A Facile and Sensitive Method for Quantification of Cyclic Nucleotide Monophosphates in Mammalian Organs: Basal Levels of Eight cNMPs and Identification of 2',3'-cIMP
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of LC-MS/MS Analytical Method
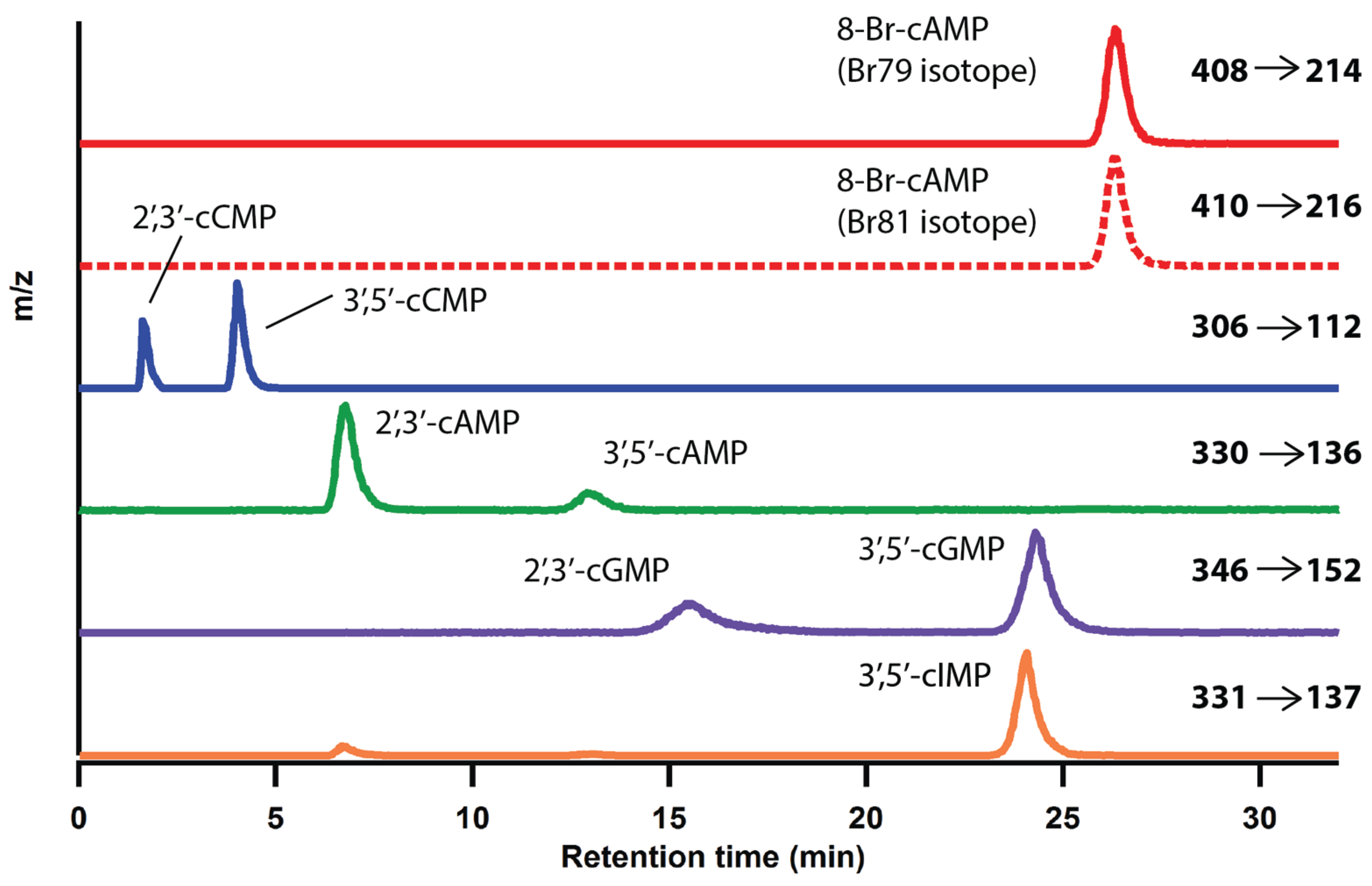
| Parameters | 2',3'-cAMP | 3',5'-cAMP | 2',3'-cCMP | 3',5'-cCMP | 2',3'-cGMP | 3',5'-cGMP | 3',5'-cIMP | 8-79Br-cAMP | 8-81Br-cAMP |
|---|---|---|---|---|---|---|---|---|---|
| [M + H]+ (m/z) | 330.1 | 330.1 | 306.1 | 306.1 | 346.1 | 346.1 | 331.0 | 408.0 | 410.0 |
| Production (m/z) | 136.1 | 136.1 | 112.1 | 112.1 | 152.1 | 152.1 | 137.1 | 214.0 | 216.0 |
| Ret. time (min) | 6.8 | 12.9 | 1.6 | 4.0 | 15.6 | 24.3 | 24.1 | 26.3 | 26.3 |
| LOD (fmol) | 273 | 153 | 171 | 455 | 94 | 487 | 219 | NA | NA |
| LOQ (fmol) | 910 | 510 | 570 | 1517 | 313 | 1623 | 730 | NA | NA |
2.2. LC-MS/MS Method Calibration and Limits of Detection
2.3. Optimization of Extraction Method


2.4. Method Validation and Reproducibility Tests

| Sample | 2',3'-cAMP | 3',5'-cAMP | 2',3'-cCMP | 3',5'-cCMP | 2',3'-cGMP | 3',5'-cGMP | 3',5'-cIMP |
|---|---|---|---|---|---|---|---|
| Portion 1 | 9.9 ± 0.2 | 88.7 ± 12.3 | 1.2 ± 0.9 | 0.9 ± 0.7 | 2.5 ± 0.01 | 10.1 ± 2.3 | 2.6 ± 0.9 |
| Portion 2 | 9.6 ± 0.3 | 92.3 ± 2.9 | ND | 0.6 ± 0.4 | 1.8 | 8.7 ± 0.3 | 2.3 ± 0.3 |
| Portion 3 | 9.7 ± 0.2 | 87.9 ± 4.8 | 1.3 | 0.6 ± 0.4 | 1.8 ± 0.5 | 8.3 ± 0.7 | 3.1 ± 0.3 |
| Average | 9.7 ± 0.2 | 89.6 ± 2.3 | 1.3 ± 0.1 | 0.7 ± 0.1 | 2.0 ± 0.4 | 9.0 ± 0.9 | 2.7 ± 0.4 |
| Inter-run Precision | 2.1 | 2.6 | 7.7 | 14.3 | 20.0 | 10.0 | 14.8 |
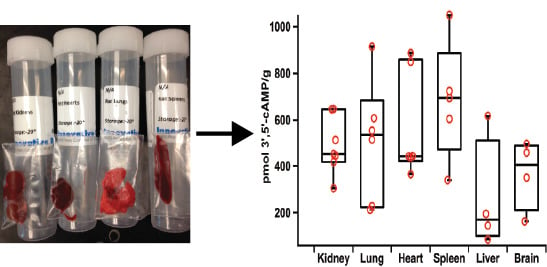
2.5. Applications




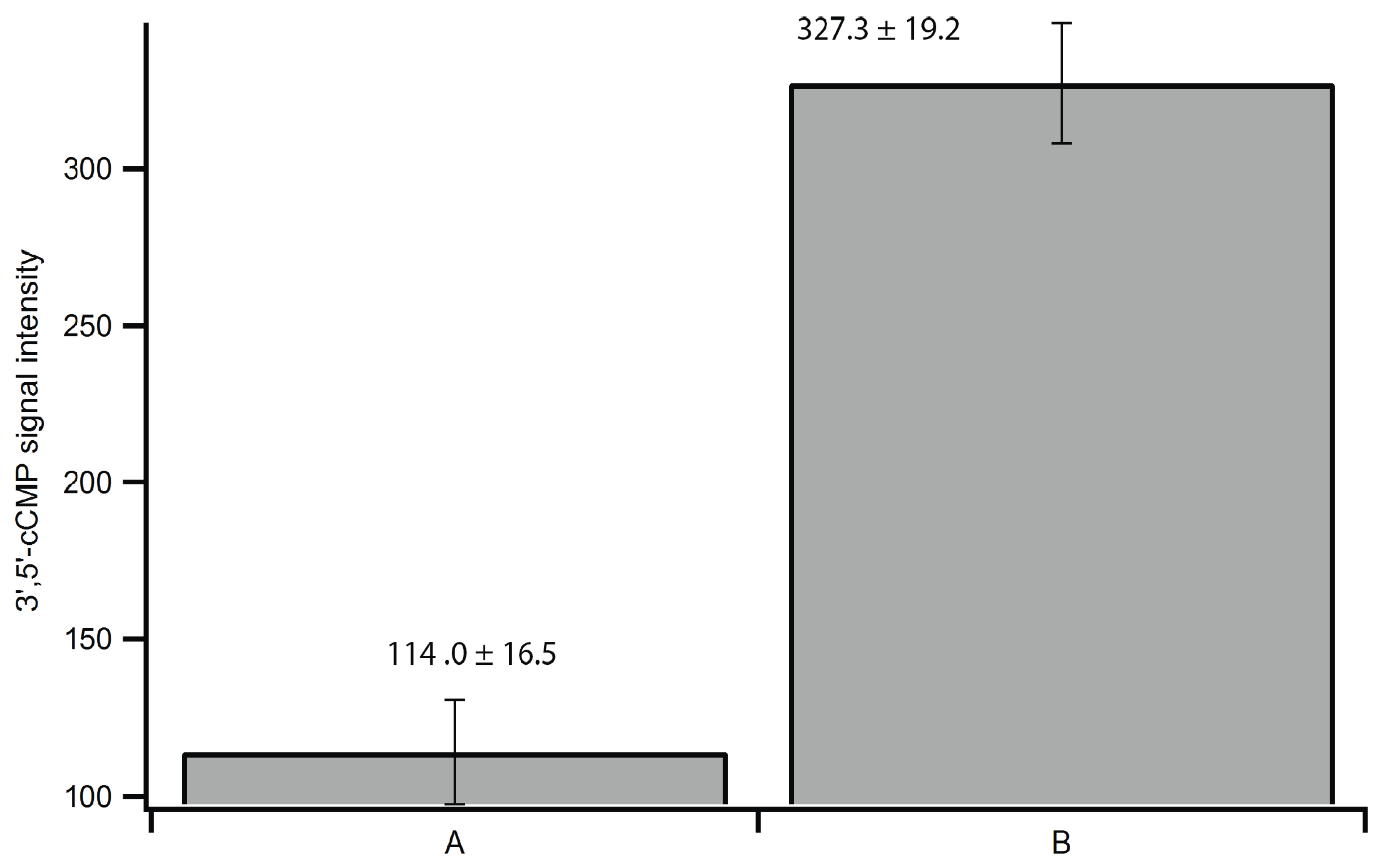
2.6. Versatility of the Method
| Replicate | 2',3'-cAMP | 3',5'-cAMP | 2',3'-cCMP | 3',5'-cCMP | 2',3'-cGMP | 3',5'-cGMP | 2',3'-cIMP | 3',5'-cIMP |
|---|---|---|---|---|---|---|---|---|
| 1 | 5.1 ± 0.3 | 4.8 ± 0.1 | 1.3 ± 0.1 | N/D | 2.1 ± 0.1 | N/D | N/D | N/D |
| 2 | N/D | 167.1 ± 35.0 | N/D | N/D | 32.0 ± 2.3 | 17.2 ± 0.3 | N/D | N/D |
| 3 | N/D | 99.8 ± 0.2 | 1.8 | N/D | 19.9 ± 0.2 | 8.0 ± 3.7 | N/D | N/D |
| Average | 5.1 | 90.6 ± 81.6 | 1.6 ± 0.3 | N/D | 18.0 ± 15.0 | 12.6 ± 4.6 | N/D | N/D |
2.7. Discussion
3. Experimental
3.1. Materials
3.2. Calibration Curves
3.3. NIH-3T3 Cell Growth
