, E.S.; Yanagida, M. Requirement of Mis6 Centromere Connector for Localizing a CENP-A-Like Protein in Fission Yeast. Science 2000, 288, 2215–2219. [Google Scholar] [CrossRef] [PubMed]
- Nakase, Y.; Murakami, H.; Suma, M.; Nagano, K.; Wakuda, A.; Kitagawa, T.; Matsumoto, T. Cdc48 and Its Co-Factor Ufd1 Extract CENP-A from Centromeric Chromatin and Can Induce Chromosome Elimination in the Fission Yeast Schizosaccharomyces pombe. Biol. Open 2024, 13, bio060287. [Google Scholar] [CrossRef]
- Kumar, S.; Jeevaraj, T.; Yunus, M.H.; Chakraborty, S.; Chakraborty, N. The Plant Cytoskeleton Takes Center Stage in Abiotic Stress Responses and Resilience. Plant Cell Environ. 2023, 46, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, N.; DeRisi, J.; Brown, P.O. New Components of a System for Phosphate Accumulation and Polyphosphate Metabolism in Saccharomyces cerevisiae Revealed by Genomic Expression Analysis. Mol. Biol. Cell 2000, 11, 4309–4321. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, P.N.; Lander, N.; Kurup, S.P.; Reiss, L.; Brewer, J.; Soares Medeiros, L.C.; Miranda, K.; Docampo, R. The Acidocalcisome Vacuolar Transporter Chaperone 4 Catalyzes the Synthesis of Polyphosphate in Insect-stages of Trypanosoma brucei and T. cruzi. J. Eukaryot. Microbiol. 2014, 61, 155–165. [Google Scholar] [CrossRef]
- Jiménez, J.; Bru, S.; Ribeiro, M.P.C.; Clotet, J. Polyphosphate: Popping up from Oblivion. Curr. Genet. 2017, 63, 15–18. [Google Scholar] [CrossRef]
- Desfougères, Y.; Saiardi, A.; Azevedo, C. Inorganic Polyphosphate in Mammals: Where’s Wally? Biochem. Soc. Trans. 2020, 48, 95–101. [Google Scholar] [CrossRef]
- Borden, E.A.; Furey, M.; Gattone, N.J.; Hambardikar, V.D.; Liang, X.H.; Scoma, E.R.; Abou Samra, A.; D-Gary, L.R.; Dennis, D.J.; Fricker, D.; et al. Is There a Link between Inorganic Polyphosphate (polyP), Mitochondria, and Neurodegeneration? Pharmacol. Res. 2021, 163, 105211. [Google Scholar] [CrossRef]
- Montilla, M.; Atienza-Navarro, I.; García-Cozar, F.J.; Castro, C.; Rodríguez-Martorell, F.J.; Ruiz, F.A. Polyphosphate Activates von Willebrand Factor Interaction with Glycoprotein Ib in the Absence of Factor VIII In Vitro. Int. J. Mol. Sci. 2022, 23, 14118. [Google Scholar] [CrossRef]
- Borghi, F.; Azevedo, C.; Johnson, E.; Burden, J.J.; Saiardi, A. A Mammalian Model Reveals Inorganic Polyphosphate Channeling into the Nucleolus and Induction of a Hyper-Condensate State. Cell Rep. Methods 2024, 4, 100814. [Google Scholar] [CrossRef]
- Garcés, P.; Amaro, A.; Montecino, M.; Van Zundert, B. Inorganic Polyphosphate: From Basic Research to Diagnostic and Therapeutic Opportunities in ALS/FTD. Biochem. Soc. Trans. 2024, 52, 123–135. [Google Scholar] [CrossRef]
- Schoeppe, R.; Waldmann, M.; Jessen, H.J.; Renné, T. An Update on Polyphosphate In Vivo Activities. Biomolecules 2024, 14, 937. [Google Scholar] [CrossRef]
- Rai, A.; Jakob, U. Polyphosphate: A Cellular Swiss Army Knife. Curr. Opin. Biotechnol. 2025, 93, 103303. [Google Scholar] [CrossRef]
