Shotgun Lipidomics Revealed Altered Profiles of Serum Lipids in Systemic Lupus Erythematosus Closely Associated with Disease Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Blood Collection
2.3. Lipid Standards and Chemicals
2.4. Preparation of Lipid Extracts from Serum Samples
2.5. Mass Spectrometric Analysis of Lipids
2.6. Data Processing
3. Results
3.1. Basic Characteristics of the Subjects
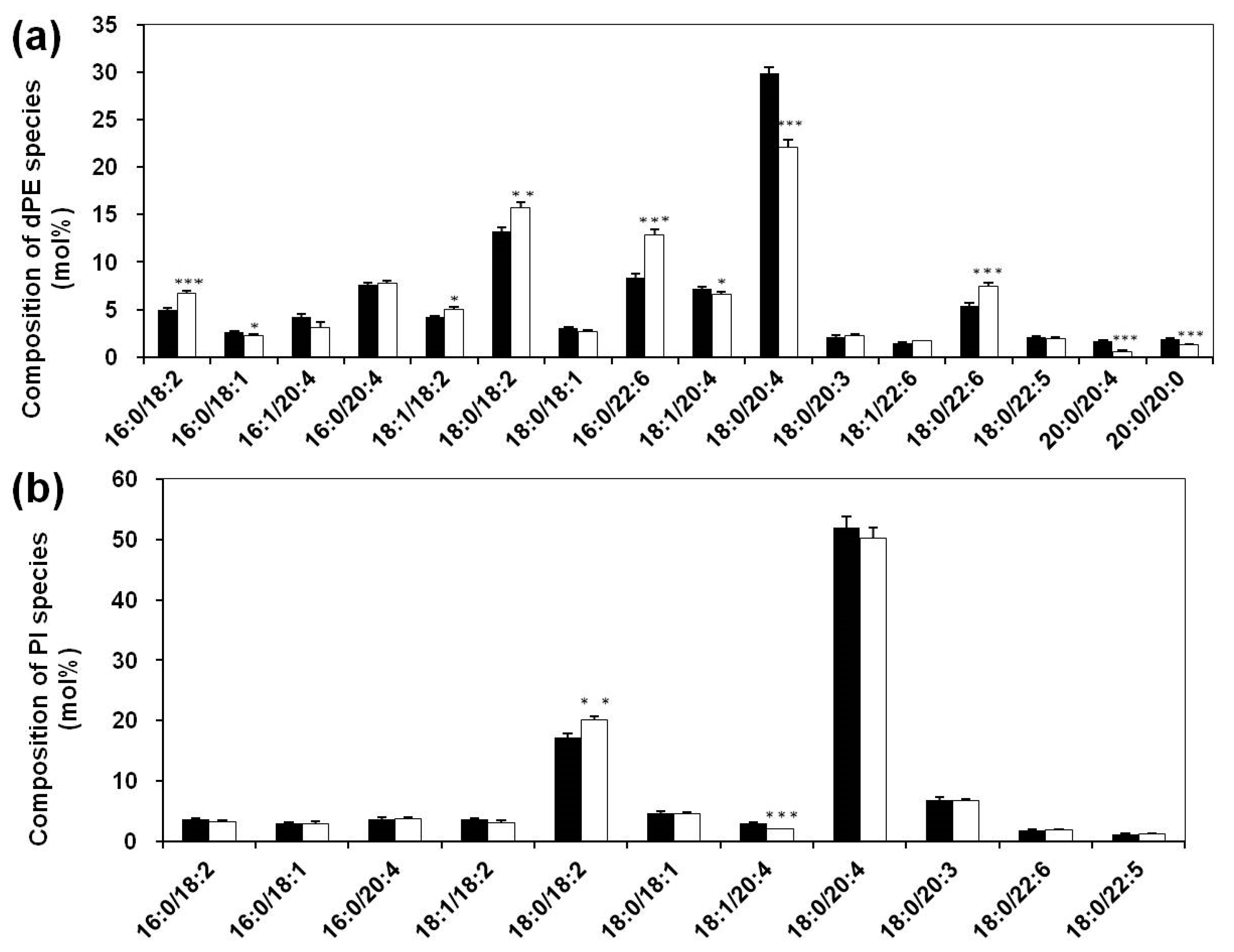
3.2. Lipidomics Analysis of Phospholipids of Serum Lipid Extracts
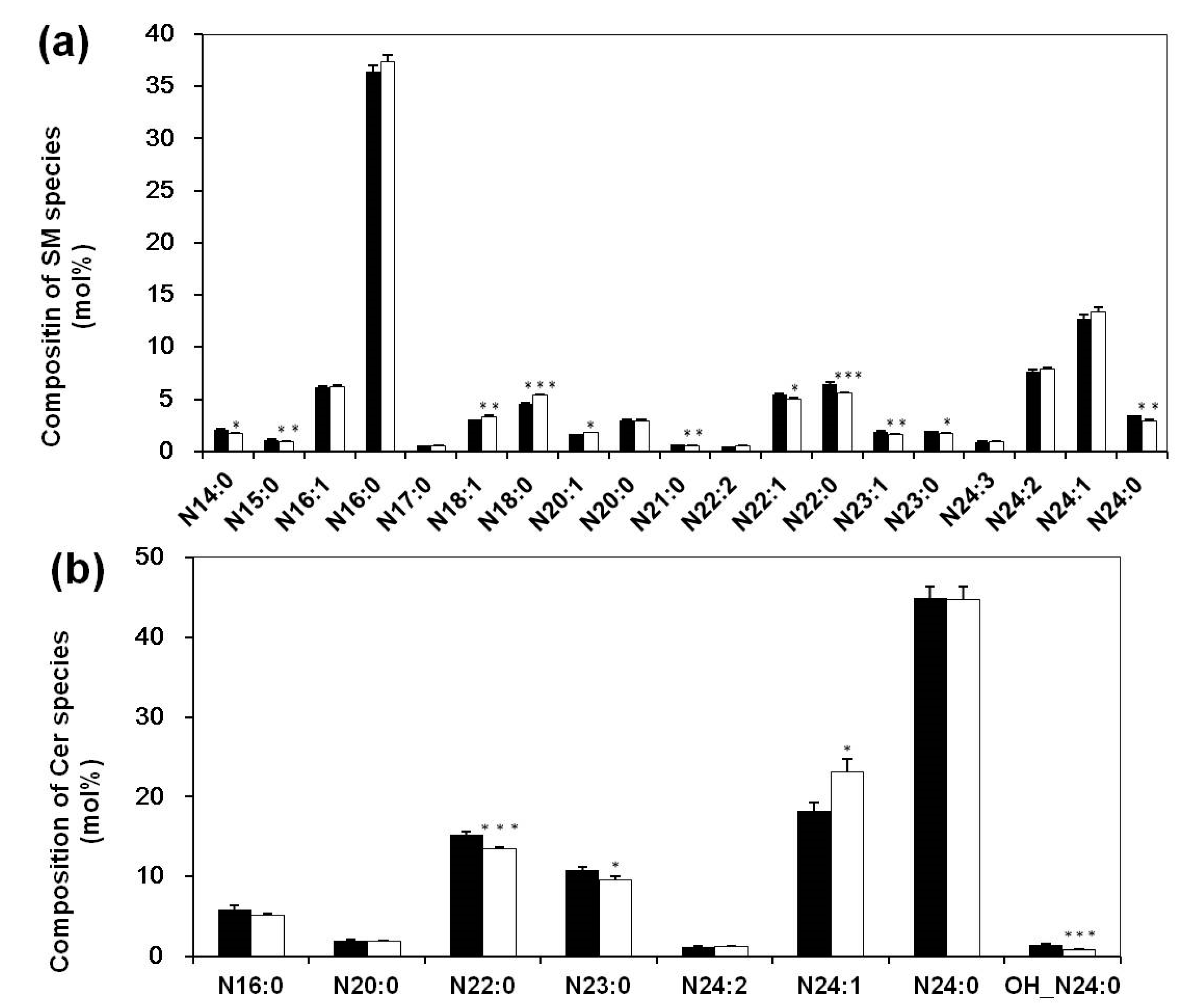
3.3. Altered Composition of Sphingolipid Species in Sera of Systemic Lupus Erythematosus Patients
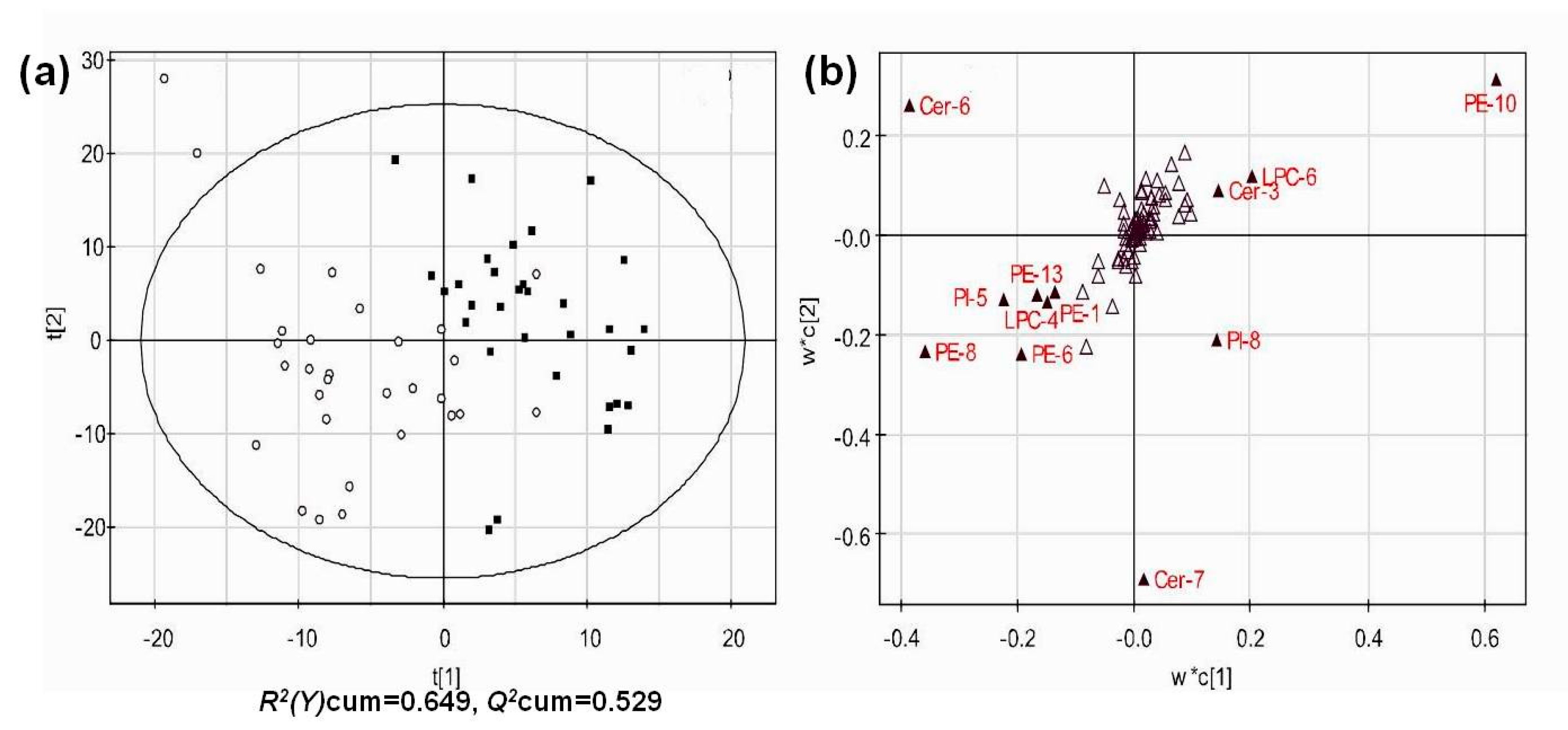
3.4. Multivariate Analysis of Lipid Profiles of Different Groups
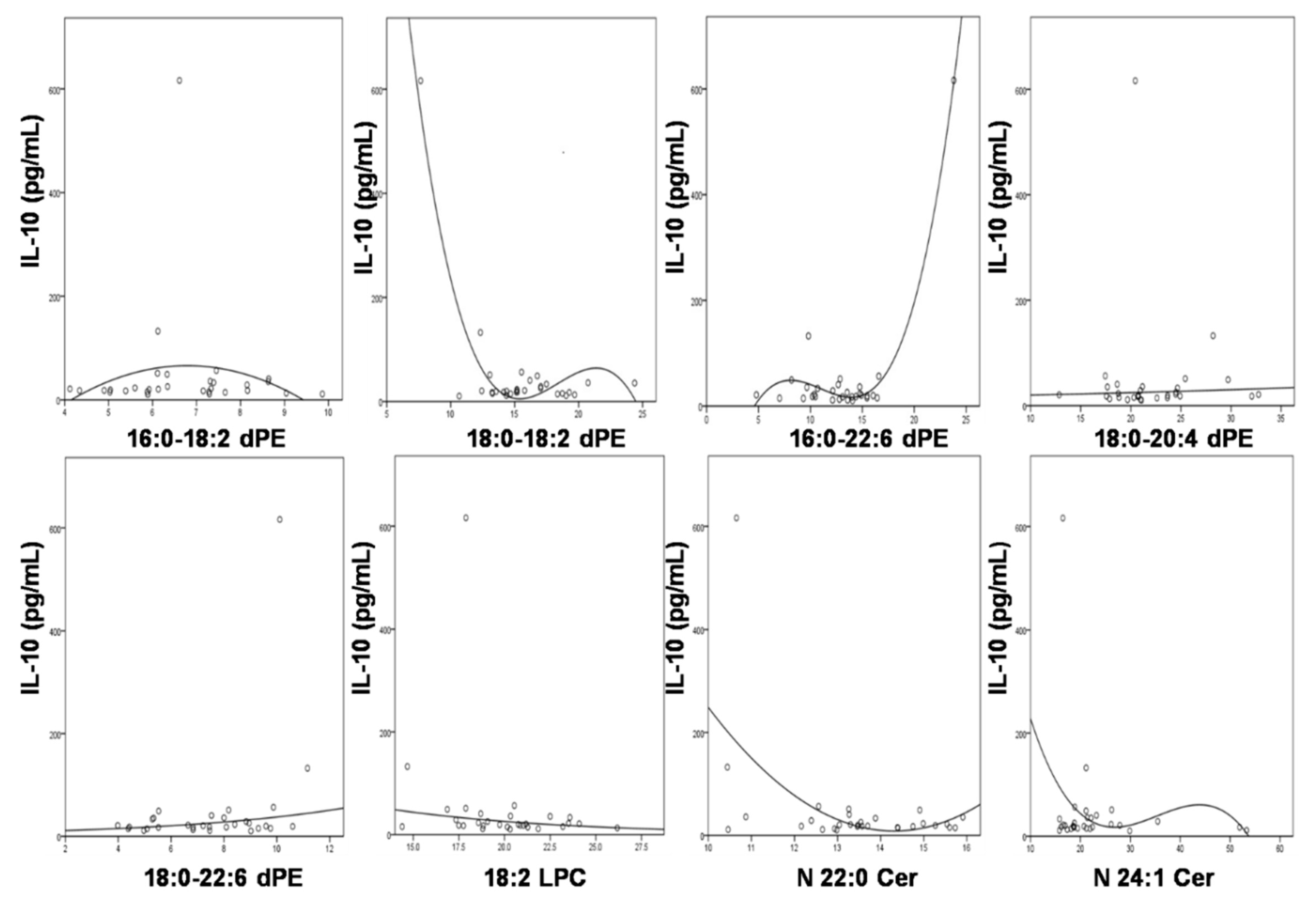
3.5. Significant Association of IL-10 with the Compositions of the Altered Lipids
3.6. Correlation of SLE Disease Activity Index with the Compositions of the Changed Lipid Species
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Cer | ceramide |
| DHA | docosahexaenicacid |
| HNE | 4-hydroxy-2(E)-nonenal |
| ox-LDL | oxidized low-density lipoprotein |
| MDMS-SL | multi-dimensional mass spectrometry-based shotgun lipidomics technology |
| PC | phosphatidylcholine |
| dPC | diacly-phosphatidylcholine |
| pPC | plasmenylcholine |
| LPC | lysoPC |
| PE | phosphatidylethanolamine |
| aPE | plasmanylethanolamine |
| dPE | diacyl-phosphatidylethanolamine |
| pPE | ethanolamine plasmalogen |
| PI | phosphatidylinositol |
| LPE | lysoPE |
| PLS-DA | partial least squares-discriminate analysis |
| PUFA | polyunsaturated fatty acyl (acid) |
| SLE | systemic lupus erythematosus |
| SLEDAI | SLE disease activity index |
| TAG | triglycerides |
References