3.4. cNMP Extraction from Rat Organs
3.5. cNMP Extraction from NIH-3T3 Cell Line
3.6. LC-MS/MS Sample Preparation
3.7. LC-MS/MS Optimized Conditions
3.8. Statistical Analysis
3.9. Method Validation Test
3.10. Method Reproducibility Test
3.11. Synthesis of 2',3'-cIMP
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sutherland, E.W. Studies on the mechanism of hormone action. Science 1972, 177, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Lucas, K.A.; Pitari, G.M.; Kazerounian, S.; Ruiz-Stewart, I.; Park, J.; Schulz, S.; Chepenik, K.P.; Waldman, S.A. Guanylyl cyclases and signaling by cyclic GMP. Pharmacol. Rev. 2000, 52, 375–414. [Google Scholar] [PubMed]
- Bahre, H.; Kaever, V. Measurement of 2',3'-cyclic nucleotides by liquid chromatography-tandem mass spectrometry in cells. J. Chromatogr. B 2014, 964, 208–211. [Google Scholar] [CrossRef]
- Newton, R.P. Cytidine 3',5'-cyclic-monophosphate: A 3rd cyclic-nucleotide secondary messenger. Nucleosides Nucleotides 1995, 14, 743–747. [Google Scholar] [CrossRef]
- Newton, R.P.; Kingston, E.E.; Hakeem, N.A.; Salih, S.G.; Beynon, J.H.; Moyse, C.D. Extraction, purification, identification and metabolism of 3',5'-cyclic UMP, 3',5'-cyclic IMP and 3',5'-cyclic dTMP from rat tissues. Biochem. J. 1986, 236, 431–439. [Google Scholar]
- Bond, A.E.; Dudley, E.; Tuytten, R.; Lemiere, F.; Smith, C.J.; Esmans, E.L.; Newton, R.P. Mass spectrometric identification of Rab23 phosphorylation as a response to challenge by cytidine 3',5'-cyclic monophosphate in mouse brain. Rapid Commun. Mass Spectrom. 2007, 21, 2685–2692. [Google Scholar] [CrossRef] [PubMed]
- Goble, A.M.; Feng, Y.; Raushel, F.M.; Cronan, J.E. Discovery of a cAMP deaminase that quenches cyclic AMP-dependent regulation. ACS Chem. Biol. 2013, 8, 2622–2629. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.K. The 2',3'-cAMP-adenosine pathway. Am. J. Physiol. Renal Physiol. 2011, 301, F1160–F1167. [Google Scholar] [PubMed]
- Newton, R.P.; Salih, S.G.; Salvage, B.J.; Kingston, E.E. Extraction, purification and identification of cytidine 3',5'-cyclic-monophosphate from rat-tissues. Biochem. J. 1984, 221, 665–673. [Google Scholar] [PubMed]
