Emerging Mechanistic Insights into AAA Complexes Regulating Proteasomal Degradation
Abstract
:1. Introduction
2. Core Particle
3. ATP-Dependent Regulators of the Archaeal Core Particle
4. Molecular Architecture of the 19S Regulatory Particle
5. Conformational Switching of the 26S Proteasome

6. Regulation of Proteasomal Degradation by Proteasome-Interacting Proteins


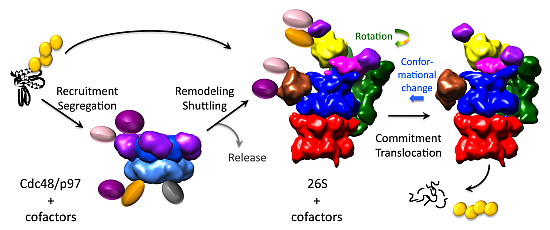
7. Cdc48/p97—A Facilitator of Proteasomal Degradation

8. Structure of Cdc48 and Its Associated Machinery
9. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A.; Varshavsky, A. Basic medical research award. The ubiquitin system. Nat. Med. 2000, 6, 1073–1081. [Google Scholar] [CrossRef]
- Hoeller, D.; Dikic, I. Targeting the ubiquitin system in cancer therapy. Nature 2009, 458, 438–444. [Google Scholar] [CrossRef]
- Lee, B.H.; Lee, M.J.; Park, S.; Oh, D.C.; Elsasser, S.; Chen, P.C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P.; et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Joazeiro, C.A. Ring domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Komander, D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009, 37, 937–953. [Google Scholar] [CrossRef]
- Finley, D.; Ulrich, H.D.; Sommer, T.; Kaiser, P. The ubiquitin-proteasome system of saccharomyces cerevisiae. Genetics 2012, 192, 319–360. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef]
- Sauer, R.T.; Baker, T.A. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 2011, 80, 587–612. [Google Scholar] [CrossRef]
- Bar-Nun, S.; Glickman, M.H. Proteasomal AAA-ATPases: Structure and function. Biochim. Biophys. Acta 2011, 1, 67–82. [Google Scholar]
- Buchberger, A.; Bukau, B.; Sommer, T. Protein quality control in the cytosol and the endoplasmic reticulum: Brothers in arms. Mol. Cell 2010, 40, 238–252. [Google Scholar] [CrossRef]
- Buchberger, A. Roles of Cdc48 in regulated protein degradation in yeast. Subcell Biochem. 2013, 66, 195–222. [Google Scholar] [CrossRef]
- Franz, A.; Ackermann, L.; Hoppe, T. Create and preserve: Proteostasis in development and aging is governed by Cdc48/p97/VCP. Biochim. Biophys. Acta 2014, 1843, 205–215. [Google Scholar] [CrossRef]
- Lowe, J.; Stock, D.; Jap, B.; Zwickl, P.; Baumeister, W.; Huber, R. Crystal structure of the 20S proteasome from the archaeon T. Acidophilum at 3.4 A resolution. Science 1995, 268, 533–539. [Google Scholar]
- Ruschak, A.M.; Religa, T.L.; Breuer, S.; Witt, S.; Kay, L.E. The proteasome antechamber maintains substrates in an unfolded state. Nature 2010, 467, 868–871. [Google Scholar] [CrossRef]
- Groll, M.; Bajorek, M.; Kohler, A.; Moroder, L.; Rubin, D.M.; Huber, R.; Glickman, M.H.; Finley, D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000, 7, 1062–1067. [Google Scholar] [CrossRef]
- Wenzel, T.; Baumeister, W. Conformational constraints in protein degradation by the 20S proteasome. Nat. Struct. Biol. 1995, 2, 199–204. [Google Scholar] [CrossRef]
- Religa, T.L.; Sprangers, R.; Kay, L.E. Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science 2010, 328, 98–102. [Google Scholar] [CrossRef]
- Latham, M.P.; Sekhar, A.; Kay, L.E. Understanding the mechanism of proteasome 20S core particle gating. Proc. Natl. Acad. Sci. USA 2014, 111, 5532–5537. [Google Scholar] [CrossRef]
- Groll, M.; Ditzel, L.; Lowe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef]
- Borissenko, L.; Groll, M. 20S proteasome and its inhibitors: Crystallographic knowledge for drug development. Chem. Rev. 2007, 107, 687–717. [Google Scholar] [CrossRef]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef]
