The Novel Serine/Threonine Protein Kinase LmjF.22.0810 from Leishmania major May Be Involved in the Resistance to Drugs such as Paromomycin
Abstract
1. Introduction
2. Materials and Methods
2.1. Database Inspection and Nucleotide and Protein Sequence Alignment
2.2. Evaluation of Secondary Structure, Conserved Domains, and Post-Translational Modifications
2.3. Phylogenetic Analysis
2.4. Screening for Homology Modeling Templates
2.5. Molecular Modeling and 3D-Structure Validation
2.6. MD Simulation
2.7. Protein Preparation, Binding Site Identification, and High-Throughput in Silico Docking
2.8. Parasite Culture Conditions
2.9. Genetic Manipulation of L. major
2.10. Fluorescence Microscopy
2.11. Metacyclic Forms Isolation
2.12. In Vitro Infections
2.13. RNA Expression Quantification
2.14. Cell Cycle Analysis by Propidium Iodide Staining and Flow Cytometry
2.15. Cytotoxicity Evaluation
2.16. Statistical Analysis
3. Results
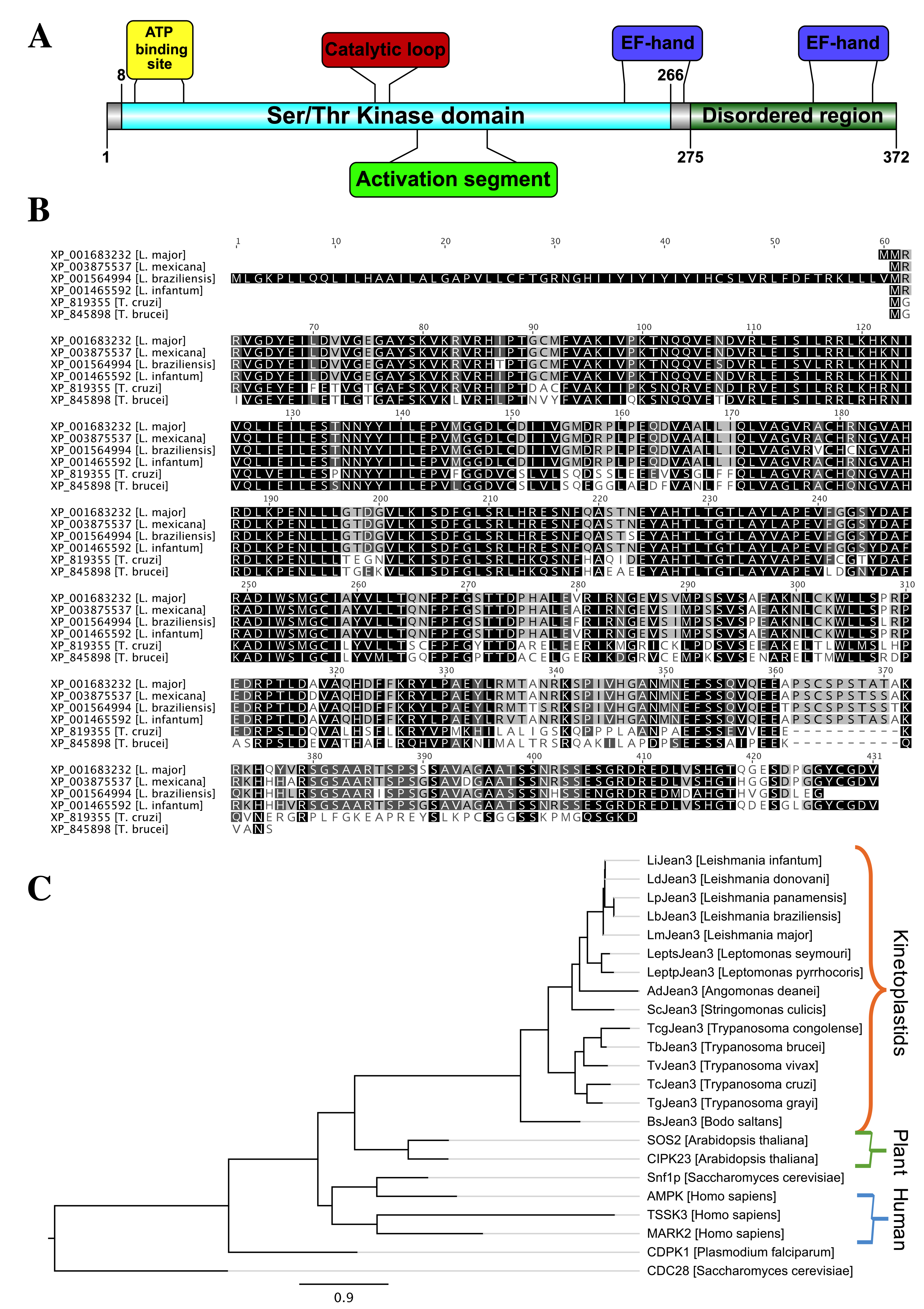
3.1. LmjF.22.0810 (LmJean3), a Novel Predicted Trypanosomatid Protein Kinase
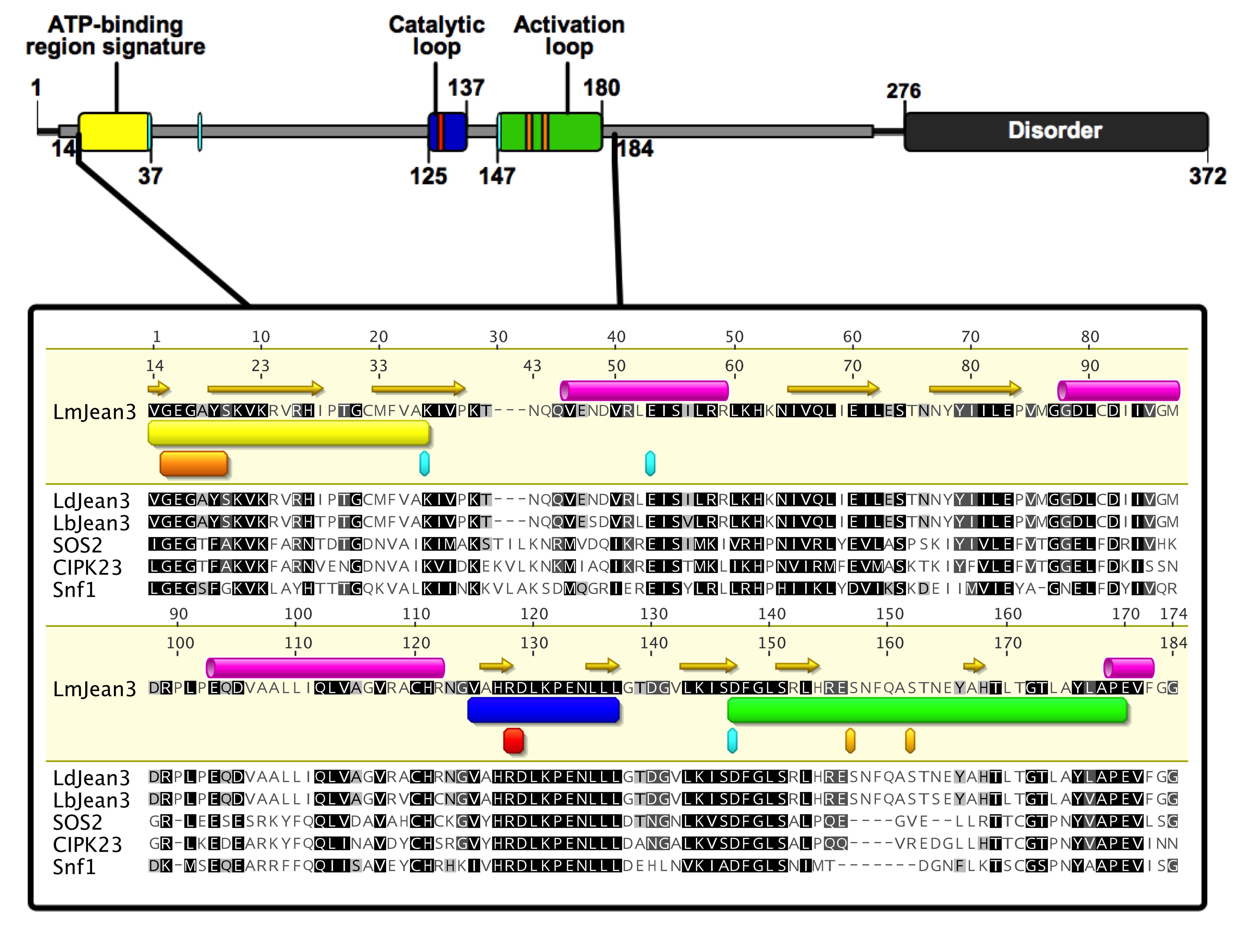
3.2. LmJean3 (LmjF.22.0810) Domains and Motifs
3.3. The Phylogeny of LmjF.22.0810 Homologues
3.4. LmJean3 was Localized in the Cytoplasm, Nucleus, and Flagellum of Leishmania Promastigotes and Was Significantly Expressed in the Infective and Amastigote Forms
3.5. The Kinase Domain of LmJean3 Is Conserved and Exhibits a High Similarity to CIPK24/SOS2, CIPK23, and Snf1 Kinase Domains
3.6. Homology Modeling, Refinement, and Validation of LmJean3 Structure
3.7. Overall Description of the Predicted Structure of LmJean3 Catalytic Domain
3.8. Molecular Dynamics Simulation
3.9. LmJean3 Binding Sites: Prediction and Analysis
3.10. Aminoglycosides Are Predicted Ligands of LmJean3.
3.11. Predicted Interactions of Paromomycin with Site B of LmJean3
3.12. Docking in Site A and Cross-Docking
3.13. Generation of LmJean3-Overexpressing Parasites (LmJ3OE)
3.14. LmJ3OE Parasites Exhibited Less Sensitivity to Paromomycin and Other Aminoglycosides