- Murphy, B.E.; Stone, J.E. Changes in the concentration of cytidine 3',5' monophosphate (cyclic CMP) in regenerating rat liver. Proc. Soc. Exp. Biol. Med. 1980, 163, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.P.; Hakeem, N.A.; Salvage, B.J.; Wassenaar, G.; Kingston, E.E. Cytidylate cyclase activity: Identification of cytidine 3',5'-cyclic monophosphate and four novel cytidine cyclic phosphates as biosynthetic products from cytidine triphosphate. Rapid Commun. Mass Spectrom. 1988, 2, 118–126. [Google Scholar] [PubMed]
- Kuo, J.F.; Brackett, N.L.; Shoji, M.; Tse, J. Cytidine 3':5'-monophosphate phosphodiesterase in mammalian tissues. Occurrence and biological involvement. J. Biol. Chem. 1978, 253, 2518–2521. [Google Scholar]
- Gaion, R.M.; Krishna, G. Cytidylate cyclase: The product isolated by the method of Cech and Ignarro is not cytidine 3',5'-monophosphate. Biochem. Biophys. Res. Commun. 1979, 86, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, X.; Ying, L.; Dou, D.; Li, Y.; Bai, Y.; Liu, J.; Liu, L.; Feng, H.; Yu, X.; et al. Cyclic IMP-synthesized by sGC as a mediator of hypoxic contraction of coronary arteries. Am. J. Physiol. Heart Circ. Physiol. 2014. [Google Scholar] [CrossRef]
- Beste, K.Y.; Burhenne, H.; Kaever, V.; Stasch, J.P.; Seifert, R. Nucleotidyl cyclase activity of soluble guanylyl cyclase α1β1. Biochemistry 2012, 51, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Seifert, R. Is cIMP a second messenger with functions opposite to those of cGMP? Naunyn Schmiedebergs Arch. Pharmacol. 2014, 387, 897–899. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Mi, Z.; Stewart, N.A.; Jackson, E.K. Identification and quantification of 2',3'-cAMP release by the kidney. J. Pharmacol. Exp. Ther. 2009, 328, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.K.; Gillespie, D.G. Extracellular 2',3'-cAMP and 3',5'-cAMP stimulate proliferation of preglomerular vascular endothelial cells and renal epithelial cells. Am. J. Physiol. Renal Physiol. 2012, 303, F954–F962. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.K.; Ren, J.; Gillespie, D.G.; Dubey, R.K. Extracellular 2',3'-cyclic adenosine 5'-monophosphate is a potent inhibitor of preglomerular vascular smooth muscle cell and mesangial cell growth. Hypertension 2010, 56, U151–U246. [Google Scholar] [CrossRef]
- Jackson, E.K.; Ren, J.; Mi, Z. Extracellular 2',3'-cAMP is a source of adenosine. J. Biol. Chem. 2009, 284, 33097–33106. [Google Scholar] [CrossRef] [PubMed]
