Bacterial Sigma Factors and Anti-Sigma Factors: Structure, Function and Distribution
Abstract
:1. Introduction
2. Structural Organisation of σ70 and Other Group 1 σ Factors


3. Structure and Function of Alternative σ Factors
3.1. Group 2 σ Factors
3.2. Group 3 σ Factors
3.3. Group 4 (ECF) σ Factors
4. Inhibition of Alternative σ Factors by Anti-σ Factors
4.1. Anti-σ Factors that Insert between σ2 and σ4

4.2. Anti-σ Factors that Wrap around σ2 and σ4
5. Mechanisms for Triggering σ Factor Release from Anti-σ Factors
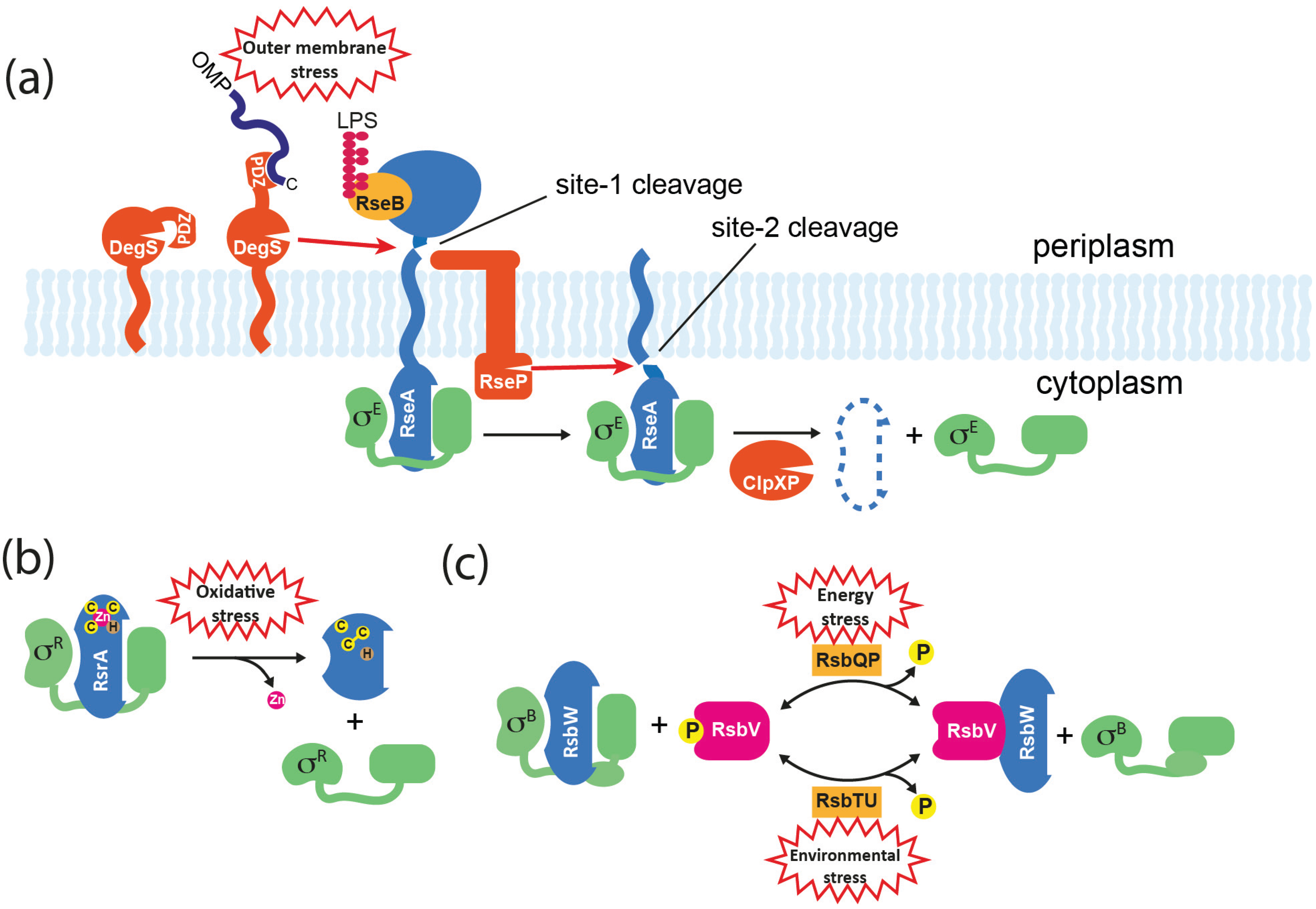
5.1. Regulated Proteolysis
5.2. Direct Sensing
5.3. Partner-Switching
6. Indirect Regulation of Alternative σ Factors by Primary σ Factor Control
7. Conclusions
Acknowledgments
Abbreviations
| ASD | Anti-sigma domain |
| σR | Sigma region |
| ZASD | Zinc binding anti-sigma domain |
| RIP | Regulated intramembrane proteolysis |
| RNAP | RNA polymerase |
| RP | RNA polymerase/ promoter complex |
| NCR | Non-conserved region of sigma |
| ECF | Extracytoplasmic function |
| OMP | Outer membrane protein |
| LPS | Lipopolysaccharide |
Conflicts of Interest
References
- Saecker, R.M.; Record, M.T.; Dehaseth, P.L. Mechanism of bacterial transcription initiation: RNA polymerase—Promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011, 412, 754–771. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Buck, M.A. Perspective on the enhancer dependent bacterial RNA polymerase. Biomolecules 2015, 5, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Perdue, S.A.; Roberts, J.W. σ70-Dependent transcription pausing in Escherichia coli. J. Mol. Biol. 2011, 412, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Raffaelle, M.; Kanin, E.I.; Vogt, J.; Burgess, R.R.; Ansari, A.Z. Holoenzyme switching and stochastic release of sigma factors from RNA polymerase in vivo. Mol. Cell 2005, 20, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Burgess, R.R.; Travers, A.A.; Dunn, J.J.; Bautz, E.K. Factor stimulating transcription by RNA polymerase. Nature 1969, 221, 43–46. [Google Scholar] [CrossRef] [PubMed]
- Haldenwang, W.G.; Losick, R. Novel RNA polymerase sigma factor from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1980, 77, 7000–7004. [Google Scholar] [CrossRef] [PubMed]
- Haldenwang, W.G.; Lang, N.; Losick, R. A sporulation-induced sigma-like regulatory protein from B. subtilis. Cell 1981, 23, 615–624. [Google Scholar] [CrossRef]
- Grossman, A.D.; Erickson, J.W.; Gross, C.A. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell 1984, 38, 383–390. [Google Scholar] [CrossRef]
- Han, K.; Li, Z.; Peng, R.; Zhu, L.; Zhou, T.; Wang, L.; Li, S.; Zhang, X.; Hu, W.; Wu, Z.; et al. Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu. Sci. Rep. 2013. [Google Scholar] [CrossRef] [PubMed]
- Lonetto, M.; Gribskov, M.; Gross, C.A. The sigma70 family: Sequence conservation and evolutionary relationships. J. Bacteriol. 1992, 174, 3843–3849. [Google Scholar] [PubMed]
- Paget, M.S.; Helmann, J.D. The sigma70 family of sigma factors. Genome Biol. 2003. [Google Scholar] [CrossRef]
- Campbell, E.A.; Muzzin, O.; Chlenov, M.; Sun, J.L.; Olson, C.A.; Weinman, O.; Trester-Zedlitz, M.L.; Darst, S.A. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell 2002, 9, 527–539. [Google Scholar] [CrossRef]