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wiwanitkit, V. Interest in paromomycin for the treatment of visceral leishmaniasis (Kala-azar). Ther. Clin. Risk Manag. 2012, 8, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Daigle, D.M.; McKay, G.A.; Thompson, P.R.; Wright, G.D. Aminoglycoside antibiotic phosphotransferases are also serine protein kinases. Chem. Biol. 1999, 6, 11–18. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside modifying enzymes. Drug Resist. Updates 2010, 13, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Stogios, P.J.; Shakya, T.; Evdokimova, E.; Savchenko, A.; Wright, G.D. Structure and function of APH(4)-Ia, a hygromycin B resistance enzyme. J. Biol. Chem. 2011, 286, 42786. [Google Scholar] [CrossRef]
- Sundar, S. Drug resistance in Indian visceral leishmaniasis. Trop. Med. Int. Health 2001, 6, 849–854. [Google Scholar] [CrossRef]
- Fairlamb, A.H.; Gow, N.A.R.; Matthews, K.R.; Waters, A.P. Drug resistance in eukaryotic microorganisms. Nat. Microbiol. 2016, 1, 16092. [Google Scholar] [CrossRef]
- Chemother, J.A. Drug resistance in. Commun. Dis. Rep. Cdr. Rev. 2000, 19, 149–150. [Google Scholar]
- Clayton, C.E. Life without transcriptional control? From fly to man and back again. EMBO J. 2002, 21, 1881–1888. [Google Scholar] [CrossRef]
- Wiese, M. A mitogen-activated protein (MAP) kinase homologue of Leishmania mexicana is essential for parasite survival in the infected host. EMBO J. 1998, 17, 2619–2628. [Google Scholar] [CrossRef]
- Nett, I.R.E.; Martin, D.M.A.; Miranda-Saavedra, D.; Lamont, D.; Barber, J.D.; Mehlert, A.; Ferguson, M.A.J. The phosphoproteome of bloodstream form Trypanosoma brucei, causative agent of African sleeping sickness. Mol. Cell. Proteom. 2009, 8, 1527–1538. [Google Scholar] [CrossRef]
- Nakayasu, E.S.; Gaynor, M.R.; Sobreira, T.J.P.; Ross, J.A.; Almeida, I.C. Phosphoproteomic analysis of the human pathogen Trypanosoma cruzi at the epimastigote stage. Proteomics 2009, 9, 3489–3506. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, R.M.L.; Charneau, S.; Mandacaru, S.C.; Schwämmle, V.; Lima, B.D.; Roepstorff, P.; Ricart, C.A.O. Quantitative proteomic and phosphoproteomic analysis of Trypanosoma cruzi amastigogenesis. Mol. Cell. Proteom. 2014, 13, 3457–3472. [Google Scholar] [CrossRef] [PubMed]