- Verrier, J.D.; Jackson, T.C.; Gillespie, D.G.; Janesko-Feldman, K.; Bansal, R.; Goebbels, S.; Nave, K.A.; Kochanek, P.M.; Jackson, E.K. Role of CNPase in the oligodendrocytic extracellular 2',3'-cAMP-adenosine pathway. Glia 2013, 61, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.K.; Gillespie, D.G.; Mi, Z.; Cheng, D.; Bansal, R.; Janesko-Feldman, K.; Kochanek, P.M. Role of 2',3'-cyclic nucleotide 3'-phosphodiesterase in the renal 2',3'-cAMP-adenosine pathway. Am. J. Physiol. Renal Physiol. 2014, 307, F14–F24. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.K.; Mi, Z. In vivo cardiovascular pharmacology of 2',3'-cAMP, 2'-AMP, and 3'-AMP in the rat. J. Pharmacol. Exp. Ther. 2013, 346, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Verrier, J.D.; Jackson, T.C.; Bansal, R.; Kochanek, P.M.; Puccio, A.M.; Okonkwo, D.O.; Jackson, E.K. The brain in vivo expresses the 2',3'-cAMP-adenosine pathway. J. Neurochem. 2012, 122, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, T.; Blancquaert, D.; Couturon, P.; van der Straeten, D.; Sandra, P.; Lynen, F. Wounding stress causes rapid increase in concentration of the naturally occurring 2',3'-isomers of cyclic guanosine- and cyclic adenosine monophosphate (cGMP and cAMP) in plant tissues. Phytochemistry 2014, 103, 59–66. [Google Scholar] [CrossRef] [PubMed]
- George, W.J.; Polson, J.B.; O’Toole, A.G.; Goldberg, N.D. Elevation of guanosine 3',5'-cyclic phosphate in rat heart after perfusion with acetylcholine. Proc. Natl. Acad. Sci. USA 1970, 66, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Beste, K.Y.; Seifert, R. cCMP, cUMP, cTMP, cIMP and cXMP as possible second messengers: Development of a hypothesis based on studies with soluble guanylyl cyclase α1β1. Biol. Chem. 2013, 394, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, N.D.; Larner, J.; Sasko, H.; O’Toole, A.G. Enzymic analysis of cyclic 3',5'-AMP in mammalian tissues and urine. Anal. Biochem. 1969, 28, 523–544. [Google Scholar] [CrossRef] [PubMed]
- Hardman, J.G.; Davis, J.W.; Sutherland, E.W. Measurement of guanosine 3',5'-monophosphate and other cyclic nucleotides. Variations in urinary excretion with hormonal state of the rat. J. Biol. Chem. 1966, 241, 4812–4815. [Google Scholar]
