Metabolomics in Radiation-Induced Biological Dosimetry: A Mini-Review and a Polyamine Study
Abstract
:1. Introduction
2. Radiation Metabolomics in Biological Dosimetry
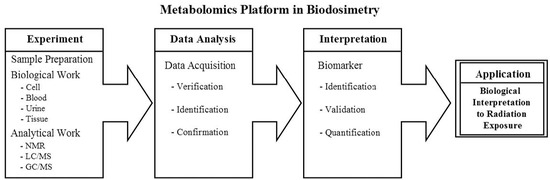
2.1. Metabolomics Platform in Biodosimetry
2.2. Metabolomics Technologies
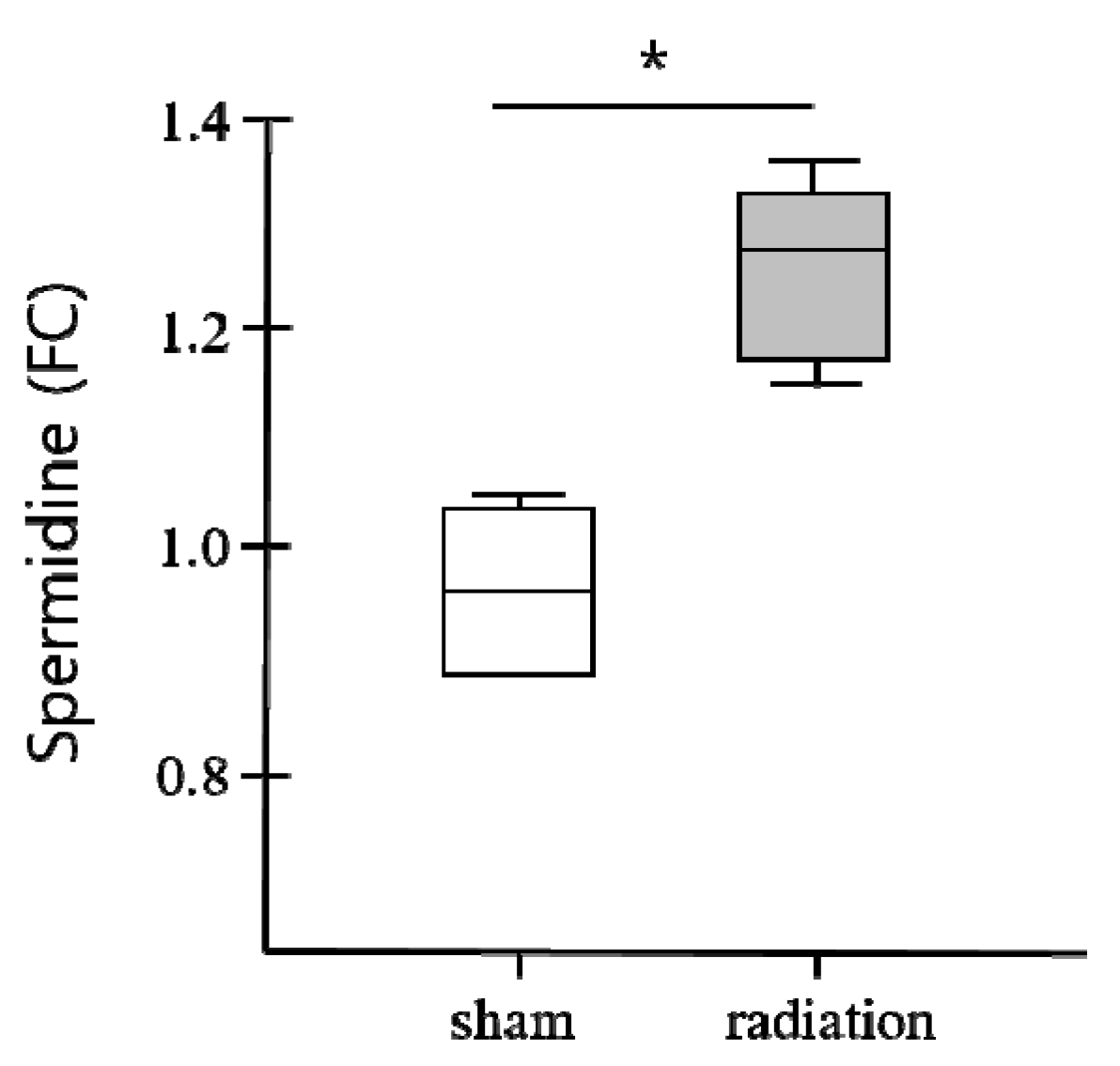
2.3. Potential Biomarkers
3. Conclusions
Acknowledgments
Conflicts of Interest
References
- Hall, E.J.; Giaccia, A.J. Radiobiology for the Radiologist, 6th ed.; Lippincott Williams & Wilkins: Philadephia, PA, USA, 2006; ISBN 0781741513. [Google Scholar]
- Chao, N.J. Accidental or intentional exposure to ionizing radiation: Biodosimetry and treatment options. Exp. Hematol. 2007, 35, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Pellmar, T.C.; Rockwell, S. Priority list of research areas for radiological nuclear threat countermeasures. Radiat. Res. 2005, 163, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.N.; Kumar, A.; Tiwar, P.; Mishra, K.P. Radiobiological basis in management of accidental radiation exposure. Int. J. Radiat. Biol. 2010, 86, 613–635. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, P.; Wardman, P. Radiation chemistry comes before radiation biology. Int. J. Radiat. Biol. 2009, 85, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Darroudi, F.; Wiegant, W.; Meijers, M.; Friedl, A.A.; van der Burg, M.; Fomina, J.; van Dongen, J.J.; van Gent, D.C.; Zdzienicka, M.Z. Role of Artemis in DSB repair and guarding chromosomal stability following exposure to ionizing radiation at different stages of cell cycle. Mutat. Res. 2007, 615, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Nuta, O.; Darroudi, F. The impact of the bystander effect on the low-dose hypersensitivity phenomenon. Radiat. Environ. Biophys. 2008, 47, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.N. Drift from DNA-centric radiation targets: A paradigm shift in Radiation Biology. Indian J. Radiat. Res. 2007, 4, 130–141. [Google Scholar]
- Lisowska, H.; Deperas-Kaminska, M.; Haghdoost, S.; Parmryd, I.; Wojcik, A. Radiation-induced DNA damage and repair in human γδ and αβ T-lymphocytes analysed by the alkaline comet assay. Genome Integr. 2010, 1, 8. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Patterson, A.D.; Krausz, K.W.; Kalinich, J.F.; Tyburski, J.B.; Kang, D.W.; Luecke, H.; Gonzalez, F.J.; Blakely, W.F.; Idle, J.R. Radiation metabolomics. 5. Identification of urinary biomarkers of ionizing radiation exposure in nonhuman primates by mass spectrometry-based metabolomics. Radiat. Res. 2012, 178, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Patterson, A.D.; Krausz, K.W.; Lanz, C.; Kang, D.W.; Luecke, H.; Gonzalez, F.J.; Idle, J.R. Radiation metabolomics. 