Dual-Family Peptidylprolyl Isomerases (Immunophilins) of Select Monocellular Organisms
Abstract
:1. Introduction
2. Dual-Family Peptidylprolyl Isomerase Structure
3. Organisms Harboring Dual-Family Peptidylprolyl Isomerases
4. Physiological Function of the Dual-Family Peptidylprolyl Isomerases
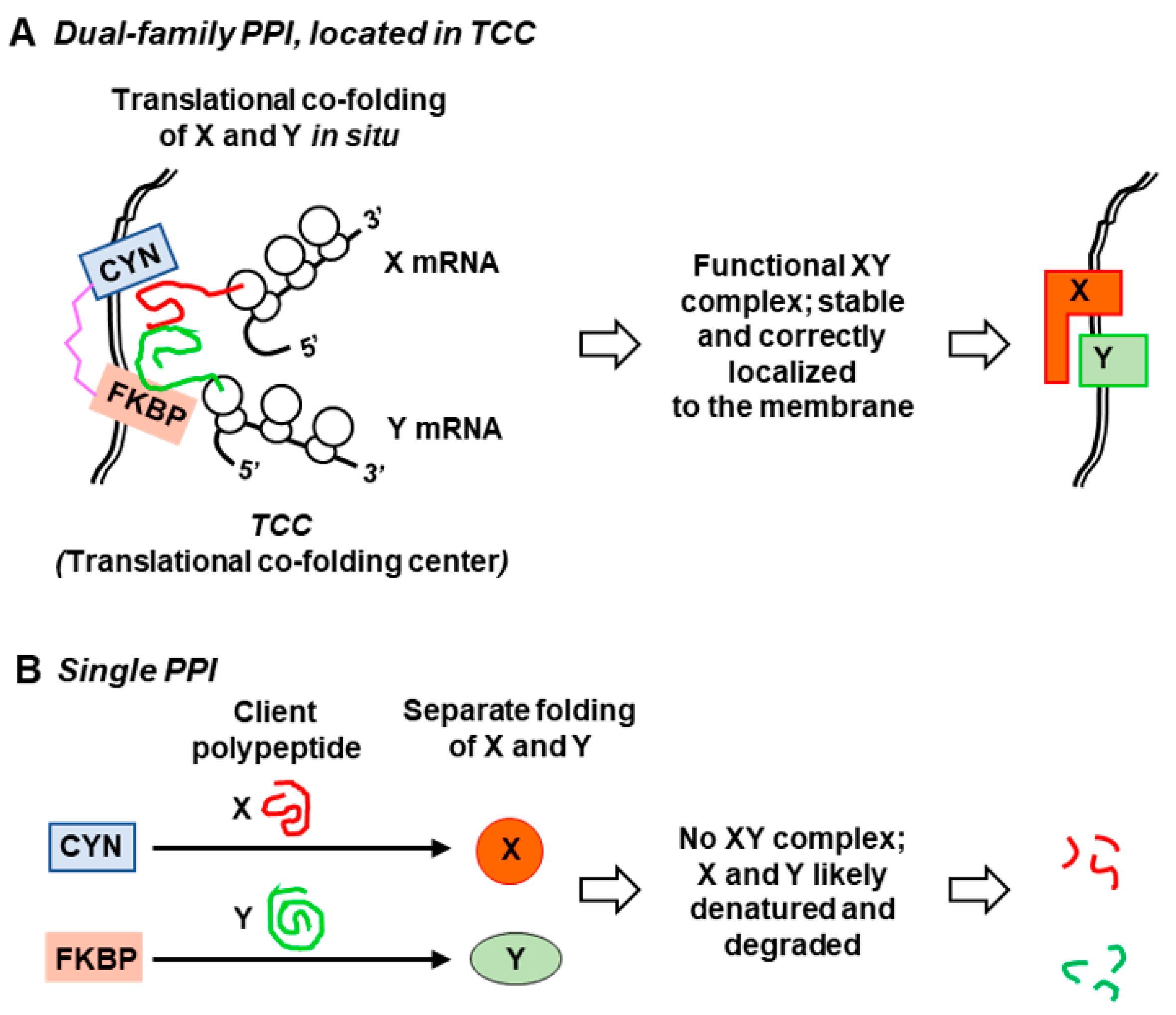
5. Available Evidence for the Translational Co-Folding Center Model
6. Phylogenetic Origin of the Dual-Family Peptidylprolyl Isomerases
7. Future Directions of Dual-Family Peptidylprolyl Isomerase Research
Funding
Acknowledgments
Conflicts of Interest
Appendix A

References
- Galat, A. Peptidylprolyl cis/trans isomerases (immunophilins): Biological diversity–targets–functions. Curr. Top. Med. Chem. 2003, 3, 1315–1347. [Google Scholar] [CrossRef] [PubMed]
- Barik, S. Immunophilins: For the love of proteins. Cell. Mol. Life. Sci. 2006, 63, 2889–2900. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W.; Sophos, N.A.; Vasiliou, V.; Nelson, D.R. Cyclophilin nomenclature problems, or, ‘a visit from the sequence police’. Hum. Genom. 2004, 1, 381–388. [Google Scholar] [CrossRef]
- Fischer, G.; Wittmann-Liebold, B.; Lang, K.; Kiefhaber, T.; Schmid, F.X. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature 1989, 337, 476–478. [Google Scholar] [CrossRef] [PubMed]
- Siekierka, J.J.; Hung, S.H.; Poe, M.; Lin, C.S.; Sigal, N.H. A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature 1989, 341, 755–757. [Google Scholar] [CrossRef] [PubMed]
- Harding, M.W.; Galat, A.; Uehling, D.E.; Schreiber, S.L. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature 1989, 341, 758–760. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Farmer, J.D., Jr.; Lane, W.S.; Friedman, J.; Weissman, I.; Schreiber, S.L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 1991, 66, 807–815. [Google Scholar] [CrossRef]
- Rao, A.; Luo, C.; Hogan, P.G. Transcription factors of the NFAT family: Regulation and function. Annu. Rev. Immunol. 1997, 15, 707–747. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.T.; Wiederrecht, G.J. Immunopharmacology of rapamycin. Annu. Rev. Immunol. 1996, 14, 483–510. [Google Scholar] [CrossRef] [PubMed]
- Kern, G.; Kern, D.; Schmid, F.X.; Fischer, G. Reassessment of the putative chaperone function of prolyl-cis/trans-isomerases. FEBS Lett. 1994, 348, 145–148. [Google Scholar] [CrossRef]
- Schmidpeter, P.A.; Schmid, F.X. Prolyl isomerization and its catalysis in protein folding and protein function. J. Mol. Biol. 2015, 427, 1609–1631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.C.; Wang, W.D.; Wang, J.S.; Pan, J.C. PPIase independent chaperone-like function of recombinant human Cyclophilin A during arginine kinase refolding. FEBS Lett. 2013, 587, 666–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, S.; Xiao, Y.; Nakaya, M.; D’Angelillo, A.; Chang, M.; Jin, J.; Hausch, F.; Masullo, M.; Feng, X.; Romano, M.F.; et al. FKBP51 employs both scaffold and isomerase functions to promote NF-κB activation in melanoma. Nucleic Acids Res. 2015, 43, 6983–6993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, Y.Y.; Her, J.; Oh, S.Y.; Chung, I.K. Hsp90-binding immunophilin FKBP52 modulates telomerase activity by promoting the cytoplasmic retrotransport of hTERT. Biochem. J. 2016, 473, 3517–3532. [Google Scholar] [CrossRef] [PubMed]