- Freimoser, F.M.; Hürlimann, H.C.; Jakob, C.A.; Werner, T.P.; Amrhein, N. Systematic Screening of Polyphosphate (Poly P) Levels in Yeast Mutant Cells Reveals Strong Interdependence with Primary Metabolism. Genome Biol. 2006, 7, R109. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Wang, C.; Shou, H.; Whelan, J. Phosphate Homeostasis in the Yeast Saccharomyces cerevisiae, the Key Role of the SPX Domain-containing Proteins. FEBS Lett. 2012, 586, 289–295. [Google Scholar] [CrossRef]
- Azevedo, C.; Livermore, T.; Saiardi, A. Protein Polyphosphorylation of Lysine Residues by Inorganic Polyphosphate. Mol. Cell 2015, 58, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Bru, S.; Martínez-Laínez, J.M.; Hernández-Ortega, S.; Quandt, E.; Torres-Torronteras, J.; Martí, R.; Canadell, D.; Ariño, J.; Sharma, S.; Jiménez, J.; et al. Polyphosphate Is Involved in Cell Cycle Progression and Genomic Stability in Saccharomyces cerevisiae. Mol. Microbiol. 2016, 101, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Solesio, M.E.; Garcia Del Molino, L.C.; Elustondo, P.A.; Diao, C.; Chang, J.C.; Pavlov, E.V. Inorganic Polyphosphate Is Required for Sustained Free Mitochondrial Calcium Elevation, Following Calcium Uptake. Cell Calcium 2020, 86, 102127. [Google Scholar] [CrossRef]
- Topolski, B.; Jakopec, V.; Künzel, N.A.; Fleig, U. Inositol Pyrophosphate Kinase Asp1 Modulates Chromosome Segregation Fidelity and Spindle Function in Schizosaccharomyces pombe. Mol. Cell. Biol. 2016, 36, 3128–3140. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pascual-Ortiz, M.; Saiardi, A.; Walla, E.; Jakopec, V.; Künzel, N.A.; Span, I.; Vangala, A.; Fleig, U. Asp1 Bifunctional Activity Modulates Spindle Function via Controlling Cellular Inositol Pyrophosphate Levels in Schizosaccharomyces pombe. Mol. Cell. Biol. 2018, 38, e00047-18. [Google Scholar] [CrossRef]
- Kuenzel, N.A.; Alcázar-Román, A.R.; Saiardi, A.; Bartsch, S.M.; Daunaraviciute, S.; Fiedler, D.; Fleig, U. Inositol Pyrophosphate-Controlled Kinetochore Architecture and Mitotic Entry in S. Pombe. J. Fungi 2022, 8, 933. [Google Scholar] [CrossRef]
- Bähler, J.; Wu, J.-Q.; Longtine, M.S.; Shah, N.G.; Mckenzie Iii, A.; Steever, A.B.; Wach, A.; Philippsen, P.; Pringle, J.R. Heterologous Modules for Efficient and Versatile PCR-Based Gene Targeting in Schizosaccharomyces pombe. Yeast 1998, 14, 943–951. [Google Scholar] [CrossRef]
- Gietz, R.D.; Woods, R.A. Transformation of Yeast by Lithium Acetate/Single-Stranded Carrier DNA/Polyethylene Glycol Method. Methods Enzymol. 2002, 350, 87–96. [Google Scholar] [CrossRef]
- Jakopec, V.; Walla, E.; Fleig, U. Versatile Use of Schizosaccharomyces pombe Plasmids in Saccharomyces cerevisiae. FEMS Yeast Res. 2011, 11, 653–655. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moreno, M.B.; Durán, A.; Ribas, J.C. A Family of Multifunctional Thiamine-Repressible Expression Vectors for Fission Yeast. Yeast 2000, 16, 861–872. [Google Scholar] [CrossRef]
