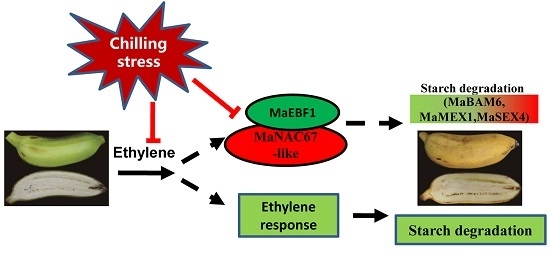
The Involvement of the Banana F-Box Protein MaEBF1 in Regulating Chilling-Inhibited Starch Degradation through Interaction with a MaNAC67-Like Protein
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Fruit Ripening and Chilling Injury Index Assessment
2.3. Starch, Fructose, Glucose, and Sucrose Content
2.4. Fruit Cell Structure Observations
2.5. Gene Expression Profiles Analysis
2.6. Construction of a cDNA Library
2.7. Y2H Assay
2.8. Bimolecular Florescence Complementation (BiFC) Assay
2.9. GST Pull-Down Assay
2.10. Subcellular Location Analysis
2.11. Dual-Luciferase Transient Expression Assays
2.12. Yeast One-Hybrid Assay
2.13. Statistical Analysis
3. Results
3.1. Change in Ripening Parameters of Fruits during Storage and Fruit Ripening
3.2. Expression Profiles of MaEBF1 and Starch Degradation-Related Genes
3.3. Protein Interactions Between MaEBF1 and MaNAC67-Like
3.4. MaNAC67-Like Acts Cooperatively with MaEBF1 to Activate Starch Degradation-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dale, J.; Paul, J.Y.; Dugdale, B.; Harding, R. Modifying Bananas: From Transgenics to Organics. Sustainability 2017, 9, 333. [Google Scholar] [CrossRef]
- Seymour, G.B.; Taylor, J.E.; Tucker, G.A. Biochemistry of Fruit Ripening. Indian J. Agric. Biochem. 1993, 18, 51–60. [Google Scholar] [CrossRef]
- Zhu, X.; Shen, L.; Fu, D.; Si, Z.; Wu, B.; Chen, W.; Li, X. Effects of the combination treatment of 1-MCP and ethylene on the ripening of harvested banana fruit. Postharvest Biol. Technol. 2015, 107, 23–32. [Google Scholar] [CrossRef]
- Kudachikar, V.B.; Kulkarni, S.G.; Prakash, M.N. Effect of modified atmosphere packaging on quality and shelf life of ’Robusta’ banana (Musa sp.) stored at low temperature. J. Food Sci. Technol. 2011, 48, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Pinto, P.M.; Spricigo, P.C.; da Silva, S.R.; Sargent, S.A.; Jacomino, A.P. Effect of 1-MCP and low-temperature storage on postharvest conservation of camu–camu. Acta Physiol. Plant. 2018, 40. [Google Scholar] [CrossRef]
- Liu, T.; Li, L.; Li, B.; Zhan, G.; Wang, Y. Evaluation of Low-Temperature Phosphine Fumigation for Control of Oriental Fruit Fly in Loquat Fruit. J. Econ. Entomol. 2018, 111, 1165–1170. [Google Scholar] [CrossRef]
- Ali, Z.M.; Chin, L.-H.; Marimuthu, M.; Lazan, H. Low temperature storage and modified atmosphere packaging of carambola fruit and their effects on ripening related texture changes, wall modification and chilling injury symptoms. Postharvest Biol. Technol. 2004, 33, 181–192. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, W.; Jiang, Y.; Luo, Y.; Jiang, W.; Joyce, D. Expression of ethylene-related expansin genes in cool-stored ripening banana fruit. Plant Sci. 2006, 170, 962–967. [Google Scholar] [CrossRef]
- Zhu, X.; Luo, J.; Li, Q.; Li, J.; Liu, T.; Wang, R.; Chen, W.; Li, X. Low temperature storage reduces aroma-related volatiles production during shelf-life of banana fruit mainly by regulating key genes involved in volatile biosynthetic pathways. Postharvest Biol. Technol. 2018, 146, 68–78. [Google Scholar] [CrossRef]