- Huber, E.M.; Basler, M.; Schwab, R.; Heinemeyer, W.; Kirk, C.J.; Groettrup, M.; Groll, M. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 2012, 148, 727–738. [Google Scholar] [CrossRef]
- Samanovic, M.I.; Li, H.; Darwin, K.H. The pup-proteasome system of mycobacterium tuberculosis. Subcell. Biochem. 2013, 66, 267–295. [Google Scholar] [CrossRef]
- Pearce, M.J.; Mintseris, J.; Ferreyra, J.; Gygi, S.P.; Darwin, K.H. Ubiquitin-like protein involved in the proteasome pathway of mycobacterium tuberculosis. Science 2008, 322, 1104–1107. [Google Scholar] [CrossRef]
- Humbard, M.A.; Miranda, H.V.; Lim, J.M.; Krause, D.J.; Pritz, J.R.; Zhou, G.; Chen, S.; Wells, L.; Maupin-Furlow, J.A. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 2010, 463, 54–60. [Google Scholar] [CrossRef]
- Miranda, H.V.; Nembhard, N.; Su, D.; Hepowit, N.; Krause, D.J.; Pritz, J.R.; Phillips, C.; Soll, D.; Maupin-Furlow, J.A. E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc. Natl. Acad. Sci. USA 2011, 108, 4417–4422. [Google Scholar] [CrossRef]
- Hepowit, N.L.; Uthandi, S.; Miranda, H.V.; Toniutti, M.; Prunetti, L.; Olivarez, O.; de Vera, I.M.; Fanucci, G.E.; Chen, S.; Maupin-Furlow, J.A. Archaeal JAB1/MPN/MOV34 metalloenzyme (HvJAMM1) cleaves ubiquitin-like small archaeal modifier proteins (SAMPs) from protein-conjugates. Mol. Microbiol. 2012, 86, 971–987. [Google Scholar] [CrossRef]
- Cope, G.A.; Suh, G.S.; Aravind, L.; Schwarz, S.E.; Zipursky, S.L.; Koonin, E.V.; Deshaies, R.J. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 2002, 298, 608–611. [Google Scholar] [CrossRef]
- Benaroudj, N.; Goldberg, A.L. Pan, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nat. Cell Biol. 2000, 2, 833–839. [Google Scholar] [CrossRef]
- Smith, D.M.; Kafri, G.; Cheng, Y.; Ng, D.; Walz, T.; Goldberg, A.L. ATP binding to pan or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol. Cell 2005, 20, 687–698. [Google Scholar]
- Djuranovic, S.; Hartmann, M.D.; Habeck, M.; Ursinus, A.; Zwickl, P.; Martin, J.; Lupas, A.N.; Zeth, K. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol. Cell 2009, 34, 580–590. [Google Scholar] [CrossRef]
- Zhang, F.; Hu, M.; Tian, G.; Zhang, P.; Finley, D.; Jeffrey, P.D.; Shi, Y. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 2009, 34, 473–484. [Google Scholar] [CrossRef]
- Smith, D.M.; Chang, S.C.; Park, S.; Finley, D.; Cheng, Y.; Goldberg, A.L. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell 2007, 27, 731–744. [Google Scholar] [CrossRef]
- Rabl, J.; Smith, D.M.; Yu, Y.; Chang, S.C.; Goldberg, A.L.; Cheng, Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 2008, 30, 360–368. [Google Scholar] [CrossRef]
- Stadtmueller, B.M.; Ferrell, K.; Whitby, F.G.; Heroux, A.; Robinson, H.; Myszka, D.G.; Hill, C.P. Structural models for interactions between the 20S proteasome and its PAN/19s activators. J. Biol. Chem. 2010, 285, 13–17. [Google Scholar]
- Yu, Y.; Smith, D.M.; Kim, H.M.; Rodriguez, V.; Goldberg, A.L.; Cheng, Y. Interactions of PAN’s C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J. 2010, 29, 692–702. [Google Scholar] [CrossRef]
- Beck, F.; Unverdorben, P.; Bohn, S.; Schweitzer, A.; Pfeifer, G.; Sakata, E.; Nickell, S.; Plitzko, J.M.; Villa, E.; Baumeister, W.; et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. USA 2012, 109, 14870–14875. [Google Scholar] [CrossRef]
- Lander, G.C.; Estrin, E.; Matyskiela, M.E.; Bashore, C.; Nogales, E.; Martin, A. Complete subunit architecture of the proteasome regulatory particle. Nature 2012, 482, 186–191. [Google Scholar]
- Lasker, K.; Förster, F.; Bohn, S.; Walzthoeni, T.; Villa, E.; Unverdorben, P.; Beck, F.; Aebersold, R.; Sali, A.; Baumeister, W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. USA 2012, 109, 1380–1387. [Google Scholar] [CrossRef]