- Moreno, S.; Klar, A.; Nurse, P. Molecular Genetic Analysis of Fission Yeast Schizosaccharomyces pombe. Methods Enzymol. 1991, 194, 795–823. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M.; Roth, S.Y.; Szent-Gyorgyi, C.; Simpson, R.T. Nucleosomes Are Positioned with Base Pair Precision Adjacent to the Alpha 2 Operator in Saccharomyces cerevisiae. EMBO J. 1991, 10, 3033–3041. [Google Scholar] [CrossRef]
- Lonetti, A.; Szijgyarto, Z.; Bosch, D.; Loss, O.; Azevedo, C.; Saiardi, A. Identification of an Evolutionarily Conserved Family of Inorganic Polyphosphate Endopolyphosphatases. J. Biol. Chem. 2011, 286, 31966–31974. [Google Scholar] [CrossRef]
- Knapp, B.D.; Huang, K.C. The Effects of Temperature on Cellular Physiology. Annu. Rev. Biophys. 2022, 51, 499–526. [Google Scholar] [CrossRef] [PubMed]
- Guan, Z.; Chen, J.; Liu, R.; Chen, Y.; Xing, Q.; Du, Z.; Cheng, M.; Hu, J.; Zhang, W.; Mei, W.; et al. The Cytoplasmic Synthesis and Coupled Membrane Translocation of Eukaryotic Polyphosphate by Signal-Activated VTC Complex. Nat. Commun. 2023, 14, 718. [Google Scholar] [CrossRef]
- Liu, W.; Wang, J.; Comte-Miserez, V.; Zhang, M.; Yu, X.; Chen, Q.; Jessen, H.J.; Mayer, A.; Wu, S.; Ye, S. Cryo-EM Structure of the Polyphosphate Polymerase VTC Reveals Coupling of Polymer Synthesis to Membrane Transit. EMBO J. 2023, 42, e113320. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Sanchez, A.M.; Schwer, B.; Prucker, I.; Jork, N.; Jessen, H.J.; Shuman, S. Activities and Genetic Interactions of Fission Yeast Aps1, a Nudix-Type Inositol Pyrophosphatase and Inorganic Polyphosphatase. mBio 2024, 15, e01084-24. [Google Scholar] [CrossRef] [PubMed]
- Pöhlmann, J.; Risse, C.; Seidel, C.; Pohlmann, T.; Jakopec, V.; Walla, E.; Ramrath, P.; Takeshita, N.; Baumann, S.; Feldbrügge, M.; et al. The Vip1 Inositol Polyphosphate Kinase Family Regulates Polarized Growth and Modulates the Microtubule Cytoskeleton in Fungi. PLoS Genet. 2014, 10, e1004586. [Google Scholar] [CrossRef]
- Lock, A.; Rutherford, K.; Harris, M.A.; Hayles, J.; Oliver, S.G.; Bähler, J.; Wood, V. PomBase 2018: User-Driven Reimplementation of the Fission Yeast Database Provides Rapid and Intuitive Access to Diverse, Interconnected Information. Nucleic Acids Res. 2019, 47, D821–D827. [Google Scholar] [CrossRef]
- Kumble, K.D.; Kornberg, A. Inorganic Polyphosphate in Mammalian Cells and Tissues. J. Biol. Chem. 1995, 270, 5818–5822. [Google Scholar] [CrossRef]
- Gerasimaitė, R.; Sharma, S.; Desfougères, Y.; Schmidt, A.; Mayer, A. Coupled Synthesis and Translocation Restrains Polyphosphate to Acidocalcisome-like Vacuoles and Prevents Its Toxicity. J. Cell Sci. 2014, 127, 5093–5104. [Google Scholar] [CrossRef]
- Sawada, N.; Ueno, S.; Takeda, K. Regulation of Inorganic Polyphosphate Is Required for Proper Vacuolar Proteolysis in Fission Yeast. J. Biol. Chem. 2021, 297, 100891. [Google Scholar] [CrossRef]
- Ariyoshi, M.; Fukagawa, T. An Updated View of the Kinetochore Architecture. Trends Genet. 2023, 39, 941–953. [Google Scholar] [CrossRef]
- Kerres, A.; Jakopec, V.; Beuter, C.; Karig, I. Fta2, an Essential Fission Yeast Kinetochore Component, Interacts Closely with the Conserved Mal2 Protein. Mol. Biol. Cell 2006, 17, 4167–4178. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Fujita, Y.; Iwasaki, O.; Adachi, Y.; Takahashi, K.; Yanagida, M. Mis16 and Mis18 Are Required for CENP-A Loading and Histone Deacetylation at Centromeres. Cell 2004, 118, 715–729. [Google Scholar] [CrossRef]