- Tsigankov, P.; Gherardini, P.F.; Helmer-Citterich, M.; Späth, G.F.; Zilberstein, D. Phosphoproteomic analysis of differentiating Leishmania parasites reveals a unique stage-specific phosphorylation motif. J. Proteome Res. 2013, 12, 3405–3412. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Nielsen, T.E.; Clausen, M.H. Small-molecule kinase inhibitors: An analysis of FDA-approved drugs. Drug Discov. Today 2016, 21, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Diaz, R.; Nguewa, P.A.; Redrado, M.; Manrique, I.; Calvo, A. Sunitinib reduces tumor hypoxia and angiogenesis, and radiosensitizes prostate cancer stem-like cells. Prostate 2015, 75, 1137–1149. [Google Scholar] [CrossRef]
- Ibanez, E.; Agliano, A.; Prior, C.; Nguewa, P.; Redrado, M.; Gonzalez-Zubeldia, I.; Plano, D.A.; Palop, J.; Sanmartin, C.; Calvo, A. The Quinoline Imidoselenocarbamate EI201 Blocks the AKT/mTOR Pathway and Targets Cancer Stem Cells Leading to a Strong Antitumor Activity. Curr. Med. Chem. 2012, 19, 3031–3043. [Google Scholar] [CrossRef]
- Nguewa, P.A.; Calvo, A.; Pullamsetti, S.S.; Banat, G.A.; Grimminger, F.; Savai, R. Tyrosine kinase inhibitors with antiangiogenic properties for the treatment of non-small cell lung cancer. Expert Opin. Investig. Drugs 2011, 20, 61–74. [Google Scholar] [CrossRef]
- Breen, M.E.; Soellner, M.B. Small molecule substrate phosphorylation site inhibitors of protein kinases: Approaches and challenges. ACS Chem. Biol. 2015, 10, 175–189. [Google Scholar] [CrossRef]
- Crowther, G.J.; Shanmugam, D.; Carmona, S.J.; Doyle, M.A.; Hertz-Fowler, C.; Berriman, M.; Nwaka, S.; Ralph, S.A.; Roos, D.S.; van Voorhis, W.C.; et al. Identification of attractive drug targets in neglected- disease pathogens using an in Silico approach. PLoS Negl. Trop. Dis. 2010, 4, e804. [Google Scholar] [CrossRef]
- Naula, C.; Parsons, M.; Mottram, J.C. Protein kinases as drug targets in trypanosomes and Leishmania. Biochim. Biophys. Acta-Proteins Proteom. 2005, 1754, 151–159. [Google Scholar] [CrossRef]