- Tosokos, G.C. Systemic Lupus Erythematosus. N. Engl. J. Med. 2011, 365, 2110–2121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koutsokeras, T.; Healy, T. Systemic lupus erythematosus and lupus nephritis. Nat. Rev. Drug Discov. 2014, 13, 173–174. [Google Scholar] [CrossRef] [PubMed]
- Lisnevskaia, L.; Murphy, G.; Isenberg, D. Systemic lupus erythematosus. Lancet 2014, 384, 1878–1888. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Wang, M.; Han, X. Shotgun lipidomics in substantiating lipid peroxidation in redox biology: Methods and applications. Redox Biol. 2017, 12, 946–955. [Google Scholar] [CrossRef] [PubMed]
- Borba, E.F.; Bonfa, E.; Vinagre, C.G.C.; Ramires, J.A.F.; Maranhao, R.C. Chylomicron metabolism is markedly altered in systemic lupus erythematosus. Arthriris Rheum. 2000, 43, 1033–1040. [Google Scholar] [CrossRef]
- Borba, E.F.; Bonfá, E. Dyslipoproteinemias in systemic lupus erythematosus: Influence of disease, activity, and anticardiolipin antibodies. Lupus 1997, 6, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Yaniv, G.; Twig, G.; Shor, D.B.; Furer, A.; Sherer, Y.; Mozes, O.; Komisar, O.; Slonimsky, E.; Klang, E.; Lotan, E.; et al. A volcanic explosion of autoantibodies in systemic lupus erythematosus: A diversity of 180 different antibodies found in SLE patients. Autoimmun. Rev. 2015, 14, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Sherer, Y.; Gorstein, A.; Fritzler, M.J.; Shoenfeld, Y. Autoantibody explosion in systemic lupus erythematosus: More than 100 different antibodies found in SLE patients. Semin. Arthritis Rheum. 2004, 34, 501–537. [Google Scholar] [CrossRef] [PubMed]