- Tian, G.; Park, S.; Lee, M.J.; Huck, B.; McAllister, F.; Hill, C.P.; Gygi, S.P.; Finley, D. An asymmetric interface between the regulatory and core particles of the proteasome. Nat. Struct. Mol. Biol. 2011, 18, 1259–1267. [Google Scholar] [CrossRef]
- Ruepp, A.; Graml, W.; Santos-Martinez, M.L.; Koretke, K.K.; Volker, C.; Mewes, H.W.; Frishman, D.; Stocker, S.; Lupas, A.N.; Baumeister, W. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 2000, 407, 508–513. [Google Scholar] [CrossRef]
- Pamnani, V.; Tamura, T.; Lupas, A.; Peters, J.; Cejka, Z.; Ashraf, W.; Baumeister, W. Cloning, sequencing and expression of VAT, a CDC48/p97 ATPase homologue from the archaeon Thermoplasma acidophilum. FEBS Lett. 1997, 404, 263–268. [Google Scholar] [CrossRef]
- Djuranovic, S.; Rockel, B.; Lupas, A.N.; Martin, J. Characterization of AMA, a new AAA protein from Archaeoglobus and methanogenic archaea. J. Struct. Biol. 2006, 156, 130–138. [Google Scholar] [CrossRef]
- Barthelme, D.; Sauer, R.T. Identification of the Cdc48*20S proteasome as an ancient AAA+ proteolytic machine. Science 2012, 337, 843–846. [Google Scholar] [CrossRef]
- Forouzan, D.; Ammelburg, M.; Hobel, C.F.; Stroh, L.J.; Sessler, N.; Martin, J.; Lupas, A.N. The archaeal proteasome is regulated by a network of AAA ATPases. J. Biol. Chem. 2012, 287, 39254–39262. [Google Scholar]
- Barthelme, D.; Chen, J.Z.; Grabenstatter, J.; Baker, T.A.; Sauer, R.T. Architecture and assembly of the archaeal Cdc48*20S proteasome. Proc. Natl. Acad. Sci. USA 2014. [Google Scholar] [CrossRef]
- Glickman, M.H.; Rubin, D.M.; Coux, O.; Wefes, I.; Pfeifer, G.; Cjeka, Z.; Baumeister, W.; Fried, V.A.; Finley, D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the Cop9-signalosome and EIF3. Cell 1998, 94, 615–623. [Google Scholar] [CrossRef]
- Förster, F.; Lasker, K.; Beck, F.; Nickell, S.; Sali, A.; Baumeister, W. An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem. Biophys. Res. Commun. 2009, 388, 228–233. [Google Scholar] [CrossRef]
- Tomko, R.J., Jr.; Funakoshi, M.; Schneider, K.; Wang, J.; Hochstrasser, M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: Implications for proteasome structure and assembly. Mol. Cell 2010, 38, 393–403. [Google Scholar] [CrossRef]
- Bohn, S.; Beck, F.; Sakata, E.; Walzthoeni, T.; Beck, M.; Aebersold, R.; Förster, F.; Baumeister, W.; Nickell, S. Structure of the 26S proteasome from schizosaccharomyces pombe at subnanometer resolution. Proc. Natl. Acad. Sci. USA 2010, 107, 20992–20997. [Google Scholar] [CrossRef]
- Unverdorben, P.; Beck, F.; Sledz, P.; Schweitzer, A.; Pfeifer, G.; Plitzko, J.M.; Baumeister, W.; Förster, F. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc. Natl. Acad. Sci. USA 2014, 111, 5544–5549. [Google Scholar] [CrossRef]
- Sakata, E.; Bohn, S.; Mihalache, O.; Kiss, P.; Beck, F.; Nagy, I.; Nickell, S.; Tanaka, K.; Saeki, Y.; Förster, F.; et al. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 1479–1484. [Google Scholar] [CrossRef]
- Pathare, G.R.; Nagy, I.; Bohn, S.; Unverdorben, P.; Hubert, A.; Korner, R.; Nickell, S.; Lasker, K.; Sali, A.; Tamura, T.; et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc. Natl. Acad. Sci. USA 2012, 109, 149–154. [Google Scholar] [CrossRef]
- Bohn, S.; Sakata, E.; Beck, F.; Pathare, G.R.; Schnitger, J.; Nagy, I.; Baumeister, W.; Förster, F. Localization of the regulatory particle subunit Sem1 in the 26S proteasome. Biochem. Biophys. Res. Commun. 2013, 435, 250–254. [Google Scholar] [CrossRef]