- Peroni-Okita, F.H.; Cardoso, M.B.; Agopian, R.G.; Louro, R.P.; Nascimento, J.R.; Purgatto, E.; Tavares, M.I.; Lajolo, F.M.; Cordenunsi, B.R. The cold storage of green bananas affects the starch degradation during ripening at higher temperature. Carbohydr. Polym. 2013, 96, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Onik, J.C.; Hu, X.; Duan, Y.; Lin, Q. Effects of (S)-Carvone and Gibberellin on Sugar Accumulation in Potatoes during Low Temperature Storage. Molecules 2018, 23, 3118. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.H.; Tian-Lai, L.I.; Meng, S.D.; Zhao, B.; Sun, L.P. Effects of Night Low Temperature on Sugar Accumulation and Sugar-Metabolizing Enzyme Activities in Melon Fruit. Sci. Agric. Sin. 2009, 42, 3592–3599. [Google Scholar]
- Jiang, Y.; Joyce, D.C.; Jiang, W.; Lu, W. Effects of Chilling Temperatures on Ethylene Binding by Banana Fruit. Plant Growth Regul. 2004, 43, 109–115. [Google Scholar] [CrossRef]
- Benavente, L.M.; Alonso, J.M. Molecular mechanisms of ethylene signaling in Arabidopsis. Mol. BioSyst. 2006, 2, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ecker, J.R. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 2003, 115, 667–677. [Google Scholar] [CrossRef]
- Potuschak, T.; Lechner, E.; Parmentier, Y.; Yanagisawa, S.; Grava, S.; Koncz, C.; Genschik, P. EIN3-Dependent Regulation of Plant Ethylene Hormone Signaling by Two Arabidopsis F Box Proteins: EBF1 and EBF2. Cell 2003, 115, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Zhang, Y.; Tu, Y.; Wang, Y.; Cheng, W.; Yang, Y. Overexpression of an EIN3-binding F-box protein2-like gene caused elongated fruit shape and delayed fruit development and ripening in tomato. Plant Sci. 2018, 272, 131–141. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, Y.; Pirrello, J.; Regad, F.; Bouzayen, M.; Deng, W.; Li, Z. Silencing Sl-EBF1 and Sl-EBF2 expression causes constitutive ethylene response phenotype, accelerated plant senescence, and fruit ripening in tomato. J. Exp. Bot. 2010, 61, 697–708. [Google Scholar] [CrossRef]
- Tacken, E.J.; Ireland, H.S.; Wang, Y.; Putterill, J.; Schaffer, R.J. Apple EIN3 BINDING F-box 1 inhibits the activity of three apple EIN3-like transcription factors. AoB Plants 2012, 2012, pls034. [Google Scholar] [CrossRef]
- Ding, X.; Zhu, X.; Ye, L.; Xiao, S.; Wu, Z.; Chen, W.; Li, X. The interaction of CpEBF1 with CpMADSs is involved in cell wall degradation during papaya fruit ripening. Hortic. Res. 2019, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Wang, W.; Yang, T.; Li, M.; Grierson, D. Transcriptome analysis identifies a zinc finger protein regulating 9 starch degradation in kiwifruit. Plant Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Wu, K.; Chen, J.; Liu, Q.; Wu, Y.; Liu, B.; Fu, X. OsSND2, a NAC family transcription factor, is involved in secondary cell wall biosynthesis through regulating MYBs expression in rice. Rice 2018, 11, 36. [Google Scholar] [CrossRef]
- Christianson, J.A.; Dennis, E.S.; Llewellyn, D.J.; Wilson, I.W. ATAF NAC transcription factors: Regulators of plant stress signaling. Plant Signal. Behav. 2010, 5, 428–432. [Google Scholar] [CrossRef] [Green Version]
- Shan, W.; Kuang, J.; Chen, L.; Xie, H.; Peng, H.; Xiao, Y.; Li, X.; Chen, W.; He, Q.; Chen, J. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 2012, 63, 5171–5187. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Wang, W.; Zeng, J.; Zhang, J.; Grierson, D.; Li, X.; Yin, X.; Chen, K. A NAC transcription factor, EjNAC1, affects lignification of loquat fruit by regulating lignin. Postharvest Biol. Technol. 2015, 102, 25–31. [Google Scholar] [CrossRef]