4. UPLC-ESI-QTOFMS-Based metabolomics for urinary biomarker discovery in gamma-irradiated rats. Radiat. Res. 2011, 175, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Lanz, C.; Patterson, A.D.; Slavík, J.; Krausz, K.W.; Ledermann, M.; Gonzalez, F.J.; Idle, J.R. Radiation metabolomics. 3. Biomarker discovery in the urine of γ-irradiated rats using a simplified metabolomics protocol of gas chromatography-mass spectrometry combined with Random Forests Machine Learning Algorithm. Radiat. Res. 2009, 172, 198–212. [Google Scholar] [CrossRef] [PubMed]
- Tyburski, J.B.; Patterson, A.D.; Krausz, K.W.; Slavík, J.; Fornace, A.J., Jr.; Gonzalez, F.J.; Idle, J.R. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal γ-radiation exposure in mice. Radiat. Res. 2009, 172, 42–57. [Google Scholar] [CrossRef] [PubMed]
- Tyburski, J.B.; Patterson, A.D.; Krausz, K.W.; Slavík, J.; Fornace, A.J., Jr.; Gonzalez, F.J.; Idle, J.R. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for γ-radiation exposure in mice. Radiat. Res. 2008, 170, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Coy, S.L.; Cheema, A.K.; Tyburski, J.B.; Laiakis, E.C.; Collins, S.P.; Fornace, A.J., Jr. Radiation metabolomics and its potential in biodosimetry. Int. J. Radiat. Biol. 2011, 87, 802–823. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Newman, V.L.; Romaine, P.L.; Hauer-Jensen, M.; Pollard, H.B. Use of biomarkers for assessing radiation injury and efficacy of countermeasures. Expert. Rev. Mol. Diagn. 2016, 16, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Orth, M.; Lauber, K.; Niyazi, M.; Friedl, A.A.; Li, M.; Maihofer, C.; Schuttrumpf, L.; Ernst, A.; Niemoller, O.M.; Belka, C. Current concepts in clinical radiation oncology. Radiat. Environ. Biophys. 2014, 53, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Menon, S.S.; Uppal, M.; Randhawa, S.; Cheema, M.S.; Aghdam, N.; Usala, R.L.; Ghosh, S.P.; Cheema, A.K.; Dritschilo, A. Radiation Metabolomics: Current Status and Future Directions. Front. Oncol. 2016, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Laiakis, E.C.; Mak, T.D.; Anizan, S.; Amundson, S.A.; Barker, C.A.; Wolden, S.L.; Brenner, D.J.; Fornace, A.J., Jr. Development of a metabolomic radiation signature in urine from patients undergoing total body irradiation. Radiat. Res. 2014, 181, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.G.; Winson, M.K.; Kell, D.B.; Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotech. 1998, 16, 373–378. [Google Scholar] [CrossRef]
- Tweeddale, H.; Notley-McRobb, L.; Ferenci, T. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“Metabolome”) analysis. J. Bacteriol. 1998, 180, 5109–5116. [Google Scholar] [PubMed]