- Saitoh, S.; Takahashi, K.; Yanagida, M. Mis6, a Fission Yeast Inner Centromere Protein, Acts during G1/S and Forms Specialized Chromatin Required for Equal Segregation. Cell 1997, 90, 131–143. [Google Scholar] [CrossRef]
- Liu, X.; McLeod, I.; Anderson, S.; Yates, J.R.; He, X. Molecular Analysis of Kinetochore Architecture in Fission Yeast. EMBO J. 2005, 24, 2919–2930. [Google Scholar] [CrossRef] [PubMed]
- Freitag, M. The Kinetochore Interaction Network (KIN) of Ascomycetes. Mycologia 2016, 108, 485–505. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Appelgren, H.; Kniola, B.; Ekwall, K. Distinct Centromere Domain Structures with Separate Functions Demonstrated in Live Fission Yeast Cells. J. Cell Sci. 2003, 116, 4035–4042. [Google Scholar] [CrossRef][Green Version]
- Jiang, K.; Akhmanova, A. Microtubule Tip-Interacting Proteins: A View from Both Ends. Curr. Opin. Cell Biol. 2011, 23, 94–101. [Google Scholar] [CrossRef]
- Pöhlmann, J.; Fleig, U. Asp1, a Conserved 1/3 Inositol Polyphosphate Kinase, Regulates the Dimorphic Switch in Schizosaccharomyces pombe. Mol. Cell. Biol. 2010, 30, 4535–4547. [Google Scholar] [CrossRef]
- Fu, C.; Jain, D.; Costa, J.; Velve-Casquillas, G.; Tran, P.T. Mmb1p Binds Mitochondria to Dynamic Microtubules. Curr. Biol. 2011, 21, 1431–1439. [Google Scholar] [CrossRef]
- Li, T.; Zheng, F.; Cheung, M.; Wang, F.; Fu, C. Fission Yeast Mitochondria Are Distributed by Dynamic Microtubules in a Motor-Independent Manner. Sci. Rep. 2015, 5, 11023. [Google Scholar] [CrossRef]
- Da Costa, R.T.; Riggs, L.M.; Solesio, M.E. Inorganic Polyphosphate and the Regulation of Mitochondrial Physiology. Biochem. Soc. Trans. 2023, 51, 2153–2161. [Google Scholar] [CrossRef] [PubMed]
- Gulli, M.-P.; Girard, J.-P.; Zabetakis, D.; Lapeyre, B.; Melese, T.; Caizergues-Ferrer, M. Gar2 Is a Nucleolar Protein from Schizosaccharomyces pombe Required for 18S rRNA and 40S Ribosomal Subunit Accumulation. Nucleic Acids Res. 1995, 23, 1912–1918. [Google Scholar] [CrossRef]
- Prakash, K.; Satishkartik, S.; Ramalingam, S.; Gangadaran, P.; Gnanavel, S.; Aruljothi, K.N. Investigating the Multifaceted Role of Nucleolin in Cellular Function and Cancer: Structure, Regulation, and Therapeutic Implications. Gene 2025, 957, 149479. [Google Scholar] [CrossRef]
- Ma, N.; Matsunaga, S.; Takata, H.; Ono-Maniwa, R.; Uchiyama, S.; Fukui, K. Nucleolin Functions in Nucleolus Formation and Chromosome Congression. J. Cell Sci. 2007, 120, 2091–2105. [Google Scholar] [CrossRef]
- Lu, M.; He, X. Ccp1 Modulates Epigenetic Stability at Centromeres and Affects Heterochromatin Distribution in Schizosaccharomyces pombe. J. Biol. Chem. 2018, 293, 12068–12080. [Google Scholar] [CrossRef]
- Stamatiou, K.; Chmielewska, A.; Ohta, S.; Earnshaw, W.C.; Vagnarelli, P. CCDC86 Is a Novel Ki-67-Interacting Protein Important for Cell Division. J. Cell Sci. 2023, 136, jcs260391. [Google Scholar] [CrossRef]