- Schwartz, E.C.; Shekhtman, A.; Dutta, K.; Pratt, M.R.; Cowburn, D.; Darst, S.; Muir, T.W. A full-length group 1 bacterial sigma factor adopts a compact structure incompatible with DNA binding. Chem. Biol. 2008, 15, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.S.; Masuda, S.; Darst, S.A. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 2002, 296, 1280–1284. [Google Scholar] [CrossRef] [PubMed]
- Vassylyev, D.G.; Sekine, S.; Laptenko, O.; Lee, J.; Vassylyeva, M.N.; Borukhov, S.; Yokoyama, S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 2002, 417, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Bae, B.; Davis, E.; Brown, D.; Campbell, E.A.; Wigneshweraraj, S.; Darst, S.A. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of σ70 domain 1.1. Proc. Natl. Acad. Sci. USA 2013, 110, 19772–19777. [Google Scholar] [CrossRef] [PubMed]
- Feklistov, A.; Darst, S.A. Structural basis for promoter −10 element recognition by the bacterial RNA polymerase σ subunit. Cell 2011, 147, 1257–1269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Feng, Y.; Chatterjee, S.; Tuske, S.; Ho, M.X.; Arnold, E.; Ebright, R.H. Structural basis of transcription initiation. Science 2012, 338, 1076–1080. [Google Scholar] [CrossRef] [PubMed]
- Haugen, S.P.; Berkmen, M.B.; Ross, W.; Gaal, T.; Ward, C.; Gourse, R.L. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: An additional recognition element for RNA polymerase. Cell 2006, 125, 1069–1082. [Google Scholar] [CrossRef] [PubMed]
- Haugen, S.P.; Ross, W.; Manrique, M.; Gourse, R.L. Fine structure of the promoter-sigma region 1.2 interaction. Proc. Natl. Acad. Sci. USA 2008, 105, 3292–3297. [Google Scholar] [CrossRef] [PubMed]
- Feklistov, A.; Barinova, N.; Sevostyanova, A.; Heyduk, E.; Bass, I.; Vvedenskaya, I.; Kuznedelov, K.; Merkiene, E.; Stavrovskaya, E.; Klimasauskas, S.; et al. A basal promoter element recognized by free RNA polymerase sigma subunit determines promoter recognition by RNA polymerase holoenzyme. Mol. Cell 2006, 23, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Steitz, T.A. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol. Cell 2015, 58, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Campagne, S.; Marsh, M.E.; Capitani, G.; Vorholt, J.A.; Allain, F.H.-T. Structural basis for −10 promoter element melting by environmentally induced sigma factors. Nat. Struct. Mol. Biol. 2014, 21, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.E.; Zheng, D.; Busby, S.J.W.; Minchin, S.D. Identification and analysis of “extended −10” promoters in Escherichia coli. Nucleic Acids Res. 2003, 31, 4689–4695. [Google Scholar] [CrossRef] [PubMed]
- Dombroski, A.J.; Walter, W.A.; Record, M.T.; Siegele, D.A.; Gross, C.A. Polypeptides containing highly conserved regions of transcription initiation factor sigma70 exhibit specificity of binding to promoter DNA. Cell 1992, 70, 501–512. [Google Scholar] [CrossRef]
- Murakami, K.S. X-ray crystal structure of Escherichia coli RNA polymerase σ70 holoenzyme. J. Biol. Chem. 2013, 288, 9126–9134. [Google Scholar] [CrossRef] [PubMed]
- Mekler, V.; Kortkhonjia, E.; Mukhopadhyay, J.; Knight, J.; Revyakin, A.; Kapanidis, A.N.; Niu, W.; Ebright, Y.W.; Levy, R.; Ebright, R.H. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell 2002, 108, 599–614. [Google Scholar] [CrossRef]