- Horning, E.C. Use of combined gas-liquid chromatography and mass spectrometry for clinical problems. Clin. Chem. 1968, 14, 777. [Google Scholar]
- Behar, K.L.; Denhollander, J.A.; Stromski, M.E.; Ogino, T.; Shulman, R.G.; Petro, V.O.A.C.; Prichard, J.W. High-resolution 1H nuclear magnetic-resonance study of cerebral hypoxia in vivo. Proc. Natl. Acad. Sci. USA 1983, 80, 4945–4948. [Google Scholar] [CrossRef] [PubMed]
- Howells, S.L.; Maxwell, R.J.; Peet, A.C.; Griffiths, J.R. An investigation of tumor 1H nuclear-magnetic-resonance spectra by the application of chemometric techniques. Magn. Reson. Med. 1992, 28, 214–236. [Google Scholar] [CrossRef] [PubMed]
- Mamas, M.; Dunn, W.B.; Neyses, L.; Goodacre, R. The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Arch. Toxicol. 2011, 85, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Yatin, M. Polyamines in living organisms. J. Cell. Mol. Biol. 2002, 1, 57–67. [Google Scholar]
- Wallace, H.M. Polyamines and their role in human disease—An introduction. Biochem. Soc. Trans. 2003, 31, 354–355. [Google Scholar] [CrossRef] [PubMed]
- Janne, J.; Alhonen, L.; Leinonen, P. Polyamines: From molecular biology to clinical applications. Trends Mol. Med. 1991, 23, 241–259. [Google Scholar] [CrossRef]
- Wallace, H.M.; Fraser, A.V.; Hughes, A. A perspective of polyamine metabolism. Biochem. J. 2003, 376, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.K.; Birchler, M.; Smith, M.K.; Rayca, K.; Mostochuk, J. Effect of elevated levels of ornithine decarboxylase on cell cycle progression in skin. Cell Growth Differ. 1999, 10, 739–748. [Google Scholar] [PubMed]
- Hebby, O. Role of Polyamines in the control of cell proliferation and differentiation. Differentiation 1981, 19, 1–20. [Google Scholar] [CrossRef]
- Tabor, C.W.; Tabor, H. Polyamines. Ann. Rev. Biochem. 1984, 53, 749–790. [Google Scholar] [CrossRef] [PubMed]
- Seiler, N.; Delcros, J.G.; Moulinoux, J.P. Polyamine transport in mammalian cells. An update. Int. J. Biochem. Cell. Biol. 1996, 28, 843–861. [Google Scholar] [CrossRef]
- Medina, M.Á.; Correa-Fiz, F.; Rodríguez-Caso, C.; Sánchez-Jiménez, F. A comprehensive view of polyamine and histamine metabolism to the light of new technologies. J. Cell. Mol. 2005, 9, 854–864. [Google Scholar] [CrossRef]
- Cohen, S.S. A Guide to Polyamines; Oxford University Press: New York, NY, USA, 1998. [Google Scholar]
- Thomas, T.; Thomas, T.J. Polyamines in cell growth and cell death: Molecular mechanisms and therapeutic applications. Cell Mol. Life Sci. 2001, 58, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.A.; Urdiales, J.L.; Rodriguez-Caso, C.; Ramirez, F.J.; Sanchez-Jimenez, F. Biogenic amines and polyamines: Similar biochemistry for different physiological missions and biomedical applications. Crit. Rev. Biochem. Mol. Biol. 2003, 38, 23–59. [Google Scholar] [CrossRef] [PubMed]