- Reichlin, M.; Fesmire, J.; Quintero-Del-Rio, A.I.; Wolfson-Reichlin, M. Autoantibodies to lipoprotein lipase and dyslipidemia in systemic lupus erythematosus. Arthritis Rheum. 2002, 46, 2957–2963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perl, A. Oxidative stress in the pathology and treatment of systemic lupus erythematosus. Nat. Rev. Rheumatol. 2013, 9, 674–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morel, L. Immunometabolism in systemic lupus erythematosus. Nat. Rev. Rheumatol. 2017, 13, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Frostegard, J.; Svenungsson, E.; Wu, R.; Gunnarsson, I.; Lundberg, I.E.; Klareskog, L.; Horkko, S.; Witztum, J.L. Lipid peroxidation is enhanced in patients with systemic lupus erythematosus and is associated with arterial and renal disease manifestations. Arthritis Rheum. 2005, 52, 192–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, M.; Grossman, J.; FitzGerald, J.; Dahlin-Lee, E.; Wallace, D.J.; Thong, B.Y.; Badsha, H.; Kalunian, K.; Charles, C.; Navab, M.; et al. Proinflammatory high-density lipoprotein as a biomarker for atherosclerosis in patients with systemic lupus erythematosus and rheumatoid arthritis. Arthriris Rheum. 2006, 54, 2541–2549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurien, B.T.; Hensley, K.; Bachmann, M.; Hal Scofield, R. Oxidatively modified autoantigens in autoimmune diseases. Free Radic. Biol. Med. 2006, 41, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xie, X.-W.; Zhou, H.; Wang, B.; Zhang, M.-J.; Tang, F.-Y. Metabolic profiling reveals new serum biomarkers of lupus nephritis. Lupus 2017, 26, 1166–1173. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, A.A.; Trygg, J.; Wuttge, D.M.; Sturfelt, G.; Theander, E.; Donten, M.; Moritz, T.; Sennbro, C.J.; Torell, F.; Lood, C.; et al. Metabolic profiling of systemic lupus erythematosus and comparison with primary Sjögren’s syndrome and systemic sclerosis. PLoS ONE 2016, 11, e0159384. [Google Scholar] [CrossRef] [PubMed]