- Hook-Barnard, I.G.; Hinton, D.M. The promoter spacer influences transcription initiation via sigma70 region 1.1 of Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. USA 2009, 106, 737–742. [Google Scholar] [CrossRef] [PubMed]
- Vuthoori, S.; Bowers, C.W.; McCracken, A.; Dombroski, A.J.; Hinton, D.M. Domain 1.1 of the sigma70 subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes. J. Mol. Biol. 2001, 309, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Leibman, M.; Hochschild, A. A sigma-core interaction of the RNA polymerase holoenzyme that enhances promoter escape. EMBO J. 2007, 26, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Buttner, M.J.; Lewis, C.G. Construction and characterization of Streptomyces coelicolor A3(2) mutants that are multiply deficient in the nonessential hrd-encoded RNA polymerase sigma factors. J. Bacteriol. 1992, 174, 5165–5167. [Google Scholar] [PubMed]
- Osanai, T.; Ikeuchi, M.; Tanaka, K. Group 2 sigma factors in cyanobacteria. Physiol. Plant. 2008, 133, 490–506. [Google Scholar] [CrossRef] [PubMed]
- Osanai, T.; Kanesaki, Y.; Nakano, T.; Takahashi, H.; Asayama, M.; Shirai, M.; Kanehisa, M.; Suzuki, I.; Murata, N.; Tanaka, K. Positive regulation of sugar catabolic pathways in the cyanobacterium Synechocystis sp. PCC 6803 by the group 2 sigma factor sigE. J. Biol. Chem. 2005, 280, 30653–30659. [Google Scholar] [CrossRef] [PubMed]
- Nair, U.; Ditty, J.L.; Min, H.; Golden, S.S. Roles for sigma factors in global circadian regulation of the cyanobacterial genome. J. Bacteriol. 2002, 184, 3530–3538. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Mutsuda, M.; Murayama, Y.; Tomita, J.; Hosokawa, N.; Terauchi, K.; Sugita, C.; Sugita, M.; Kondo, T.; Iwasaki, H. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc. Natl. Acad. Sci. USA 2009, 106, 14168–14173. [Google Scholar] [CrossRef] [PubMed]
- Battesti, A.; Majdalani, N.; Gottesman, S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011, 65, 189–213. [Google Scholar] [CrossRef] [PubMed]
- Ihssen, J.; Egli, T. Specific growth rate and not cell density controls the general stress response in Escherichia coli. Microbiology 2004, 150, 1637–1648. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Polen, T.; Heuveling, J.; Wendisch, V.F.; Hengge, R. Genome-wide analysis of the general stress response network in Escherichia coli: SigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 2005, 187, 1591–1603. [Google Scholar] [CrossRef] [PubMed]
- Becker, G.; Hengge-Aronis, R. What makes an Escherichia coli promoter sigmaS dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 of sigmaS. Mol. Microbiol. 2001, 39, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.-M.; Rhodius, V.A.; Campbell, E.A.; Gross, C.A. Mutational analysis of Escherichia coli sigma28 and its target promoters reveals recognition of a composite −10 region, comprised of an “extended −10” motif and a core −10 element. Mol. Microbiol. 2009, 72, 830–843. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.-M.; Rhodius, V.A.; Campbell, E.A.; Gross, C.A. Dissection of recognition determinants of Escherichia coli sigma32 suggests a composite −10 region with an “extended −10” motif and a core −10 element. Mol. Microbiol. 2009, 72, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Wösten, M.M. Eubacterial sigma-factors. FEMS Microbiol. Rev. 1998, 22, 127–150. [Google Scholar] [CrossRef]