- Ishikawa, E.; Ishikawa, S.; Davis, J.W.; Sutherland, E.W. Determination of guanosine 3',5'-monophosphate in tissues and of guanyl cyclase in rat intestine. J. Biol. Chem. 1969, 244, 6371–6376. [Google Scholar] [PubMed]
- Steiner, A.L.; Parker, C.W.; Kipnis, D.M. Radioimmunoassay for cyclic nucleotides. I. Preparation of antibodies and iodinated cyclic nucleotides. J. Biol. Chem. 1972, 247, 1106–1113. [Google Scholar]
- Steiner, A.L.; Pagliara, A.S.; Chase, L.R.; Kipnis, D.M. Radioimmunoassay for cyclic nucleotides. II. Adenosine 3',5'-monophosphate and guanosine 3',5'-monophosphate in mammalian tissues and body fluids. J. Biol. Chem. 1972, 247, 1114–1120. [Google Scholar]
- Kingan, T.G. A competitive enzyme-linked immunosorbent assay: Applications in the assay of peptides, steroids, and cyclic nucleotides. Anal. Biochem. 1989, 183, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Wellard, J.; Blind, B.; Hamprech, B. An enzyme-linked immunosorbent assay for the rapid quantification of intracellular and extracellular guanosine 3',5'-cyclic monophosphate in cultured glial cells. Neurochem. Res. 2004, 29, 2177–2187. [Google Scholar] [CrossRef] [PubMed]
- Engvall, E.; Jonsson, K.; Perlmann, P. Enzyme-linked immunosorbent assay. II. Quantitative assay of protein antigen, immunoglobulin G, by means of enzyme-labelled antigen and antibody-coated tubes. Biochim. Biophys. Acta 1971, 251, 427–434. [Google Scholar]
- Faupel-Badger, J.M.; Fuhrman, B.J.; Xu, X.; Falk, R.T.; Keefer, L.K.; Veenstra, T.D.; Hoover, R.N.; Ziegler, R.G. Comparison of liquid chromatography-tandem mass spectrometry, RIA, and ELISA methods for measurement of urinary estrogens. Cancer Epidemiol. Biomark. Prev. 2010, 19, 292–300. [Google Scholar] [CrossRef]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. Trends Anal. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef]
- Esmans, E.L.; Broes, D.; Hoes, I.; Lemiere, F.; Vanhoutte, K. Liquid chromatography-mass spectrometry in nucleoside, nucleotide and modified nucleotide characterization. J. Chromatogr. A 1998, 794, 109–127. [Google Scholar] [CrossRef]
- Prakash, C.; Shaffer, C.L.; Nedderman, A. Analytical strategies for identifying drug metabolites. Mass Spectrom. Rev. 2007, 26, 340–369. [Google Scholar] [CrossRef] [PubMed]