- Lagadari, M.; Zgajnar, N.R.; Gallo, L.I.; Galigniana, M.D. Hsp90-binding immunophilin FKBP51 forms complexes with hTERT enhancing telomerase activity. Mol. Oncol. 2016, 10, 1086–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, B.; Musiyenko, A.; Kumar, R.; Barik, S. A novel class of dual-family immunophilins. J. Biol. Chem. 2005, 280, 24308–24314. [Google Scholar] [CrossRef] [PubMed]
- Barik, S. On the role, ecology, phylogeny, and structure of dual-family immunophilins. Cell Stress Chaperones 2017, 22, 833–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krücken, J.; Greif, G.; von Samson-Himmelstjerna, G. In silico analysis of the cyclophilin repertoire of apicomplexan parasites. Parasit. Vectors 2009, 2, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barik, S. Bioinformatic analysis reveals conservation of intrinsic disorder in the linker sequences of prokaryotic dual-family immunophilin chaperones. Comput. Struct. Biotechnol. J. 2017, 16, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Deller, M.C.; Kong, L.; Rupp, B. Protein stability: A crystallographer’s perspective. Acta Crystallogr. F Struct. Biol. Commun. 2016, 72, 72–95. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J. Making sense of intrinsically disordered proteins. Biophys. J. 2016, 110, 1013–1016. [Google Scholar] [CrossRef] [PubMed]
- Kriwacki, R.W.; Hengst, L.; Tennant, L.; Reed, S.I.; Wright, P.E. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: Conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. USA 1996, 93, 11504–11509. [Google Scholar] [CrossRef] [PubMed]
- Sheiner, L.; Soldati-Favre, D. Protein trafficking inside Toxoplasma gondii. Traffic 2008, 9, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Charon, N.W.; Cockburn, A.; Li, C.; Liu, J.; Miller, K.A.; Miller, M.R.; Motaleb, M.A.; Wolgemuth, C.W. The unique paradigm of spirochete motility and chemotaxis. Annu. Rev. Microbiol. 2012, 66, 349–370. [Google Scholar] [CrossRef] [PubMed]
- McBride, M.J. Cytophaga-flavobacterium gliding motility. J. Mol. Microbiol. Biotechnol. 2004, 7, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Frénal, K.; Dubremetz, J.F.; Lebrun, M.; Soldati-Favre, D. Gliding motility powers invasion and egress in Apicomplexa. Nat. Rev. Microbiol. 2017, 15, 645–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, B.; Frickel, E.M. The Toxoplasma parasitophorous vacuole: An evolving host-parasite frontier. Trends Parasitol. 2017, 33, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Ossorio, P.N.; Dubremetz, J.F.; Joiner, K.A. A soluble secretory protein of the intracellular parasite Toxoplasma gondii associates with the parasitophorous vacuole membrane through hydrophobic interactions. J. Biol. Chem. 1994, 27, 15350–15357. [Google Scholar]