- Tomko, R.J., Jr.; Hochstrasser, M. The intrinsically disordered Sem1 protein functions as a molecular tether during proteasome lid biogenesis. Mol. Cell 2014, 53, 433–443. [Google Scholar] [CrossRef]
- Estrin, E.; Lopez-Blanco, J.R.; Chacon, P.; Martin, A. Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure 2013, 21, 1624–1635. [Google Scholar] [CrossRef]
- Pathare, G.R.; Nagy, I.; Sledz, P.; Anderson, D.J.; Zhou, H.J.; Pardon, E.; Steyaert, J.; Forster, F.; Bracher, A.; Baumeister, W. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc. Natl. Acad. Sci. USA 2014, 111, 2984–2989. [Google Scholar] [CrossRef]
- Worden, E.J.; Padovani, C.; Martin, A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat. Struct. Mol. Biol. 2014, 21, 220–227. [Google Scholar] [CrossRef]
- Yao, T.; Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 2002, 419, 403–407. [Google Scholar] [CrossRef]
- Verma, R.; Aravind, L.; Oania, R.; McDonald, W.H.; Yates, J.R., 3rd; Koonin, E.V.; Deshaies, R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 2002, 298, 611–615. [Google Scholar] [CrossRef]
- Förster, F.; Unverdorben, P.; Sledz, P.; Baumeister, W. Unveiling the long-held secrets of the 26S proteasome. Structure 2013, 21, 1551–1562. [Google Scholar] [CrossRef]
- Lander, G.C.; Martin, A.; Nogales, E. The proteasome under the microscope: The regulatory particle in focus. Curr. Opin. Struct. Biol. 2013, 23, 243–251. [Google Scholar] [CrossRef]
- Da Fonseca, P.C.; He, J.; Morris, E.P. Molecular model of the human 26S proteasome. Mol. Cell 2012, 46, 54–66. [Google Scholar] [CrossRef]
- Arai, S.; Saijo, S.; Suzuki, K.; Mizutani, K.; Kakinuma, Y.; Ishizuka-Katsura, Y.; Ohsawa, N.; Terada, T.; Shirouzu, M.; Yokoyama, S.; et al. Rotation mechanism of enterococcus hirae V1-ATPase based on asymmetric crystal structures. Nature 2013, 493, 703–707. [Google Scholar] [CrossRef]
- Itsathitphaisarn, O.; Wing, R.A.; Eliason, W.K.; Wang, J.; Steitz, T.A. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 2012, 151, 267–277. [Google Scholar] [CrossRef]
- Thomsen, N.D.; Berger, J.M. Running in reverse: The structural basis for translocation polarity in hexameric helicases. Cell 2009, 139, 523–534. [Google Scholar] [CrossRef]
- Sledz, P.; Unverdorben, P.; Beck, F.; Pfeifer, G.; Schweitzer, A.; Förster, F.; Baumeister, W. Structure of the 26S proteasome with ATP-gammas bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc. Natl. Acad. Sci. USA 2013, 110, 7264–7269. [Google Scholar]
- Matyskiela, M.E.; Lander, G.C.; Martin, A. Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 2013, 20, 781–788. [Google Scholar] [CrossRef]
- Lee, M.J.; Lee, B.H.; Hanna, J.; King, R.W.; Finley, D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol. Cell. Proteomics 2011. [Google Scholar] [CrossRef]
- Prakash, S.; Inobe, T.; Hatch, A.J.; Matouschek, A. Substrate selection by the proteasome during degradation of protein complexes. Nat. Chem. Biol. 2009, 5, 29–36. [Google Scholar]
- Inobe, T.; Fishbain, S.; Prakash, S.; Matouschek, A. Defining the geometry of the two-component proteasome degron. Nat. Chem. Biol. 2011, 7, 161–167. [Google Scholar] [CrossRef]
- Peth, A.; Uchiki, T.; Goldberg, A.L. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol. Cell 2010, 40, 671–681. [Google Scholar] [CrossRef]
- Nyquist, K.; Martin, A. Marching to the beat of the ring: Polypeptide translocation by AAA+ proteases. Trends Biochem. Sci. 2013, 39, 53–60. [Google Scholar] [CrossRef]
- Sen, M.; Maillard, R.A.; Nyquist, K.; Rodriguez-Aliaga, P.; Presse, S.; Martin, A.; Bustamante, C. The ClpXP protease unfolds substrates using a constant rate of pulling but different gears. Cell 2013, 155, 636–646. [Google Scholar] [CrossRef]