- Merritt, C.; Silva, L.E.; Tanner, A.L.; Stuart, K.; Pollastri, M.P. Kinases as druggable targets in trypanosomatid protozoan parasites. Chem. Rev. 2014, 114, 11280–11304. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.; Worthey, E.A.; Ward, P.N.; Mottram, J.C. Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genom. 2005, 6, 127. [Google Scholar] [CrossRef] [PubMed]
- Klug, D.M.; Gelb, M.H.; Pollastri, M.P. Repurposing strategies for tropical disease drug discovery. Bioorganic Med. Chem. Lett. 2016, 26, 2569–2576. [Google Scholar] [CrossRef] [PubMed]
- Kufareva, I.; Abagyan, R. Type-II kinase inhibitor docking, screening, and profiling using modified structures of active kinase states. J. Med. Chem. 2008, 51, 7921–7932. [Google Scholar] [CrossRef] [PubMed]
- Danilenko, V.N.; Simonov, A.Y.; Lakatosh, S.A.; Kubbutat, M.H.G.; Totzke, F.; Schächtele, C.; Elizarov, S.M.; Bekker, O.B.; Printsevskaya, S.S.; Luzikov, Y.N.; et al. Search for inhibitors of bacterial and human protein kinases among derivatives of diazepines[1,4] annelated with maleimide and indole cycles. J. Med. Chem. 2008, 51, 7731–7736. [Google Scholar] [CrossRef] [PubMed]
- Diaz, R.; Nguewa, P.A.; Parrondo, R.; Perez-Stable, C.; Manrique, I.; Redrado, M.; Catena, R.; Collantes, M.; Peñuelas, I.; Díaz-González, J.A.; et al. Antitumor and antiangiogenic effect of the dual EGFR and HER-2 tyrosine kinase inhibitor lapatinib in a lung cancer model. BMC Cancer 2010, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Diaz, R.; Nguewa, P.A.; Diaz-Gonzalez, J.A.; Hamel, E.; Gonzalez-Moreno, O.; Catena, R.; Serrano, D.; Redrado, M.; Sherris, D.; Calvo, A. The novel Akt inhibitor Palomid 529 (P529) enhances the effect of radiotherapy in prostate cancer. Br. J. Cancer 2009, 100, 932–940. [Google Scholar] [CrossRef]
- Flórez, A.F.; Park, D.; Bhak, J.; Kim, B.C.; Kuchinsky, A.; Morris, J.H.; Espinosa, J.; Muskus, C. Protein network prediction and topological analysis in Leishmania major as a tool for drug target selection. BMC Bioinform. 2010, 11, 484. [Google Scholar] [CrossRef]
- Fang, Z.; Grütter, C.; Rauh, D. Strategies for the selective regulation of kinases with allosteric modulators: Exploiting exclusive structural features. ACS Chem. Biol. 2013, 8, 58–70. [Google Scholar] [CrossRef]