- Chen, Y.F.; Helmann, J.D. Restoration of motility to an Escherichia coli fliA flagellar mutant by a Bacillus subtilis sigma factor. Proc. Natl. Acad. Sci. USA 1992, 89, 5123–5127. [Google Scholar] [CrossRef] [PubMed]
- Chater, K.F.; Bruton, C.J.; Plaskitt, K.A.; Buttner, M.J.; Mendez, C.; Helmann, J.D. The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility sigma factor of B. subtilis. Cell 1989, 59, 133–143. [Google Scholar] [CrossRef]
- Yu, H.H.; Tan, M. Sigma28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia. Mol. Microbiol. 2003, 50, 577–584. [Google Scholar] [PubMed]
- Hecker, M.; Pané-Farré, J.; Völker, U. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 2007, 61, 215–236. [Google Scholar] [CrossRef] [PubMed]
- Nannapaneni, P.; Hertwig, F.; Depke, M.; Hecker, M.; Mäder, U.; Völker, U.; Steil, L.; van Hijum, S.A.F.T. Defining the structure of the general stress regulon of Bacillus subtilis using targeted microarray analysis and random forest classification. Microbiology 2012, 158, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Viollier, P.H.; Kelemen, G.H.; Dale, G.E.; Nguyen, K.T.; Buttner, M.J.; Thompson, C.J. Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Mol. Microbiol. 2003, 47, 699–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potuckova, L.; Kelemen, G.H.; Findlay, K.C.; Lonetto, M.A.; Buttner, M.J.; Kormanec, J. A new RNA polymerase sigma factor, sigmaF, is required for the late stages of morphological differentiation in Streptomyces spp. Mol. Microbiol. 1995, 17, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Dalton, K.A.; Thibessard, A.; Hunter, J.I.B.; Kelemen, G.H. A novel compartment, the “subapical stem” of the aerial hyphae, is the location of a sigN-dependent, developmentally distinct transcription in Streptomyces coelicolor. Mol. Microbiol. 2007, 64, 719–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilbert, D.W.; Piggot, P.J. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 2004, 68, 234–262. [Google Scholar] [CrossRef] [PubMed]
- Lonetto, M.A.; Brown, K.L.; Rudd, K.E.; Buttner, M.J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA 1994, 91, 7573–7577. [Google Scholar] [CrossRef] [PubMed]
- Staroń, A.; Sofia, H.J.; Dietrich, S.; Ulrich, L.E.; Liesegang, H.; Mascher, T. The third pillar of bacterial signal transduction: Classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 2009, 74, 557–581. [Google Scholar] [CrossRef] [PubMed]
- Jogler, C.; Waldmann, J.; Huang, X.; Jogler, M.; Glöckner, F.O.; Mascher, T.; Kolter, R. Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in Planctomycetes by comparative genomics. J. Bacteriol. 2012, 194, 6419–6430. [Google Scholar] [CrossRef] [PubMed]
- Mascher, T.; Hachmann, A.-B.; Helmann, J.D. Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function factors. J. Bacteriol. 2007, 189, 6919–6927. [Google Scholar] [CrossRef] [PubMed]
- Rhodius, V.A.; Segall-Shapiro, T.H.; Sharon, B.D.; Ghodasara, A.; Orlova, E.; Tabakh, H.; Burkhardt, D.H.; Clancy, K.; Peterson, T.C.; Gross, C.A.; et al. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol. Syst. Biol. 2013. [Google Scholar] [CrossRef]