- Perl, A.; Hanczko, R.; Lai, Z.W.; Oaks, Z.; Kelly, R.; Borsuk, R.; Asara, J.M.; Phillips, P.E. Comprehensive metabolome analyses reveal N-acetylcysteine-responsive accumulation of kynurenine in systemic lupus erythematosus: Implications for activation of the mechanistic target of rapamycin. Metabolomics 2015, 11, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Guleria, A.; Pratap, A.; Dubey, D.; Rawat, A.; Chaurasia, S.; Sukesh, E.; Phatak, S.; Ajmani, S.; Kumar, U.; Khetrapal, C.L.; et al. NMR based serum metabolomics reveals a distinctive signature in patients with Lupus Nephritis. Sci. Rep. 2016, 6, 35309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, B.; Huang, J.; Zhang, C.; Hu, X.; Gao, M.; Shi, A.; Zha, W.; Shi, L.; Huang, C.; Yang, L. Serum metabolomic profiling in patients with systemic lupus erythematosus by GC/MS. Mod. Rheumatol. 2016, 26, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zhou, J.; Yang, S.; Li, H.; Wang, C.; Fang, X.; Fan, Y.; Zhang, J.; Han, X.; Wen, C. Oxidative stress leads to reduction of plasmalogen serving as a novel biomarker for systemic lupus erythematosus. Free Radic. Biol. Med. 2016, 101, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Hochberg, M.C. Updating the American College of Rheumatology Revised Criteria for the Classification of Systemic Lupus Erythematosus. Arthritis Rheum. 1997, 40, 1725. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Guan, S.; Han, X. Abundance of triacylglycerols in ganglia and their depletion in diabetic mice: Implications for the role of altered triacylglycerols in diabetic neuropathy. J. Neurochem. 2006, 97, 1288–1300. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Rozen, S.; Boyle, S.H.; Hellegers, C.; Cheng, H.; Burke, J.R.; Welsh-Bohmer, K.A.; Doraiswamy, P.M.; Kaddurah-Daouk, R. Metabolomics in early alzheimer’s disease: Identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS ONE 2011, 6, e21643. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Cheng, H.; Gross, R.W.; Han, X. Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal. Chem. 2009, 81, 4356–4368. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yang, K.; Gross, R.W. Microfluidics-based electrospray ionization enhances the intrasource seperation of lipid classes and extends identification of individual molecular species through multi-dimensional mass apectrometry: Development of an automated high-throughput platform for shotgun lipidomics. Rapid Commun. Mass Spectrom. 2008, 22, 2115–2124. [Google Scholar] [PubMed]
- Hu, C.; Wang, Y.; Fan, Y.; Li, H.; Wang, C.; Zhang, J.; Zhang, S.; Han, X.; Wen, C. Lipidomics revealed idiopathic pulmonary fibrosis-induced hepatic lipid disorders corrected with treatment of baicalin in a murine model. AAPS J. 2015, 17, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Han, X. Lipidomics: Comprehensive Mass Spectrometry of Lipids; Wiley: Hoboken, NJ, USA, 2016; 496p. [Google Scholar]
- Watson, A.D. Lipidomics: A global approach to lipid analysis in biological systems. J. Lipid Res. 2006, 47, 11. [Google Scholar] [CrossRef] [PubMed]
- Quehenberger, Q.; Dennis, E.A. the human plasma lipidome. N. Engl. J. Med. 2011, 365, 1812–1823. [Google Scholar] [CrossRef] [PubMed]
- Echard, A. Phosphoinositides and cytokinesis: The “PIP” of the iceberg. Cytoskeleton 2012, 69, 893–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, O.T.T.; Nguyen, T.T.N.; Lee, S.Y. Phosphoinositide turnover in Toll-like receptor signaling and trafficking. BMB Rep. 2014, 47, 361–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maceyka, M.; Spiegel, S. Sphingolipid