- Roskoski, R. Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol. Res. 2016, 103, 26–48. [Google Scholar] [CrossRef]
- Müller, S.; Chaikuad, A.; Gray, N.S.; Knapp, S. The ins and outs of selective kinase inhibitor development. Nat. Chem. Biol. 2015, 11, 818–821. [Google Scholar] [CrossRef]
- Lamba, V.; Ghosh, I. New Directions in Targeting Protein Kinases: Focusing Upon True Allosteric and Bivalent Inhibitors. Curr. Pharm. Des. 2012, 18, 2936–2945. [Google Scholar] [CrossRef]
- Docampo, R.; Huang, G. Calcium signaling in trypanosomatid parasites. Cell Calcium 2015, 57, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Aslett, M.; Aurrecoechea, C.; Berriman, M.; Brestelli, J.; Brunk, B.P.; Carrington, M.; Depledge, D.P.; Fischer, S.; Gajria, B.; Gao, X.; et al. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2009, 38, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, L.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; Deweese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; et al. CDD: Specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009, 37, D205–D210. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef]
- Yachdav, G.; Kloppmann, E.; Kajan, L.; Hecht, M.; Goldberg, T.; Hamp, T.; Hönigschmid, P.; Schafferhans, A.; Roos, M.; Bernhofer, M.; et al. PredictProtein—An open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014, 42, W337–W343. [Google Scholar] [CrossRef]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C.; Bateman, A.; Bork, P.; Bridge, A.J.; Chang, H.Y.; Dosztanyi, Z.; El-Gebali, S.; Fraser, M.; et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, W.; Kirberger, M.; Lee, H.W.; Ayalasomayajula, G.; Yang, J.J. Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand proteins. Proteins Struct. Funct. Genet. 2006, 65, 643–655. [Google Scholar] [CrossRef]
- Pierleoni, A.; Martelli, P.L.; Casadio, R. PredGPI: A GPI-anchor predictor. BMC Bioinform. 2008, 9, 392. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999, 294, 1351–1362. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Harrington, B.; Engelen, J. Inkscape. Available online: http://www.inkscape.Org (accessed on 20 Ocotrber 2009).
- Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Chenna, R.; Sugawara, H.; Koike, T.; Lopez, R.; Gibson, T.J.; Higgins, D.G.; Thompson, J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The protein data bank. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Chaves-Sanjuan, A.; Sanchez-Barrena, M.J.; Gonzalez-Rubio, J.M.; Moreno, M.; Ragel, P.; Jimenez, M.; Pardo, J.M.; Martinez-Ripoll, M.; Quintero, F.J.; Albert, A. Structural basis of the regulatory mechanism of the plant CIPK family of protein kinases controlling ion homeostasis and abiotic stress. Proc. Natl. Acad. Sci. USA 2014, 111, E4532–E4541. [Google Scholar] [CrossRef]
- Vinet, L.; Zhedanov, A. A “missing” family of classical orthogonal polynomials. J. Phys. A Math. Theor. 2011, 44, 085201. [Google Scholar] [CrossRef]
- Krieger, E.; Joo, K.; Lee, J.; Lee, J.; Raman, S.; Thompson, J.; Tyka, M.; Baker, D.; Karplus, K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins Struct. Funct. Bioinform. 2009, 77, 114–122. [Google Scholar] [CrossRef]
- Eisenberg, D.; Lüthy, R.; Bowie, J.U. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997, 277, 396–404. [Google Scholar]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Benkert, P.; Künzli, M.; Schwede, T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009, 37, W510–W514. [Google Scholar] [CrossRef]
- Benkert, P.; Tosatto, S.C.E.; Schomburg, D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins Struct. Funct. Genet. 2008, 71, 261–277. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Schwede, T.; Tosatto, S.C. QMEANclust: Estimation of protein model quality by combining a composite scoring function with structural density information. BMC Struct. Biol. 2009, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Melo, F.; Devos, D.; Depiereux, E.; Feytmans, E. ANOLEA: A www server to assess protein structures. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997, 5, 187–190. [Google Scholar] [PubMed]
- Dong, Q.W.; Wang, X.L.; Lin, L. Novel knowledge-based mean force potential at the profile level. BMC Bioinform. 2006, 7, 207–222. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Melo, F.; Feytmans, E. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 1998, 277, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- van Gunsteren, W.F.; Billeter, S.R.; Eising, A.A.; Hünenberger, P.H.; Krüger, P.; Mark, A.E.; Scott, W.R.P.; Tironi, I.G. Biomolecular Simulation: The {GROMOS96} Manual and User Guide; Vdf Hochschulverlag AG an der ETH Zürich: Zürich, Switzerland, 1996. [Google Scholar]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen--bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Hutchinson, E.G.; Thornton, J.M. PROMOTIF-A program to identify and analyze structural motifs in proteins. Protein Sci. 2008, 5, 212–220. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Sippl, M.J. Recognition of errors in three--dimensional structures of proteins. Proteins Struct. Funct. Bioinform. 1993, 17, 355–362. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces; Springer, Dordrecht: Dordrecht, The Netherlands, 1981; pp. 331–342. [Google Scholar]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 14101. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Rostkowski, M.; Olsson, M.H.; Søndergaard, C.R.; Jensen, J.H. Graphical analysis of pH-dependent properties of proteins predicted using PROPKA. BMC Struct. Biol. 2011, 11, 6. [Google Scholar] [CrossRef]
- Halgren, T. New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 2007, 69, 146–148. [Google Scholar] [CrossRef]
- Halgren, T.A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009, 49, 377–389. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. ZINC—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pKa prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, J.R.; Calkins, D.; Sullivan, A.P.; Shelley, J.C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput. Aided Mol. Des. 2010, 24, 591–604. [Google Scholar] [CrossRef]