- Henriquez, F.L.; Nickdel, M.B.; McLeod, R.; Lyons, R.E.; Lyons, K.; Dubremetz, J.F.; Grigg, M.E.; Samuel, B.U.; Roberts, C.W. Toxoplasma gondii dense granule protein 3 (GRA3) is a type I transmembrane protein that possesses a cytoplasmic dilysine (KKXX) endoplasmic reticulum (ER) retrieval motif. Parasitology 2005, 131, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Ruiz, J.M. Protein kinetic stability. Biophys. Chem. 2010, 148, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chebotareva, N.A.; Roman, S.G.; Kurganov, B.I. Dissociative mechanism for irreversible thermal denaturation of oligomeric proteins. Biophys. Rev. 2016, 8, 397–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaremko, M.; Jaremko, Ł.; Kim, H.Y.; Cho, M.K.; Schwieters, C.D.; Giller, K.; Becker, S.; Zweckstetter, M. Cold denaturation of a protein dimer monitored at atomic resolution. Nat. Chem. Biol. 2013, 9, 264–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheison, S.C.; Kulozik, U. Impact of the environmental conditions and substrate pre-treatment on whey protein hydrolysis: A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 418–453. [Google Scholar] [CrossRef] [PubMed]
- Runde, S.; Molière, N.; Heinz, A.; Maisonneuve, E.; Janczikowski, A.; Elsholz, A.K.; Gerth, U.; Hecker, M.; Turgay, K. The role of thiol oxidative stress response in heat-induced protein aggregate formation during thermotolerance in Bacillus subtilis. Mol. Microbiol. 2014, 91, 1036–1052. [Google Scholar] [CrossRef] [PubMed]
- Kellis, J.; Todd, R.; Arnold, F. Protein stabilization by engineered metal chelation. Nat. Biotechnol. 1991, 9, 994–995. [Google Scholar] [CrossRef]
- Loose, J.S.M.; Arntzen, M.Ø.; Bissaro, B.; Ludwig, R.; Eijsink, V.G.H.; Vaaje-Kolstad, G. Multi-point precision binding of substrate protects LPMOs from self-destructive off-pathway processes. Biochemistry 2018. [Google Scholar] [CrossRef] [PubMed]
- Thakur, V.; Morley, S.; Manor, D. Hepatic α-Tocopherol transfer protein: Ligand-induced protection from proteasomal degradation. Biochemistry 2010, 49, 9339–9344. [Google Scholar] [CrossRef] [PubMed]
- Pruteanu, M.; Neher, S.B.; Baker, T.A. Ligand-controlled proteolysis of the Escherichia coli transcriptional regulator ZntR. J. Bacteriol. 2007, 189, 3017–3025. [Google Scholar] [CrossRef] [PubMed]
- Feller, G. Protein folding at extreme temperatures: Current issues. Semin. Cell. Dev. Biol. 2017, 108, 30125–30128. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Tyedmers, J.; Mogk, A.; Bukau, B. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell. Biol. 2010, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Dimou, M.; Venieraki, A.; Katinakis, P. Microbial cyclophilins: Specialized functions in virulence and beyond. World J. Microbiol. Biotechnol. 2017, 33, 164. [Google Scholar] [CrossRef] [PubMed]
- Huber, F.M.; Hoelz, A. Molecular basis for protection of ribosomal protein L4 from cellular degradation. Nat. Commun. 2017, 8, 14354. [Google Scholar] [CrossRef] [PubMed]
- Minamino, T.; Imada, K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 2015, 23, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Chevance, F.F.V.; Hughes, K.T. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 2008, 6, 455–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, M.J.; Levenson, R.; Kim, E.A.; Sircar, R.; Blair, D.F.; Dahlquist, F.W.; Crane, B.R. Co-Folding of a FliF-FliG split domain forms the basis of the MS:C ring interface within the bacterial flagellar motor. Structure 2017, 25, 317–328. [Google Scholar] [CrossRef] [PubMed]