- Kim, M.S.; Dufour, Y.S.; Yoo, J.S.; Cho, Y.B.; Park, J.H.; Nam, G.B.; Kim, H.M.; Lee, K.L.; Donohue, T.J.; Roe, J.H. Conservation of thiol-oxidative stress responses regulated by SigR orthologues in actinomycetes. Mol. Microbiol. 2012, 85, 326–344. [Google Scholar] [CrossRef] [PubMed]
- Kallifidas, D.; Thomas, D.; Doughty, P.; Paget, M.S. The sigmaR regulon of Streptomyces coelicolor A32 reveals a key role in protein quality control during disulphide stress. Microbiology 2010, 156, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- Wecke, T.; Halang, P.; Staroń, A.; Dufour, Y.S.; Donohue, T.J.; Mascher, T. Extracytoplasmic function σ factors of the widely distributed group ECF41 contain a fused regulatory domain. Microbiologyopen 2012, 1, 194–213. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Westblade, L.F.; Darst, S.A. Regulation of bacterial RNA polymerase sigma factor activity: A structural perspective. Curr. Opin. Microbiol. 2008, 11, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Greenwell, R.; Anthony, J.R.; Wang, S.; Lim, L.; Das, K.; Sofia, H.J.; Donohue, T.J.; Darst, S.A. A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol. Cell 2007, 27, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Tupy, J.L.; Gruber, T.M.; Wang, S.; Sharp, M.M.; Gross, C.A.; Darst, S.A. Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol. Cell 2003, 11, 1067–1078. [Google Scholar] [CrossRef]
- Paget, M.S.; Bae, J.B.; Hahn, M.Y.; Li, W.; Kleanthous, C.; Roe, J.H.; Buttner, M.J. Mutational analysis of RsrA, a zinc-binding anti-sigma factor with a thiol-disulphide redox switch. Mol. Microbiol. 2001, 39, 1036–1047. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, M.K.; Ray, S.S.; Darst, S.A. Crystal structure of the flagellar sigma/anti-sigma complex sigma(28)/FlgM reveals an intact sigma factor in an inactive conformation. Mol. Cell 2004, 14, 127–138. [Google Scholar] [CrossRef]
- Maillard, A.P.; Girard, E.; Ziani, W.; Petit-Härtlein, I.; Kahn, R.; Covès, J. The crystal structure of the anti-σ factor CnrY in complex with the σ factor CnrH shows a new structural class of anti-σ factors targeting extracytoplasmic function σ factors. J. Mol. Biol. 2014, 426, 2313–2327. [Google Scholar] [CrossRef] [PubMed]
- Campagne, S.; Damberger, F.F.; Kaczmarczyk, A.; Francez-Charlot, A.; Allain, F.H.-T.; Vorholt, J.A. Structural basis for sigma factor mimicry in the general stress response of Alphaproteobacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E1405–E1414. [Google Scholar] [CrossRef] [PubMed]
- Herrou, J.; Rotskoff, G.; Luo, Y.; Roux, B.; Crosson, S. Structural basis of a protein partner switch that regulates the general stress response of α-proteobacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E1415–E1423. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.; Ye, J.; Rawson, R.; Goldstein, J. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell 2000, 100, 391–398. [Google Scholar] [CrossRef]
- Ades, S.E.; Connolly, L.E.; Alba, B.M.; Gross, C.A. The Escherichia coli sigmaE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999, 13, 2449–2461. [Google Scholar] [CrossRef] [PubMed]