- Vacas, A.; Sugden, C.; Velasco-Rodriguez, Ó.; Algarabel-Olona, M.; Peña-Guerrero, J.; Larrea, E.; Fernández-Rubio, C.; Nguewa, P.A. Construction of Two mCherry Plasmids (pXG-mCherry) for Transgenic Leishmania: Valuable Tools for Future Molecular Analysis. J. Parasitol. Res. 2017, 2017, 1964531. [Google Scholar] [CrossRef]
- Medina-Acosta, E.; Cross, G.A.M. Rapid isolation of DNA from trypanosomatid protozoa using a simple “mini-prep” procedure. Mol. Biochem. Parasitol. 1993, 59, 327–329. [Google Scholar] [CrossRef]
- Cruz, A.; Coburn, C.M.; Beverley, S.M. Double targeted gene replacement for creating null mutants. Proc. Natl. Acad. Sci. USA 1991, 88, 7170–7174. [Google Scholar] [CrossRef]
- Sacks, D.L.; Hieny, S.; Sher, A. Identification of cell surface carbohydrate and antigenic changes between noninfective and infective developmental stages of Leishmania major promastigotes. J. Immunol. 1985, 135, 564–569. [Google Scholar]
- Fernández-Rubio, C.; Campbell, D.; Vacas, A.; Ibañez, E.; Moreno, E.; Espuelas, S.; Calvo, A.; Palop, J.A.; Plano, D.; Sanmartin, C.; et al. Leishmanicidal activities of novel methylseleno-imidocarbamates. Antimicrob. Agents Chemother. 2015, 59, 5705–5713. [Google Scholar] [CrossRef][Green Version]
- Dame, N. GraphPad Prism, Version 7.0, Software for Technical Computation; GraphPad Software, Inc.: San Diego, CA, USA, 2015. [Google Scholar]
- Wang, J.; Pan, C.; Wang, Y.; Ye, L.; Wu, J.; Chen, L.; Zou, T.; Lu, G. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genomics 2015, 16, 386. [Google Scholar] [CrossRef] [PubMed]
- Nolen, B.; Taylor, S.; Ghosh, G. Regulation of protein kinases: Controlling activity through activation segment conformation. Mol. Cell 2004, 15, 661–675. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S.; Henikoff, J.G. Amino acid substitution matrices. Adv. Protein Chem. 2000, 54, 73–97. [Google Scholar] [PubMed]
- Bansal, A.; Singh, S.; More, K.R.; Hans, D.; Nangalia, K.; Yogavel, M.; Sharma, A.; Chitnis, C.E. Erratum: Characterization of plasmodium falciparum calcium-dependent protein kinase 1 (PfCDPK1) and its role in microneme secretion during erythrocyte invasion. J. Biol. Chem. 2013, 288, 19643. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, J.C.; Hou, L.; Cao, P.R.; Wu, L.; Zhang, Q.S.; Yang, H.Y.; Zang, Y.; Ding, J.P.; Li, J. Functional role of histidine in the conserved His-x-Asp motif in the catalytic core of protein kinases. Sci. Rep. 2015, 5, 10115. [Google Scholar] [CrossRef]
- Taylor, S.S.; Keshwani, M.M.; Steichen, J.M.; Kornev, A.P. Evolution of the eukaryotic protein kinases as dynamic molecular switches. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 2517–2528. [Google Scholar] [CrossRef]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. DOG 1.0: Illustrator of protein domain structures. Cell Res. 2009, 19, 271–273. [Google Scholar] [CrossRef]
- Negi, A.; Bhushan, S.; Gupta, P.; Garg, P.; Kumar, R. Cystathionine β -Lyase-Like Protein with Pyridoxal Binding Domain Characterized in Leishmania major by Comparative Sequence Analysis and Homology Modelling. ISRN Comput. Biol. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Taylor, S.S.; Kornev, A.P. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011, 36, 65–77. [Google Scholar] [CrossRef]
- Fabbro, D.; Cowan-Jacob, S.W.; Moebitz, H. Ten things you should know about protein kinases: IUPHAR Review 14. Br. J. Pharmacol. 2015, 172, 2675–2700. [Google Scholar] [CrossRef]
- Xu, M.; Yu, L.; Wan, B.; Yu, L.; Huang, Q. Predicting inactive conformations of protein kinases using active structures: Conformational selection of Type-II inhibitors. PLoS ONE 2011, 6, e22644. [Google Scholar] [CrossRef] [PubMed]