- Fu, C.; Han, Y.; Fan, Z.; Chen, J.; Chen, W.; Lu, W.; Kuang, J. The Papaya Transcription Factor CpNAC1 Modulates Carotenoid Biosynthesis through Activating Phytoene Desaturase Genes CpPDS2/4 during Fruit Ripening. J. Agric. Food Chem. 2016, 64, 5454–5463. [Google Scholar] [CrossRef]
- Kou, X.; Liu, C.; Han, L.; Wang, S.; Xue, Z. NAC transcription factors play an important role in ethylene biosynthesis, reception and signaling of tomato fruit ripening. Mol. Genet. Genom. 2016, 291, 1205–1217. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, G.; Zhou, S.; Tu, Y.; Wang, Y.; Dong, T.; Hu, Z. A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 2014, 55, 119–135. [Google Scholar] [CrossRef]
- Kumar, R.; Tamboli, V.; Sharma, R.; Sreelakshmi, Y. NAC-NOR mutations in tomato Penjar accessions attenuate multiple metabolic processes and prolong the fruit shelf life. Food Chem. 2018, 259, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zuo, J.; Hou, X.; Yan, Y.; Wei, Y.; Liu, J.; Li, M.; Xu, B.; Jin, Z. The auxin response factor gene family in banana: Genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front Plant Sci. 2015, 6, 742. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Long, X.; Wang, W.; Huang, Y.; Huang, W.; Yao, J. Preliminary report on introduction experiment of nine banana cultivars in Guangxi. Guangdong Agric. Sci. 2013, 10, 30–32. [Google Scholar]
- Shiga, T.M.; Soares, C.A.; Nascimento, J.R.; Purgatto, E.; Lajolo, F.M.; Cordenunsi, B.R. Ripening-associated changes in the amounts of starch and non-starch polysaccharides and their contributions to fruit softening in three banana cultivars. J. Sci. Food Agric. 2011, 91, 1511–1516. [Google Scholar] [CrossRef]
- Kondo, S.; Kittikorn, M.; Kanlayanarat, S. Preharvest antioxidant activities of tropical fruit and the effect of low temperature storage on antioxidants and jasmonates. Postharvest Biol. Technol. 2005, 36, 309–318. [Google Scholar] [CrossRef]
- Xiao, Y.; Kuang, J.; Qi, X.; Ye, Y.; Wu, Z.; Chen, J.; Lu, W. A comprehensive investigation of starch degradation process and identification of a transcriptional activator MabHLH6 during banana fruit ripening. Plant Biotechnol. J. 2018, 16, 151–164. [Google Scholar] [CrossRef]
- Suo, J.; Li, H.; Ban, Q.; Han, Y.; Meng, K.; Jin, M.; Zhang, Z.; Rao, J. Characteristics of chilling injury-induced lignification in kiwifruit with different sensitivities to low temperatures. Postharvest Biol. Technol. 2018, 135, 8–18. [Google Scholar] [CrossRef]
- Song, Z.; Li, F.; Guan, H.; Xu, Y.; Fu, Q.; Li, D. Combination of nisin and ε-polylysine with chitosan coating inhibits the white blush of fresh-cut carrots. Food Control 2017, 74, 34–44. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, L.; Zhong, H.Y.; Kuang, J.F.; Li, J.G.; Lu, W.J.; Chen, J.Y. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 2011, 234, 377–390. [Google Scholar] [CrossRef]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef] [PubMed]
- Nardozza, S.; Boldingh, H.L.; Osorio, S.; Hohne, M.; Wohlers, M.; Gleave, A.P.; MacRae, E.A.; Richardson, A.C.; Atkinson, R.G.; Sulpice, R.; et al. Metabolic analysis of kiwifruit (Actinidia deliciosa) berries from extreme genotypes reveals hallmarks for fruit starch metabolism. J. Exp. Bot. 2013, 64, 5049–5063. [Google Scholar] [CrossRef] [PubMed]