- Wang, X.; Huang, L. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol. Cell. Proteomics 2008, 7, 46–57. [Google Scholar] [CrossRef]
- Jacobson, A.D.; Macfadden, A.; Wu, Z.; Peng, J.; Liu, C.W. Autoregulation of the 26S proteasome by in situ ubiquitination. Mol. Biol. Cell 2014. [Google Scholar] [CrossRef]
- Wilkinson, C.R.; Seeger, M.; Hartmann-Petersen, R.; Stone, M.; Wallace, M.; Semple, C.; Gordon, C. Proteins containing the Uba domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 2001, 3, 939–943. [Google Scholar] [CrossRef]
- Bertolaet, B.L.; Clarke, D.J.; Wolff, M.; Watson, M.H.; Henze, M.; Divita, G.; Reed, S.I. Uba domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 2001, 8, 417–422. [Google Scholar] [CrossRef]
- Verma, R.; Oania, R.; Graumann, J.; Deshaies, R.J. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 2004, 118, 99–110. [Google Scholar] [CrossRef]
- Gomez, T.A.; Kolawa, N.; Gee, M.; Sweredoski, M.J.; Deshaies, R.J. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 2011. [Google Scholar] [CrossRef]
- Schmidt, M.; Hanna, J.; Elsasser, S.; Finley, D. Proteasome-associated proteins: Regulation of a proteolytic machine. Biol. Chem. 2005, 386, 725–737. [Google Scholar]
- Leggett, D.S.; Hanna, J.; Borodovsky, A.; Crosas, B.; Schmidt, M.; Baker, R.T.; Walz, T.; Ploegh, H.; Finley, D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 2002, 10, 495–507. [Google Scholar] [CrossRef]
- Inobe, T.; Matouschek, A. Paradigms of protein degradation by the proteasome. Curr. Opin. Struct. Biol. 2014, 24, 156–164. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Bronner, V.; Zhang, D.; Fushman, D.; Glickman, M.H. Rpn1 and Rpn2 coordinate ubiquitin processing factors at the proteasome. J. Biol. Chem. 2012, 287, 14659–14671. [Google Scholar] [CrossRef]
- Elsasser, S.; Gali, R.R.; Schwickart, M.; Larsen, C.N.; Leggett, D.S.; Muller, B.; Feng, M.T.; Tubing, F.; Dittmar, G.A.; Finley, D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 2002, 4, 725–730. [Google Scholar] [CrossRef]
- Saeki, Y.; Sone, T.; Toh-e, A.; Yokosawa, H. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem. Biophys. Res. Commun. 2002, 296, 813–819. [Google Scholar] [CrossRef]
- Hiyama, H.; Yokoi, M.; Masutani, C.; Sugasawa, K.; Maekawa, T.; Tanaka, K.; Hoeijmakers, J.H.; Hanaoka, F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26S proteasome. J. Biol. Chem. 1999, 274, 28019–28025. [Google Scholar] [CrossRef]
- Walters, K.J.; Kleijnen, M.F.; Goh, A.M.; Wagner, G.; Howley, P.M. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 2002, 41, 1767–1777. [Google Scholar] [CrossRef]
- Peth, A.; Besche, H.C.; Goldberg, A.L. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol. Cell 2009, 36, 794–804. [Google Scholar] [CrossRef]
- Hanna, J.; Hathaway, N.A.; Tone, Y.; Crosas, B.; Elsasser, S.; Kirkpatrick, D.S.; Leggett, D.S.; Gygi, S.P.; King, R.W.; Finley, D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 2006, 127, 99–111. [Google Scholar] [CrossRef]
- Chen, X.; Lee, B.H.; Finley, D.; Walters, K.J. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein Hrpn2. Mol. Cell 2010, 38, 404–415. [Google Scholar] [CrossRef]
- Lam, Y.A.; Xu, W.; DeMartino, G.N.; Cohen, R.E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 1997, 385, 737–740. [Google Scholar] [CrossRef]
- Holzl, H.; Kapelari, B.; Kellermann, J.; Seemuller, E.; Sumegi, M.; Udvardy, A.; Medalia, O.; Sperling, J.; Muller, S.A.; Engel, A.; et al. The regulatory complex of drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J. Cell Biol. 2000, 150, 119–130. [Google Scholar] [CrossRef] [Green Version]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef]