- Gerner, E.W.; Meyskens, F.L., Jr. Polyamines and cancer: Old molecules, new understanding. Nat. Rev. Cancer 2004, 4, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Aziz, S.M.; Yatin, M.; Worthen, D.R.; Lipke, D.W.; Crooks, P.A. A novel technique for visualizing the intracellular localization and distribution of transported polyamines in cultured pulmonary artery smooth muscle cells. J. Pharm. Biomed. Anal. 1998, 17, 307–320. [Google Scholar] [CrossRef]
- Cullis, P.M.; Green, R.E.; Merson-Davies, L.; Travis, N. Probing the mechanism of transport and compartmentalization of polyamines in mammalian cells. Chem. Biol. 1999, 6, 717–729. [Google Scholar] [CrossRef]
- Casero, R.A., Jr.; Marton, L.J. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 2007, 6, 373–390. [Google Scholar] [CrossRef] [PubMed]
- Roh, C.; Yu, D.K.; Kim, I.; Jo, S.K. The biological response of spermidine induced by ionization radiation. Molecules 2012, 17, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.K.; Krausz, K.W.; Bonzo, J.A.; Idle, J.R.; Gonzalez, F.J. Metabolomics reveals aging-associated attenuation of noninvasive radiation biomarkers in mice: Potential role of polyamine catabolism and incoherent DNA damage-repair. J. Proteome Res. 2013, 12, 2269–2281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porciani, S.; Lanini, A.; Balzi, M.; Faraoni, P.; Becciolini, A. Polyamines as biochemical indicators of radiation injury. Phys. Med. 2001, 17, 187–188. [Google Scholar] [PubMed]
- Kafy, A.M.L.; Haigh, C.G.; Lewis, D.A. In vitro interactions between endogenous polyamines and superoxide anion. Agents Actions 1986, 18, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Gaboriau, F.; Vaultier, M.; Moulinoux, J.P.; Delcros, J.G. Antioxidative properties of natural polyamines and dimethylsilane analogues. Redox Rep. 2005, 10, 9–18. [Google Scholar] [CrossRef] [PubMed]



© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roh, C. Metabolomics in Radiation-Induced Biological Dosimetry: A Mini-Review and a Polyamine Study. Biomolecules 2018, 8, 34. https://doi.org/10.3390/biom8020034
Roh C. Metabolomics in Radiation-Induced Biological Dosimetry: A Mini-Review and a Polyamine Study. Biomolecules. 2018; 8(2):34. https://doi.org/10.3390/biom8020034
Chicago/Turabian StyleRoh, Changhyun. 2018. "Metabolomics in Radiation-Induced Biological Dosimetry: A Mini-Review and a Polyamine Study" Biomolecules 8, no. 2: 34. https://doi.org/10.3390/biom8020034
APA StyleRoh, C. (2018). Metabolomics in Radiation-Induced Biological Dosimetry: A Mini-Review and a Polyamine Study. Biomolecules, 8(2), 34. https://doi.org/10.3390/biom8020034