- Richards, H.; Das, S.; Smith, C.J.; Pereira, L.; Geisbrecht, A.; Devitt, N.J.; Games, D.E.; van Geyschem, J.; Gareth Brenton, A.; Newton, R.P. Cyclic nucleotide content of tobacco BY-2 cells. Phytochemistry 2002, 61, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Witters, E.; Vanhoutte, K.; Dewitte, W.; Machackova, I.; Benkova, E.; van Dongen, W.; Esmans, E.L.; van Onckelen, H.A. Analysis of cyclic nucleotides and cytokinins in minute plant samples using phase-system switching capillary electrospray-liquid chromatography-tandem mass spectrometry. Phytochem. Anal. 1999, 10, 143–151. [Google Scholar]
- Becker, S.; Kortz, L.; Helmschrodt, C.; Thiery, J.; Ceglarek, U. LC-MS-based metabolomics in the clinical laboratory. J. Chromatogr. B 2012. [Google Scholar] [CrossRef]
- Oeckl, P.; Ferger, B. Simultaneous LC-MS/MS analysis of the biomarkers cAMP and cGMP in plasma, CSF and brain tissue. J. Neurosci. Methods 2012, 203, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Martens-Lobenhoffer, J.; Dautz, C.; Bode-Boger, S.M. Improved method for the determination of cyclic guanosine monophosphate (cGMP) in human plasma by LC-MS/MS. J. Chromatogr. B 2010, 878, 487–491. [Google Scholar] [CrossRef]
- Van Damme, T.; Zhang, Y.; Lynen, F.; Sandra, P. Determination of cyclic guanosine- and cyclic adenosine monophosphate (cGMP and cAMP) in human plasma and animal tissues by solid phase extraction on silica and liquid chromatography-triple quadrupole mass spectrometry. J. Chromatogr. B 2012, 909, 14–21. [Google Scholar] [CrossRef]
- Zhang, Y.; Dufield, D.; Klover, J.; Li, W.; Szekely-Klepser, G.; Lepsy, C.; Sadagopan, N. Development and validation of an LC-MS/MS method for quantification of cyclic guanosine 3',5'-monophosphate (cGMP) in clinical applications: A comparison with a EIA method. J. Chromatogr. B 2009, 877, 513–520. [Google Scholar] [CrossRef]
- Van Damme, T.; Lachova, M.; Lynen, F.; Szucs, R.; Sandra, P. Solid-phase extraction based on hydrophilic interaction liquid chromatography with acetone as eluent for eliminating matrix effects in the analysis of biological fluids by LC-MS. Anal. Bioanal. Chem. 2014, 406, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Van Eeckhaut, A.; Lanckmans, K.; Sarre, S.; Smolders, I.; Michotte, Y. Validation of bioanalytical LC-MS/MS assays: Evaluation of matrix effects. J. Chromatogr. B 2009, 877, 2198–2207. [Google Scholar]