- Qiu, X.B.; Ouyang, S.Y.; Li, C.J.; Miao, S.; Wang, L.; Goldberg, A.L. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006, 25, 5742–5753. [Google Scholar] [CrossRef]
- Hamazaki, J.; Iemura, S.; Natsume, T.; Yashiroda, H.; Tanaka, K.; Murata, S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006, 25, 4524–4536. [Google Scholar] [CrossRef]
- Crosas, B.; Hanna, J.; Kirkpatrick, D.S.; Zhang, D.P.; Tone, Y.; Hathaway, N.A.; Buecker, C.; Leggett, D.S.; Schmidt, M.; King, R.W.; et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 2006, 127, 1401–1413. [Google Scholar] [CrossRef]
- Jarosch, E.; Taxis, C.; Volkwein, C.; Bordallo, J.; Finley, D.; Wolf, D.H.; Sommer, T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 2002, 4, 134–139. [Google Scholar] [CrossRef]
- Ye, Y.; Meyer, H.H.; Rapoport, T.A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 2001, 414, 652–656. [Google Scholar] [CrossRef]
- Rabinovich, E.; Kerem, A.; Frohlich, K.U.; Diamant, N.; Bar-Nun, S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 2002, 22, 626–634. [Google Scholar] [CrossRef]
- Xu, S.; Peng, G.; Wang, Y.; Fang, S.; Karbowski, M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol. Biol. Cell 2011, 22, 291–300. [Google Scholar] [CrossRef]
- Verma, R.; Oania, R.S.; Kolawa, N.J.; Deshaies, R.J. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife 2013, 2, e00308. [Google Scholar] [CrossRef]
- Dantuma, N.P.; Hoppe, T. Growing sphere of influence: Cdc48/p97 orchestrates ubiquitin-dependent extraction from chromatin. Trends Cell Biol. 2012, 22, 483–491. [Google Scholar] [CrossRef]
- Johnson, E.S.; Ma, P.C.; Ota, I.M.; Varshavsky, A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 1995, 270, 17442–17456. [Google Scholar]
- Lee, R.J.; Liu, C.W.; Harty, C.; McCracken, A.A.; Latterich, M.; Romisch, K.; DeMartino, G.N.; Thomas, P.J.; Brodsky, J.L. Uncoupling retro-translocation and degradation in the ER-associated degradation of a soluble protein. EMBO J. 2004, 23, 2206–2215. [Google Scholar] [CrossRef]
- Wahlman, J.; DeMartino, G.N.; Skach, W.R.; Bulleid, N.J.; Brodsky, J.L.; Johnson, A.E. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell 2007, 129, 943–955. [Google Scholar] [CrossRef]
- Lipson, C.; Alalouf, G.; Bajorek, M.; Rabinovich, E.; Atir-Lande, A.; Glickman, M.; Bar-Nun, S. A proteasomal ATPase contributes to dislocation of endoplasmic reticulum-associated degradation (ERAD) substrates. J. Biol. Chem. 2008, 283, 7166–7175. [Google Scholar]
- Braun, S.; Matuschewski, K.; Rape, M.; Thoms, S.; Jentsch, S. Role of the ubiquitin-selective Cdc48UFD1/NPl4 chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002, 21, 615–621. [Google Scholar] [CrossRef]
- Hitchcock, A.L.; Krebber, H.; Frietze, S.; Lin, A.; Latterich, M.; Silver, P.A. The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol. Biol. Cell 2001, 12, 3226–3241. [Google Scholar] [CrossRef]
- Rape, M.; Hoppe, T.; Gorr, I.; Kalocay, M.; Richly, H.; Jentsch, S. Mobilization of processed, membrane-tethered Spt23 transcription factor by Cdc48UFD1/NPL4, a ubiquitin-selective chaperone. Cell 2001, 107, 667–677. [Google Scholar] [CrossRef]
- Richly, H.; Rape, M.; Braun, S.; Rumpf, S.; Hoege, C.; Jentsch, S. A series of ubiquitin binding factors connects Cdc48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 2005, 120, 73–84. [Google Scholar] [CrossRef]
- Rumpf, S.; Jentsch, S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol. Cell 2006, 21, 261–269. [Google Scholar] [CrossRef]