- Walsh, N.P.; Alba, B.M.; Bose, B.; Gross, C.A.; Sauer, R.T. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 2003, 113, 61–71. [Google Scholar] [CrossRef]
- Kanehara, K.; Ito, K.; Akiyama, Y. YaeL (EcfE) activates the sigmaE pathway of stress response through a site-2 cleavage of anti-sigmaE, RseA. Genes Dev. 2002, 16, 2147–2155. [Google Scholar] [CrossRef] [PubMed]
- Alba, B.M.; Leeds, J.A.; Onufryk, C.; Lu, C.Z.; Gross, C.A. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigmaE-dependent extracytoplasmic stress response. Genes Dev. 2002, 16, 2156–2168. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Levchenko, I.; Sauer, R.T.; Baker, T.A. Modulating substrate choice: The SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev. 2004, 18, 2292–2301. [Google Scholar] [CrossRef] [PubMed]
- Grigorova, I.L.; Chaba, R.; Zhong, H.J.; Alba, B.M.; Rhodius, V.; Herman, C.; Gross, C.A. Fine-tuning of the Escherichia coli sigmaE envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev. 2004, 18, 2686–2697. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.; Guo, M.S.; Chaba, R.; Gross, C.A.; Sauer, R.T. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 2013, 340, 837–841. [Google Scholar] [CrossRef] [PubMed]
- Ellermeier, C.D.; Losick, R. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 2006, 20, 1911–1922. [Google Scholar] [CrossRef] [PubMed]
- Schöbel, S.; Zellmeier, S.; Schumann, W.; Wiegert, T. The Bacillus subtilis sigmaW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 2004, 52, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, J.; Hein, K.; Wiegert, T. Two proteolytic modules are involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW. Mol. Microbiol. 2009, 74, 1412–1426. [Google Scholar] [CrossRef] [PubMed]
- Zellmeier, S.; Schumann, W.; Wiegert, T. Involvement of Clp protease activity in modulating the Bacillus subtilis sigmaW stress response. Mol. Microbiol. 2006, 61, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Beare, P.A.; For, R.J.; Martin, L.W.; Lamont, I.L. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: Divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 2003, 47, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Draper, R.C.; Martin, L.W.; Beare, P.A.; Lamont, I.L. Differential proteolysis of sigma regulators controls cell-surface signalling in Pseudomonas aeruginosa. Mol. Microbiol. 2011, 82, 1444–1453. [Google Scholar] [CrossRef] [PubMed]
- Llamas, M.A.; Imperi, F.; Visca, P.; Lamont, I.L. Cell-surface signaling in Pseudomonas: Stress responses, iron transport, and pathogenicity. FEMS Microbiol. Rev. 2014, 38, 569–597. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.G.; Paget, M.S.; Seok, Y.J.; Hahn, M.Y.; Bae, J.B.; Hahn, J.S.; Kleanthous, C.; Buttner, M.J.; Roe, J.H. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 1999, 18, 4292–4298. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Bottrill, A.R.; Bibb, M.J.; Buttner, M.J.; Paget, M.S.; Kleanthous, C. The Role of zinc in the disulphide stress-regulated anti-sigma factor RsrA from Streptomyces coelicolor. J. Mol. Biol. 2003, 333, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Masloboeva, N.; Reutimann, L.; Stiefel, P.; Follador, R.; Leimer, N.; Hennecke, H.; Mesa, S.; Fischer, H.-M. Reactive oxygen species-inducible ECF σ factors of Bradyrhizobium japonicum. PLoS ONE 2012, 7, e43421. [Google Scholar] [CrossRef] [PubMed]