metabolites in inflammatory disease. Nature 2014, 510, 58–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, W.J.; Park, J.W. The effect of altered sphingolipid acyl chain length on various disease models. Biol. Chem. 2015, 396, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Kitatani, K.; Idkowiak-Baldys, J.; Hannun, Y.A. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell. Signal. 2008, 20, 1010–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 2016, 12, 668–679. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, E.M.; Smith, S.; Lee, R.Z.; Cunnane, G.; Doran, M.F.; Donnelly, S.; Howard, D.; O’Connell, P.; Kearns, G.; Ni Gabhann, J.; et al. The association of cytokines with disease activity and damage scores in systemic lupus erythematosus patients. Rheumatology 2014, 53, 1586–1594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Hishikawa, D.; Hashidate, T.; Shimizu, T.; Shindou, H. Diversity and function of membrane glycerophospholipids generated by the remodeling pathway in mammalian cells. J. Lipid Res. 2014, 55, 799–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serhan, C.N.; Chiang, N.; Van Dyke, T.E. Resolving inflammation: Dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol. 2008, 8, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Ariel, A.; Serhan, C.N. Resolvins and protectins in the termination program of acute inflammation. Trends Immunol. 2007, 28, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Gorjao, R.; Azevedo-Martins, A.K.; Rodrigues, H.G.; Abdulkader, F.; Arcisio-Miranda, M.; Procopio, J.; Curi, R. Comparative effects of DHA and EPA on cell function. Pharmacol. Ther. 2009, 122, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N.; Petasis, N.A. Resolvins and protectins in inflammation resolution. Chem. Rev. 2011, 111, 5922–5943. [Google Scholar] [CrossRef] [PubMed]