- Hanzelmann, P.; Stingele, J.; Hofmann, K.; Schindelin, H.; Raasi, S. The yeast E4 ubiquitin ligase Ufd2 interacts with the ubiquitin-like domains of Rad23 and Dsk2 via a novel and distinct ubiquitin-like binding domain. J. Biol. Chem. 2010, 285, 20390–20398. [Google Scholar] [CrossRef]
- Jentsch, S.; Rumpf, S. Cdc48 (p97): A “molecular gearbox” in the ubiquitin pathway? Trends Biochem. Sci. 2007, 32, 6–11. [Google Scholar] [CrossRef]
- DeLaBarre, B.; Brunger, A.T. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 2003, 10, 856–863. [Google Scholar] [CrossRef]
- Davies, J.M.; Brunger, A.T.; Weis, W.I. Improved structures of full-length p97, an AAA ATPase: Implications for mechanisms of nucleotide-dependent conformational change. Structure 2008, 16, 715–726. [Google Scholar] [CrossRef]
- Rothballer, A.; Tzvetkov, N.; Zwickl, P. Mutations in p97/VCP induce unfolding activity. FEBS Lett. 2007, 581, 1197–1201. [Google Scholar] [CrossRef]
- Barthelme, D.; Sauer, R.T. Bipartite determinants mediate an evolutionarily conserved interaction between Cdc48 and the 20S peptidase. Proc. Natl. Acad. Sci. USA 2013, 110, 3327–3332. [Google Scholar] [CrossRef]
- Davies, J.M.; Tsuruta, H.; May, A.P.; Weis, W.I. Conformational changes of p97 during nucleotide hydrolysis determined by small-angle X-ray scattering. Structure 2005, 13, 183–195. [Google Scholar] [CrossRef]
- Briggs, L.C.; Baldwin, G.S.; Miyata, N.; Kondo, H.; Zhang, X.; Freemont, P.S. Analysis of nucleotide binding to p97 reveals the properties of a tandem AAA hexameric ATPase. J. Biol. Chem. 2008, 283, 13745–13752. [Google Scholar] [CrossRef]
- Song, C.; Wang, Q.; Li, C.C. ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J. Biol. Chem. 2003, 278, 3648–3655. [Google Scholar] [CrossRef]
- Smith, D.M.; Fraga, H.; Reis, C.; Kafri, G.; Goldberg, A.L. ATP binds to proteasomal ATPases inpairs with distinct functional effects, implying an ordered reaction cycle. Cell 2011, 144, 526–538. [Google Scholar] [CrossRef]
- Noi, K.; Yamamoto, D.; Nishikori, S.; Arita-Morioka, K.; Kato, T.; Ando, T.; Ogura, T. High-speed atomic force microscopic observation of ATP-dependent rotation of the AAA+ chaperone p97. Structure 2013, 21, 1992–2002. [Google Scholar] [CrossRef]
- Yeung, H.O.; Forster, A.; Bebeacua, C.; Niwa, H.; Ewens, C.; McKeown, C.; Zhang, X.; Freemont, P.S. Inter-ring rotations of AAA ATPase p97 revealed by electron cryomicroscopy. Open Biol. 2014. [Google Scholar] [CrossRef]
- Rouiller, I.; DeLaBarre, B.; May, A.P.; Weis, W.I.; Brunger, A.T.; Milligan, R.A.; Wilson-Kubalek, E.M. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat. Struct. Biol. 2002, 9, 950–957. [Google Scholar] [CrossRef]
- Meyer, H.H.; Kondo, H.; Warren, G. The p47 co-factor regulates the ATPase activity of the membrane fusion protein, p97. FEBS Lett. 1998, 437, 255–257. [Google Scholar] [CrossRef]
- Coles, M.; Diercks, T.; Liermann, J.; Groger, A.; Rockel, B.; Baumeister, W.; Koretke, K.K.; Lupas, A.; Peters, J.; Kessler, H. The solution structure of VAT-N reveals a “missing link” in the evolution of complex enzymes from a simple betaalphabetabeta element. Curr. Biol. 1999, 9, 1158–1168. [Google Scholar] [CrossRef]
- Dreveny, I.; Kondo, H.; Uchiyama, K.; Shaw, A.; Zhang, X.; Freemont, P.S. Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. EMBO J. 2004, 23, 1030–1039. [Google Scholar] [CrossRef]
- Schuberth, C.; Buchberger, A. Ubx domain proteins: Major regulators of the AAA ATPase Cdc48/p97. Cell. Mol. Life Sci. 2008, 65, 2360–2371. [Google Scholar] [CrossRef]