- Cordenunsi-Lysenko, B.R.; Nascimento, J.R.O.; Castro-Alves, V.C.; Purgatto, E.; Fabi, J.P.; Peroni-Okyta, F.H.G. The Starch Is (Not) Just Another Brick in the Wall: The Primary Metabolism of Sugars During Banana Ripening. Front Plant Sci. 2019, 10, 391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saraiva, L.D.A.; Florence Polegato, C.; Renata, S.; Hassimotto, N.M.A.; Eduardo, P.; Marc, C.; Beatriz Rosana, C. Black leaf streak disease affects starch metabolism in banana fruit. J. Agric. Food Chem. 2013, 61, 5582–5589. [Google Scholar] [CrossRef]
- Wang, J.; Tang, X.; Chen, P.; Huang, H. Changes in resistant starch from two banana cultivars during postharvest storage. Food Chem. 2014, 156, 319–325. [Google Scholar] [CrossRef]
- Kojima, K.; Sakurai, N.; Kuraishi, S. Fruit softening in banana: Correlation among stress-relaxation parameters, cell wall components and starch during ripening. Physiol. Plant. 2010, 90, 772–778. [Google Scholar] [CrossRef]
- Hu, X.; Kuang, S.; Zhang, A.; Zhang, W.; Chen, M.; Yin, X.; Chen, K. Characterization of Starch Degradation Related Genes in Postharvest Kiwifruit. Int. J. Mol. Sci. 2016, 17, 2112. [Google Scholar] [CrossRef]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.; Ba, L.; Shan, W.; Xiao, Y.; Lu, W.; Kuang, J.; Chen, J. A banana R2R3-MYB transcription factor MaMYB3 is involved in fruit ripening through modulation of starch degradation by repressing starch degradation-related genes and MabHLH6. Plant J. 2018, 96, 1191–1205. [Google Scholar] [CrossRef]
- Giovannoni, J.J.; Nguyen, C.; Ampofo, B.; Zhong, S.; Fei, Z. Epigenome and Transcriptional Dynamics of Fruit Ripening. Annu. Rev. Plant Biol. 2017, 68. [Google Scholar] [CrossRef]
- Ma, N.; Feng, H.; Meng, X.; Li, D.; Yang, D.; Wu, C.; Meng, Q. Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 2014, 14, 351. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Pirrello, J.; Chen, Y.; Li, N.; Zhu, S.; Chirinos, X.; Bouzayen, M.; Liu, Y.; Liu, M. A novel tomato F-box protein, SlEBF3, is involved in tuning ethylene signaling during plant development and climacteric fruit ripening. Plant J. 2018, 95, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Kuang, J.; Chen, L.; Shan, W.; Yang, S.; Lu, W.; Chen, J. Molecular characterization of two banana ethylene signaling component MaEBFs during fruit ripening. Postharvest Biol. Technol. 2013, 85, 94–101. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Z.; Qin, J.; Zheng, Q.; Ding, X.; Chen, W.; Lu, W.; Li, X.; Zhu, X. The Involvement of the Banana F-Box Protein MaEBF1 in Regulating Chilling-Inhibited Starch Degradation through Interaction with a MaNAC67-Like Protein. Biomolecules 2019, 9, 552. https://doi.org/10.3390/biom9100552
Song Z, Qin J, Zheng Q, Ding X, Chen W, Lu W, Li X, Zhu X. The Involvement of the Banana F-Box Protein MaEBF1 in Regulating Chilling-Inhibited Starch Degradation through Interaction with a MaNAC67-Like Protein. Biomolecules. 2019; 9(10):552. https://doi.org/10.3390/biom9100552
Chicago/Turabian StyleSong, Zunyang, Jiajia Qin, Qiuli Zheng, Xiaochun Ding, Weixin Chen, Wangjin Lu, Xueping Li, and Xiaoyang Zhu. 2019. "The Involvement of the Banana F-Box Protein MaEBF1 in Regulating Chilling-Inhibited Starch Degradation through Interaction with a MaNAC67-Like Protein" Biomolecules 9, no. 10: 552. https://doi.org/10.3390/biom9100552
APA StyleSong, Z., Qin, J., Zheng, Q., Ding, X., Chen, W., Lu, W., Li, X., & Zhu, X. (2019). The Involvement of the Banana F-Box Protein MaEBF1 in Regulating Chilling-Inhibited Starch Degradation through Interaction with a MaNAC67-Like Protein. Biomolecules, 9(10), 552. https://doi.org/10.3390/biom9100552