- Trepreau, J.; Girard, E.; Maillard, A.P.; de Rosny, E.; Petit-Haertlein, I.; Kahn, R.; Covès, J. Structural basis for metal sensing by CnrX. J. Mol. Biol. 2011, 408, 766–779. [Google Scholar] [CrossRef] [PubMed]
- Hastie, J.L.; Williams, K.B.; Sepúlveda, C.; Houtman, J.C.; Forest, K.T.; Ellermeier, C.D. Evidence of a bacterial receptor for lysozyme: Binding of lysozyme to the anti-σ factor RsiV controls activation of the ECF σ factor σV. PLoS Genet. 2014, 10, e1004643. [Google Scholar] [CrossRef] [PubMed]
- Marles-Wright, J.; Grant, T.; Delumeau, O.; van Duinen, G.; Firbank, S.J.; Lewis, P.J.; Murray, J.W.; Newman, J.A.; Quin, M.B.; Race, P.R.; et al. Molecular architecture of the “stressosome,” a signal integration and transduction Hub. Science 2008, 322, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Brody, M.S.; Stewart, V.; Price, C.W. Bypass suppression analysis maps the signalling pathway within a multidomain protein: The RsbP energy stress phosphatase 2C from Bacillus subtilis. Mol. Microbiol. 2009, 72, 1221–1234. [Google Scholar] [CrossRef] [PubMed]
- Gourion, B.; Rossignol, M.; Vorholt, J.A. A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc. Natl. Acad. Sci. USA 2006, 103, 13186–13191. [Google Scholar] [CrossRef] [PubMed]
- Francez-Charlot, A.; Frunzke, J.; Reichen, C.; Ebneter, J.Z.; Gourion, B.; Vorholt, J.A. Sigma factor mimicry involved in regulation of general stress response. Proc. Natl. Acad. Sci. USA 2009, 106, 3467–3472. [Google Scholar] [CrossRef] [PubMed]
- Herrou, J.; Foreman, R.; Fiebig, A.; Crosson, S. A structural model of anti-anti-σ inhibition by a two-component receiver domain: The PhyR stress response regulator. Mol. Microbiol. 2010, 78, 290–304. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarczyk, A.; Hochstrasser, R.; Vorholt, J.A.; Francez-Charlot, A. Complex two-component signaling regulates the general stress response in Alphaproteobacteria. Proc. Natl. Acad. Sci. USA 2014, 111, E5196–E5204. [Google Scholar] [CrossRef] [PubMed]
- Grigorova, I.L.; Phleger, N.J.; Mutalik, V.K.; Gross, C.A. Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. Proc. Natl. Acad. Sci. USA 2006, 103, 5332–5337. [Google Scholar] [CrossRef] [PubMed]
- Piper, S.E.; Mitchell, J.E.; Lee, D.J.; Busby, S.J. A global view of Escherichia coli Rsd protein and its interactions. Mol. Biosyst. 2009, 5, 1943–1947. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Fujita, N.; Ishihama, A. Competition among seven Escherichia coli sigma subunits: Relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 2000, 28, 3497–3503. [Google Scholar] [CrossRef] [PubMed]
- Mauri, M.; Klumpp, S. A model for sigma factor competition in bacterial cells. PLoS Comput. Biol. 2014, 10, e1003845. [Google Scholar] [CrossRef] [PubMed]
- Jishage, M.; Ishihama, A. A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase. Proc. Natl. Acad. Sci. USA 1998, 95, 4953–4958. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.H.; Gregory, B.D.; Sharp, J.S.; McCleary, K.D.; Dove, S.L.; Hochschild, A. Rsd family proteins make simultaneous interactions with regions 2 and 4 of the primary sigma factor. Mol. Microbiol. 2008, 70, 1136–1151. [Google Scholar] [CrossRef] [PubMed]