- Tatebe, H.; Goshima, G.; Takeda, K.; Nakagawa, T.; Kinoshita, K.; Yanagida, M. Fission Yeast Living Mitosis Visualized by GFP-Tagged Gene Products. Micron 2001, 32, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Hothorn, M.; Neumann, H.; Lenherr, E.D.; Wehner, M.; Rybin, V.; Hassa, P.O.; Uttenweiler, A.; Reinhardt, M.; Schmidt, A.; Seiler, J.; et al. Catalytic Core of a Membrane-Associated Eukaryotic Polyphosphate Polymerase. Science 2009, 324, 513–516. [Google Scholar] [CrossRef] [PubMed]
- Chabert, V.; Kim, G.-D.; Qiu, D.; Liu, G.; Mayer, L.M.; Muhammed Jamsheer, K.; Jessen, H.J.; Mayer, A. Inositol Pyrophosphate Dynamics Reveals Control of the Yeast Phosphate Starvation Program Through 1,5-IP8 and the SPX Domain of Pho81. eLife 2023, 12, RP87956. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.J.; Jakob, U. Oxidative Stress Protection by Polyphosphate—New Roles for an Old Player. Curr. Opin. Microbiol. 2015, 24, 1–6. [Google Scholar] [CrossRef]
- Kus, F.; Smolenski, R.T.; Tomczyk, M. Inorganic Polyphosphate—Regulator of Cellular Metabolism in Homeostasis and Disease. Biomedicines 2022, 10, 913. [Google Scholar] [CrossRef]
- Morano, K.A.; Grant, C.M.; Moye-Rowley, W.S. The Response to Heat Shock and Oxidative Stress in Saccharomyces cerevisiae. Genetics 2012, 190, 1157–1195. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, P.S.; Larkin, M.; Thillainadesan, G.; Dhakshnamoorthy, J.; Balachandran, V.; Xiao, H.; Wellman, C.; Chatterjee, R.; Wheeler, D.; Grewal, S.I.S. Iron Homeostasis Regulates Facultative Heterochromatin Assembly in Adaptive Genome Control. Nat. Struct. Mol. Biol. 2018, 25, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Chaba, Z.; Jain, I.; Tran, P.T. Fission Yeast Spindle Dynamics and Chromosome Segregation Fidelity Show Distinct Thermosensitivity. MicroPubl. Biol. 2024, 10, 17912. [Google Scholar] [CrossRef]
- Robert, V.A.; Casadevall, A. Vertebrate Endothermy Restricts Most Fungi as Potential Pathogens. J. Infect. Dis. 2009, 200, 1623–1626. [Google Scholar] [CrossRef]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef]
- Lee, A.J.X.; Endesfelder, D.; Rowan, A.J.; Walther, A.; Birkbak, N.J.; Futreal, P.A.; Downward, J.; Szallasi, Z.; Tomlinson, I.P.M.; Howell, M.; et al. Chromosomal Instability Confers Intrinsic Multidrug Resistance. Cancer Res. 2011, 71, 1858–1870. [Google Scholar] [CrossRef]
- Jimenez-Nunez, M.D.; Moreno-Sanchez, D.; Hernandez-Ruiz, L.; Benitez-Rondan, A.; Ramos-Amaya, A.; Rodriguez-Bayona, B.; Medina, F.; Brieva, J.A.; Ruiz, F.A. Myeloma Cells Contain High Levels of Inorganic Polyphosphate Which Is Associated with Nucleolar Transcription. Haematologica 2012, 97, 1264–1271. [Google Scholar] [CrossRef]
- Xie, L.; Rajpurkar, A.; Quarles, E.; Taube, N.; Rai, A.S.; Erba, J.; Sliwinski, B.; Markowitz, M.; Jakob, U.; Knoefler, D. Accumulation of Nucleolar Inorganic Polyphosphate Is a Cellular Response to Cisplatin-Induced Apoptosis. Front. Oncol. 2019, 9, 1410. [Google Scholar] [CrossRef]