- Trufelli, H.; Palma, P.; Famiglini, G.; Cappiello, A. An overview of matrix effects in liquid chromatography-mass spectrometry. Mass Spectrom. Rev. 2011, 30, 491–509. [Google Scholar] [CrossRef] [PubMed]
- King, R.; Bonfiglio, R.; Fernandez-Metzler, C.; Miller-Stein, C.; Olah, T. Mechanistic investigation of ionization suppression in electrospray ionization. J. Am. Soc. Mass Spectrom. 2000, 11, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Witters, E.; van Dongen, W.; Esmans, E.L.; van Onckelen, H.A. Ion-pair liquid chromatography-electrospray mass spectrometry for the analysis of cyclic nucleotides. J. Chromatogr. B 1997, 694, 55–63. [Google Scholar] [CrossRef]
- Lorenzetti, R.; Lilla, S.; Donato, J.L.; de Nucci, G. Simultaneous quantification of GMP, AMP, cyclic GMP and cyclic AMP by liquid chromatography coupled to tandem mass spectrometry. J. Chromatogr. B 2007, 859, 37–41. [Google Scholar]
- O’Kane, A.A.; Chevallier, O.P.; Graham, S.F.; Elliott, C.T.; Mooney, M.H. Metabolomic profiling of in vivo plasma responses to dioxin-associated dietary contaminant exposure in rats: Implications for identification of sources of animal and human exposure. Environ. Sci. Technol. 2013, 47, 5409–5418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wu, X.; Ke, C.; Yin, M.; Li, Z.; Fan, L.; Zhang, W.; Zhang, H.; Zhao, F.; Zhou, X.; et al. Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling. J. Proteome Res. 2013, 12, 505–512. [Google Scholar]
- Buszewski, B.; Noga, S. Hydrophilic interaction liquid chromatography (HILIC)--a powerful separation technique. Anal. Bioanal. Chem. 2012, 402, 231–247. [Google Scholar] [CrossRef] [PubMed]
- Kehr, J.; Chavko, M. Separation of nucleotides by reversed-phase high-performance liquid-chromatography-advantages and limitations. Fresen Z Anal. Chem 1986, 325, 466–469. [Google Scholar] [CrossRef]
- Shrivastava, A.; Gupta, V.B. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron. Young Sci. 2011, 2, 21–25. [Google Scholar] [CrossRef]
- ICH of Technical Requirements for Registration of Pharmaceuticals for Human Use. Validation of Analytical Procedures: Text and Methodology Q2(R1). Available online: http://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Quality/Q2_R1/Step4/Q2_R1__Guideline.pdf (accessed on 26 November 2014).