- McBride, M.J.; Braun, T.F. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J. Bacteriol. 2004, 186, 2295–2302. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.F.; McBride, M.J. Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J. Bacteriol. 2005, 187, 2628–2637. [Google Scholar] [CrossRef] [PubMed]
- Hunnicutt, D.W.; Kempf, M.J.; McBride, M.J. Mutations in Flavobacterium johnsoniae gldF and gldG disrupt gliding motility and interfere with membrane localization of GldA. J. Bacteriol. 2002, 184, 2370–2378. [Google Scholar] [CrossRef] [PubMed]
- McBride, M.J.; Zhu, Y. Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J. Bacteriol. 2013, 195, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, R.G.; Nelson, S.S.; Pochiraju, S.; McBride, M.J. Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF. J. Bacteriol. 2011, 193, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Heitlinger, E.; Spork, S.; Lucius, R.; Dieterich, C. The genome of Eimeria falciformis—Reduction and specialization in a single host apicomplexan parasite. BMC Genom. 2014, 15, 696. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.M.; Bannai, H.; Xuan, X.; Nishikawa, Y. Toxoplasma gondii cyclophilin 18-mediated production of nitric oxide induces Bradyzoite conversion in a CCR5-dependent manner. Infect. Immun. 2009, 77, 3686–3695. [Google Scholar] [CrossRef] [PubMed]
- Heller, G.T.; Aprile, F.A.; Vendruscolo, M. Methods of probing the interactions between small molecules and disordered proteins. Cell. Mol. Life Sci. 2017, 74, 3225–3243. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Organism (Kingdom/Family) | Number of DFPPIs Found |
|---|---|
| Bacteria (prokaryotes); total CFBP | 278 |
| Bacteroides (Flavobacteria) | 235 |
| Spirochetes | 26 |
| Proteobacteria | 7 |
| Others | 10 |
| Eukarya (all Monocellular); total FCBP | 44 |
| Apicomplexa | 13 |
| Oomycetes | 11 |
| Ciliophora | 8 |
| Diatom | 4 |
| Dinoflagellates | 2 |
| Others | 6 |
| Organism | DFPPI/Single PPI | GenBank # | Size (Amino Acids) |
|---|---|---|---|
| Toxoplasma gondii | |||
| FCBP | AAX51680.1 | 521 [16] | |
| CYN | XP_018637313.1 | 106 | |
| CYN | XP_018636397.1 | 165 | |
| CYN | XP_002369951.1 | 179 [53] | |
| CYN | XP_002365722.1 | 195 | |
| CYN | XP_002369214.1 | 211 | |
| CYN | XP_002367963.2 | 237 | |
| CYN | XP_018637703.1 | 283 | |
| CYN | XP_002365354.1 | 311 | |
| CYN | XP_002367801.1 | 348 | |
| CYN | XP_002366733.1 | 575 | |
| CYN | XP_002367918.1 | 587 | |
| CYN | XP_002366408.1 | 592 | |
| CYN | XP_002370366.1 | 612 | |
| CYN | XP_002369921.1 | 764 | |
| Flavobacterium johnsoniae | |||
| CFBP | WP_012024258.1 | 310 | |
| CFBP | WP_012024434.1 | 357 | |
| CFBP | WP_012024433.1 | 372 | |
| FKBP | WP_071634832.1 | 151 | |
| FKBP | WP_066033786.1 | 157 | |
| FKBP | WP_071638913.1 | 208 | |
| FKBP | WP_066033733.1 | 380 | |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barik, S. Dual-Family Peptidylprolyl Isomerases (Immunophilins) of Select Monocellular Organisms. Biomolecules 2018, 8, 148. https://doi.org/10.3390/biom8040148
Barik S. Dual-Family Peptidylprolyl Isomerases (Immunophilins) of Select Monocellular Organisms. Biomolecules. 2018; 8(4):148. https://doi.org/10.3390/biom8040148
Chicago/Turabian StyleBarik, Sailen. 2018. "Dual-Family Peptidylprolyl Isomerases (Immunophilins) of Select Monocellular Organisms" Biomolecules 8, no. 4: 148. https://doi.org/10.3390/biom8040148