- Delinassios, J.G.; Hoffman, R.M.; Koumakis, G.; Palitskaris, D.; Poulatsidou, K.-N.; Delinasios, G.J. Sub-Toxic Cisplatin Concentrations Induce Extensive Chromosomal, Nuclear and Nucleolar Abnormalities Associated with High Malignancy before Acquired Resistance Develops: Implications for Clinical Caution. PLoS ONE 2024, 19, e0311976. [Google Scholar] [CrossRef]
- Sanchez, A.M.; Garg, A.; Schwer, B.; Shuman, S. Inorganic Polyphosphate Abets Silencing of a Sub-Telomeric Gene Cluster in Fission Yeast. MicroPubl. Biol. 2023, 10, 17912. [Google Scholar] [CrossRef]
- Beaufay, F.; Amemiya, H.M.; Guan, J.; Basalla, J.; Meinen, B.A.; Chen, Z.; Mitra, R.; Bardwell, J.C.A.; Biteen, J.S.; Vecchiarelli, A.G.; et al. Polyphosphate Drives Bacterial Heterochromatin Formation. Sci. Adv. 2021, 7, eabk0233. [Google Scholar] [CrossRef] [PubMed]
- Chawla, R.; Tom, J.K.A.; Boyd, T.; Tu, N.H.; Bai, T.; Grotjahn, D.A.; Park, D.; Deniz, A.A.; Racki, L.R. Reentrant DNA Shells Tune Polyphosphate Condensate Size. Nat. Commun. 2024, 15, 9258. [Google Scholar] [CrossRef] [PubMed]
- Shiroiwa, Y.; Hayashi, T.; Fujita, Y.; Villar-Briones, A.; Ikai, N.; Takeda, K.; Ebe, M.; Yanagida, M. Mis17 Is a Regulatory Module of the Mis6-Mal2-Sim4 Centromere Complex That Is Required for the Recruitment of CenH3/CENP-A in Fission Yeast. PLoS ONE 2011, 6, e17761. [Google Scholar] [CrossRef]
- Catania, S.; Allshire, R.C. Anarchic Centromeres: Deciphering Order from Apparent Chaos. Curr. Opin. Cell Biol. 2014, 26, 41–50. [Google Scholar] [CrossRef]
- Yamagishi, Y.; Sakuno, T.; Goto, Y.; Watanabe, Y. Kinetochore Composition and Its Function: Lessons from Yeasts. FEMS Microbiol. Rev. 2014, 38, 185–200. [Google Scholar] [CrossRef]
- Dong, Q.; Yin, F.-X.; Gao, F.; Shen, Y.; Zhang, F.; Li, Y.; He, H.; Gonzalez, M.; Yang, J.; Zhang, S.; et al. Ccp1 Homodimer Mediates Chromatin Integrity by Antagonizing CENP-A Loading. Mol. Cell 2016, 64, 79–91. [Google Scholar] [CrossRef] [PubMed][Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bollé, S.; Koc, E.; Saiardi, A.; Juhran, L.; Walla, E.; Fleig, U.; Alcázar-Román, A. Inorganic Polyphosphate Modulates Chromosome Transmission Fidelity in the Fission Yeast Schizosaccharomyces pombe. Biomolecules 2025, 15, 1331. https://doi.org/10.3390/biom15091331
Bollé S, Koc E, Saiardi A, Juhran L, Walla E, Fleig U, Alcázar-Román A. Inorganic Polyphosphate Modulates Chromosome Transmission Fidelity in the Fission Yeast Schizosaccharomyces pombe. Biomolecules. 2025; 15(9):1331. https://doi.org/10.3390/biom15091331
Chicago/Turabian StyleBollé, Sarune, Elisa Koc, Adolfo Saiardi, Lisa Juhran, Eva Walla, Ursula Fleig, and Abel Alcázar-Román. 2025. "Inorganic Polyphosphate Modulates Chromosome Transmission Fidelity in the Fission Yeast Schizosaccharomyces pombe" Biomolecules 15, no. 9: 1331. https://doi.org/10.3390/biom15091331
APA StyleBollé, S., Koc, E., Saiardi, A., Juhran, L., Walla, E., Fleig, U., & Alcázar-Román, A. (2025). Inorganic Polyphosphate Modulates Chromosome Transmission Fidelity in the Fission Yeast Schizosaccharomyces pombe. Biomolecules, 15(9), 1331. https://doi.org/10.3390/biom15091331