- Andrews, S.C.; Guest, J.R. Nucleotide sequence of the gene encoding the GMP reductase of Escherichia coli K12. Biochem. J. 1988, 255, 35–43. [Google Scholar]
- US Department of Health and Human Services; F.D.A.; Center for Drug Evaluation and Research (CDER). Guidance for industry, Bioanalytical Method Validation, BP. Available online: http://www.fda.gov/downloads/Drugs/Guidances/ucm070107.pdf (accessed on 26 November 2014).
- Bloch, A. Cytidine 3',5'-monophosphate (cyclic CMP). I. Isolation from extracts of leukemia L-1210 Cells. Biochem. Biophys. Res. Commun. 1974, 58, 652–659. [Google Scholar]
- Patton, G.C.; Stenmark, P.; Gollapalli, D.R.; Sevastik, R.; Kursula, P.; Flodin, S.; Schuler, H.; Swales, C.T.; Eklund, H.; Himo, F.; et al. Cofactor mobility determines reaction outcome in the IMPDH and GMPR β-α8 barrel enzymes. Nat. Chem. Biol. 2011, 7, 950–958. [Google Scholar]
- Lin, T.S.; Cheng, J.C.; Ishiguro, K.; Sartorelli, A.C. Purine and 8-substituted purine arabinofuranosyl and ribofuranosyl nucleoside derivatives as potential inducers of the differentiation of the Friend erythroleukemia. J. Med. Chem. 1985, 28, 1481–1485. [Google Scholar] [CrossRef] [PubMed]
- Kotra, L.P.; Manouilov, K.K.; Cretton-Scott, E.; Sommadossi, J.P.; Boudinot, F.D.; Schinazi, R.F.; Chu, C.K. Synthesis, biotransformation, and pharmacokinetic studies of 9-(β-D-arabinofuranosyl)-6-azidopurine: A prodrug for ara-A designed to utilize the azide reduction pathway. J. Med. Chem. 1996, 39, 5202–5207. [Google Scholar] [CrossRef] [PubMed]
- Banoub, J.H.; Limbach, P.A. Mass Spectrometry of Nucleosides and Nucleic Acids; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Bahre, H.; Danker, K.Y.; Stasch, J.P.; Kaever, V.; Seifert, R. Nucleotidyl cyclase activity of soluble guanylyl cyclase in intact cells. Biochem. Biophys. Res. Commun. 2014, 443, 1195–1199. [Google Scholar] [CrossRef] [PubMed]
- Beste, K.Y.; Spangler, C.M.; Burhenne, H.; Koch, K.W.; Shen, Y.; Tang, W.J.; Kaever, V.; Seifert, R. Nucleotidyl cyclase activity of particulate guanylyl cyclase A: Comparison with particulate guanylyl cyclases E and F, soluble guanylyl cyclase and bacterial adenylyl cyclases CyaA and edema factor. PLOS ONE 2013, 8, e70223. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, X.; Fontaine, B.M.; Strobel, F.; Weinert, E.E. A Facile and Sensitive Method for Quantification of Cyclic Nucleotide Monophosphates in Mammalian Organs: Basal Levels of Eight cNMPs and Identification of 2',3'-cIMP. Biomolecules 2014, 4, 1070-1092. https://doi.org/10.3390/biom4041070
Jia X, Fontaine BM, Strobel F, Weinert EE. A Facile and Sensitive Method for Quantification of Cyclic Nucleotide Monophosphates in Mammalian Organs: Basal Levels of Eight cNMPs and Identification of 2',3'-cIMP. Biomolecules. 2014; 4(4):1070-1092. https://doi.org/10.3390/biom4041070
Chicago/Turabian StyleJia, Xin, Benjamin M. Fontaine, Fred Strobel, and Emily E. Weinert. 2014. "A Facile and Sensitive Method for Quantification of Cyclic Nucleotide Monophosphates in Mammalian Organs: Basal Levels of Eight cNMPs and Identification of 2',3'-cIMP" Biomolecules 4, no. 4: 1070-1092. https://doi.org/10.3390/biom4041070