- Patikoglou, G.A.; Westblade, L.F.; Campbell, E.A.; Lamour, V.; Lane, W.J.; Darst, S.A. Crystal structure of the Escherichia coli regulator of sigma70, Rsd, in complex with sigma70 domain 4. J. Mol. Biol. 2007, 372, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-H.; Lee, C.-R.; Choe, M.; Seok, Y.-J. HPr antagonizes the anti-σ70 activity of Rsd in Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 21142–21147. [Google Scholar] [CrossRef] [PubMed]
- Deretic, V.; Konyecsni, W.M. Control of mucoidy in Pseudomonas aeruginosa: Transcriptional regulation of algR and identification of the second regulatory gene, algQ. J. Bacteriol. 1989, 171, 3680–3688. [Google Scholar] [PubMed]
- Schurr, M.J.; Martin, D.W.; Mudd, M.H.; Deretic, V. Gene cluster controlling conversion to alginate-overproducing phenotype in Pseudomonas aeruginosa: Functional analysis in a heterologous host and role in the instability of mucoidy. J. Bacteriol. 1994, 176, 3375–3382. [Google Scholar] [PubMed]
- Xie, Z.D.; Hershberger, C.D.; Shankar, S.; Ye, R.W.; Chakrabarty, A.M. Sigma factor-anti-sigma factor interaction in alginate synthesis: Inhibition of AlgT by MucA. J. Bacteriol. 1996, 178, 4990–4996. [Google Scholar] [PubMed]
- Schurr, M.J.; Yu, H.; Martinez-Salazar, J.M.; Boucher, J.C.; Deretic, V. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J. Bacteriol. 1996, 178, 4997–5004. [Google Scholar] [PubMed]
- Kato, J.; Chu, L.; Kitano, K.; DeVault, J.D.; Kimbara, K.; Chakrabarty, A.M.; Misra, T.K. Nucleotide sequence of a regulatory region controlling alginate synthesis in Pseudomonas aeruginosa: Characterization of the algR2 gene. Gene 1989, 84, 31–38. [Google Scholar] [CrossRef]
- Dove, S.L.; Hochschild, A. Bacterial two-hybrid analysis of interactions between region 4 of the sigma70 subunit of RNA polymerase and the transcriptional regulators Rsd from Escherichia coli and AlgQ from Pseudomonas aeruginosa. J. Bacteriol. 2001, 183, 6413–6421. [Google Scholar] [CrossRef] [PubMed]
- Ambrosi, C.; Tiburzi, F.; Imperi, F.; Putignani, L.; Visca, P. Involvement of AlgQ in transcriptional regulation of pyoverdine genes in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2005, 187, 5097–5107. [Google Scholar] [CrossRef] [PubMed]
- Mascher, T. Signaling diversity and evolution of extracytoplasmic function (ECF) σ factors. Curr. Opin. Microbiol. 2013, 16, 148–155. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paget, M.S. Bacterial Sigma Factors and Anti-Sigma Factors: Structure, Function and Distribution. Biomolecules 2015, 5, 1245-1265. https://doi.org/10.3390/biom5031245
Paget MS. Bacterial Sigma Factors and Anti-Sigma Factors: Structure, Function and Distribution. Biomolecules. 2015; 5(3):1245-1265. https://doi.org/10.3390/biom5031245
Chicago/Turabian StylePaget, Mark S. 2015. "Bacterial Sigma Factors and Anti-Sigma Factors: Structure, Function and Distribution" Biomolecules 5, no. 3: 1245-1265. https://doi.org/10.3390/biom5031245
APA StylePaget, M. S. (2015). Bacterial Sigma Factors and Anti-Sigma Factors: Structure, Function and Distribution. Biomolecules, 5(3), 1245-1265. https://doi.org/10.3390/biom5031245