- Tsokos, G.C.; Lo, M.S.; Costa Reis, P.; Sullivan, K.E. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat. Rev. Rheumatol. 2016, 12, 716–730. [Google Scholar] [CrossRef] [PubMed]
- Anolik, J.H.; Aringer, M. New treatments for SLE: Cell-depleting and anti-cytokine therapies. Best Pract. Res. Clin. Rheumatol. 2005, 19, 859–878. [Google Scholar] [CrossRef] [PubMed]
- Mesicek, J.; Lee, H.; Feldman, T.; Jiang, X.; Skobeleva, A.; Berdyshev, E.V.; Haimovitz-Friedman, A.; Fuks, Z.; Kolesnick, R. Ceramide synthases 2, 5, and 6 confer distinct roles in radiation-induced apoptosis in HeLa cells. Cell. Signal. 2010, 22, 1300–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gergely, P.J.; Niland, B.; Gonchoroff, N.; Pullmann, R.J.; Phillips, P.; Perl, A. Persistent mitochondrial hyperpolarization, increased reactive oxygen intermediate production, and cytoplasmic alkalinization characterize altered IL-10 signaling in patients with systemic lupus erythematosus. J. Immunol. 2002, 169, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Gergely, P.J.; Grossman, C.; Niland, B.; Puskas, F.; Neupane, H.; Allam, F.; Banki, K.; Phillips, P.; Perl, A. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthriris Rheum. 2002, 46, 175–190. [Google Scholar] [CrossRef] [Green Version]
| Parameters | Patients (N = 30) | Controls (N = 30) |
|---|---|---|
| Age (years) | 35.0 ± 1.4 | 35.4 ± 1.5 |
| Gender (% women) | 100 | 100 |
| Disease duration (years) | 10.2 ± 1.3 | — |
| Body mass index (Kg/m2) | 22.6 ± 0.4 | 23.1 ± 0.5 |
| Anti-dsDNA antibody positivity | 20/30 | — |
| Low C3/C4 | 23/30 | — |
| Proteinuria | 13/30 | — |
| SLEDAI | Inactive (0–11): 23/30 Active (>11): 7/30 | — |
| Lipid Specie | Equation | R2 | F | df1 | df2 | p |