- Golbik, R.; Lupas, A.N.; Koretke, K.K.; Baumeister, W.; Peters, J. The Janus face of the archaeal Cdc48/p97 homologue VAT: Protein folding vs. unfolding. Biol. Chem. 1999, 380, 1049–1062. [Google Scholar]
- Park, S.; Isaacson, R.; Kim, H.T.; Silver, P.A.; Wagner, G. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure 2005, 13, 995–1005. [Google Scholar] [CrossRef]
- Bruderer, R.M.; Brasseur, C.; Meyer, H.H. The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism. J. Biol. Chem. 2004, 279, 49609–49616. [Google Scholar] [CrossRef]
- Isaacson, R.L.; Pye, V.E.; Simpson, P.; Meyer, H.H.; Zhang, X.; Freemont, P.S.; Matthews, S. Detailed structural insights into the p97-Npl4-Ufd1 interface. J. Biol. Chem. 2007, 282, 21361–21369. [Google Scholar]
- Wang, B.; Alam, S.L.; Meyer, H.H.; Payne, M.; Stemmler, T.L.; Davis, D.R.; Sundquist, W.I. Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4. J. Biol. Chem. 2003, 278, 20225–20234. [Google Scholar]
- Bebeacua, C.; Forster, A.; McKeown, C.; Meyer, H.H.; Zhang, X.; Freemont, P.S. Distinct conformations of the protein complex p97-Ufd1-Npl4 revealed by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 1098–1103. [Google Scholar] [CrossRef]
- Bohm, S.; Lamberti, G.; Fernandez-Saiz, V.; Stapf, C.; Buchberger, A. Cellular functions of Ufd2 and Ufd3 in proteasomal protein degradation depend on Cdc48 binding. Mol. Cell. Biol. 2011, 31, 1528–1539. [Google Scholar] [CrossRef]
- Tu, D.; Li, W.; Ye, Y.; Brunger, A.T. Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc. Natl. Acad. Sci. USA 2007, 104, 15599–15606. [Google Scholar] [CrossRef]
- Koegl, M.; Hoppe, T.; Schlenker, S.; Ulrich, H.D.; Mayer, T.U.; Jentsch, S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999, 96, 635–644. [Google Scholar] [CrossRef]
- Boeddrich, A.; Gaumer, S.; Haacke, A.; Tzvetkov, N.; Albrecht, M.; Evert, B.O.; Muller, E.C.; Lurz, R.; Breuer, P.; Schugardt, N.; et al. An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of Ataxin-3 fibrillogenesis. EMBO J. 2006, 25, 1547–1558. [Google Scholar] [CrossRef]
- Qiu, L.; Pashkova, N.; Walker, J.R.; Winistorfer, S.; Allali-Hassani, A.; Akutsu, M.; Piper, R.; Dhe-Paganon, S. Structure and function of the Plaa/Ufd3-p97/Cdc48 complex. J. Biol. Chem. 2010, 285, 365–372. [Google Scholar] [CrossRef]
- Zhao, G.; Li, G.; Schindelin, H.; Lennarz, W.J. An armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation. Proc. Natl. Acad. Sci. USA 2009, 106, 16197–16202. [Google Scholar] [CrossRef]
- Kim, S.J.; Cho, J.H.; Song, E.J.; Kim, S.J.; Kim, H.M.; Lee, K.E.; Suh, S.W.; Kim, E.E. Structural basis for ovarian tumor domain-containing protein 1 (OTU1) binding to p97/valosin-containing protein (VCP). J. Biol. Chem. 2014, 289, 12264–12274. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Förster, F.; Schuller, J.M.; Unverdorben, P.; Aufderheide, A. Emerging Mechanistic Insights into AAA Complexes Regulating Proteasomal Degradation. Biomolecules 2014, 4, 774-794. https://doi.org/10.3390/biom4030774
Förster F, Schuller JM, Unverdorben P, Aufderheide A. Emerging Mechanistic Insights into AAA Complexes Regulating Proteasomal Degradation. Biomolecules. 2014; 4(3):774-794. https://doi.org/10.3390/biom4030774
Chicago/Turabian StyleFörster, Friedrich, Jan M. Schuller, Pia Unverdorben, and Antje Aufderheide. 2014. "Emerging Mechanistic Insights into AAA Complexes Regulating Proteasomal Degradation" Biomolecules 4, no. 3: 774-794. https://doi.org/10.3390/biom4030774
APA StyleFörster, F., Schuller, J. M., Unverdorben, P., & Aufderheide, A. (2014). Emerging Mechanistic Insights into AAA Complexes Regulating Proteasomal Degradation. Biomolecules, 4(3), 774-794. https://doi.org/10.3390/biom4030774