|---|---|---|---|---|---|---|
| 16:0/18:2 dPE | Quadratic | 0.081 | 1.186 | 2 | 27 | 0.321 |
| 18:0/18:2 dPE | Quadratic | 0.226 | 3.951 | 2 | 27 | 0.031 |
| 16:0/22:6 dPE | Quadratic | 0.087 | 1.294 | 2 | 27 | 0.291 |
| 18:0/20:4 dPE | S | 0.056 | 1.671 | 1 | 28 | 0.207 |
| 18:0/22:6 dPE | Cubic | 0.115 | 1.127 | 3 | 26 | 0.356 |
| 18:2 LPC | Cubic | 0.12 | 1.848 | 2 | 27 | 0.177 |
| N22:0 Cer | Logarithmic | 0.004 | 0.118 | 1 | 28 | 0.747 |
| N24:1 Cer | Quadratic | 0.028 | 0.392 | 2 | 27 | 0.679 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, L.; Hu, C.; Zhao, Y.; He, L.; Zhou, J.; Li, H.; Du, Y.; Wang, Y.; Wen, C.; Han, X.; et al. Shotgun Lipidomics Revealed Altered Profiles of Serum Lipids in Systemic Lupus Erythematosus Closely Associated with Disease Activity. Biomolecules 2018, 8, 105. https://doi.org/10.3390/biom8040105
Lu L, Hu C, Zhao Y, He L, Zhou J, Li H, Du Y, Wang Y, Wen C, Han X, et al. Shotgun Lipidomics Revealed Altered Profiles of Serum Lipids in Systemic Lupus Erythematosus Closely Associated with Disease Activity. Biomolecules. 2018; 8(4):105. https://doi.org/10.3390/biom8040105
Chicago/Turabian StyleLu, Lu, Changfeng Hu, Yanxia Zhao, Lijiao He, Jia Zhou, Haichang Li, Yu Du, Yonghua Wang, Chengping Wen, Xianlin Han, and et al. 2018. "Shotgun Lipidomics Revealed Altered Profiles of Serum Lipids in Systemic Lupus Erythematosus Closely Associated with Disease Activity" Biomolecules 8, no. 4: 105. https://doi.org/10.3390/biom8040105
APA StyleLu, L., Hu, C., Zhao, Y., He, L., Zhou, J., Li, H., Du, Y., Wang, Y., Wen, C., Han, X., & Fan, Y. (2018). Shotgun Lipidomics Revealed Altered Profiles of Serum Lipids in Systemic Lupus Erythematosus Closely Associated with Disease Activity. Biomolecules, 8(4), 105. https://doi.org/10.3390/biom8040105