- Gräter, F.; Shen, J.; Jiang, H.; Gautel, M.; Grubmüller, H. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys. J. 2005, 88, 790–804. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol. Res. 2015, 100, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol. Res. 2015, 94, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Oruganty, K.; Talathi, N.S.; Wood, Z.A.; Kannan, N. Identification of a hidden strain switch provides clues to an ancient structural mechanism in protein kinases. Proc. Natl. Acad. Sci. USA 2013, 110, 924–929. [Google Scholar] [CrossRef] [PubMed]
- Shalev, M.; Kondo, J.; Kopelyanskiy, D.; Jaffe, C.L.; Adir, N.; Baasov, T. Identification of the molecular attributes required for aminoglycoside activity against Leishmania. Proc. Natl. Acad. Sci. USA 2013, 110, 13333–13338. [Google Scholar] [CrossRef]
- Pfister, P.; Hobbie, S.; Brüll, C.; Corti, N.; Vasella, A.; Westhof, E.; Böttger, E.C. Mutagenesis of 16S rRNA C1409-G1491 base-pair differentiates between 6′OH and 6′NH3+ aminoglycosides. J. Mol. Biol. 2005, 346, 467–475. [Google Scholar] [CrossRef]
- Davidson, R.N.; den Boer, M.; Ritmeijer, K. Paromomycin. Trans. R. Soc. Trop. Med. Hyg. 2009, 103, 653–660. [Google Scholar] [CrossRef]
- El-On, J.; Jacobs, G.P.; Witztum, E.; Greenblatt, C.L. Development of topical treatment for cutaneous leishmaniasis caused by Leishmania major in experimental animals. Antimicrob. Agents Chemother. 1984, 26, 745–751. [Google Scholar] [CrossRef]
- Oruganty, K.; Talevich, E.E.; Neuwald, A.F.; Kannan, N. Identification and classification of small molecule kinases: Insights into substrate recognition and specificity Genome evolution and evolutionary systems biology. BMC Evol. Biol. 2016, 16, 7. [Google Scholar] [CrossRef]
- Kannan, N.; Neuwald, A.F. Did protein kinase regulatory mechanisms evolve through elaboration of a simple structural component? J. Mol. Biol. 2005, 351, 956–972. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Y.; Dai, D.Q.; Yan, H. PRL-dock: Protein-ligand docking based on hydrogen bond matching and probabilistic relaxation labeling. Proteins Struct. Funct. Bioinform. 2012, 80, 2137–2153. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.J.; Rather, P.N.; Hare, R.S.; Miller, G.H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 1993, 57, 138–163. [Google Scholar] [PubMed]
- Thompson, P.R.; Boehr, D.D.; Berghuis, A.M.; Wright, G.D. Mechanism of aminoglycoside antibiotic kinase APH(3′)-IIIa: Role of the nucleotide positioning loop. Biochemistry 2002, 41, 7001–7007. [Google Scholar] [CrossRef] [PubMed]
- Menard, R.; Molinas, C.; Arthur, M.; Duval, J.; Courvalin, P.; Leclercq, R. Overproduction of 3′-aminoglycoside phosphotransferase type I confers resistance to tobramycin in Escherichia coli. Antimicrob. Agents Chemother. 1993, 37, 78–83. [Google Scholar] [CrossRef][Green Version]
- Magnet, S.; Smith, T.A.; Zheng, R.; Nordmann, P.; Blanchard, J.S. Aminoglycoside resistance resulting from tight drug binding to an altered aminoglycoside acetyltransferase. Antimicrob. Agents Chemother. 2003, 47, 1577–1583. [Google Scholar] [CrossRef]
- Maarouf, M.; De Kouchkovsky, Y.; Brown, S.; Petit, P.X.; Robert-Gero, M. In vivo interference of paromomycin with mitochondrial activity of Leishmania. Exp. Cell Res. 1997, 232, 339–348. [Google Scholar] [CrossRef]
- Bhandari, V.; Sundar, S.; Dujardin, J.C.; Salotra, P. Elucidation of cellular mechanisms involved in experimental paromomycin resistance in leishmania donoVani. Antimicrob. Agents Chemother. 2014, 58, 2580–2585. [Google Scholar] [CrossRef]
- Chawla, B.; Jhingran, A.; Panigrahi, A.; Stuart, K.D.; Madhubala, R. Paromomycin affects translation and vesicle-mediated trafficking as revealed by proteomics of paromomycin -susceptible -resistant leishmania donovani. PLoS ONE 2011, 6, e26660. [Google Scholar] [CrossRef]
- El Fadili, K.; Messier, N.; Leprohon, P.; Roy, G.; Guimond, C.; Trudel, N.; Saravia, N.G.; Papadopoulou, B.; Légaré, D.; Ouellette, M. Role of the ABC transporter MRPA (PGPA) in antimony resistance in Leishmania infantum axenic and intracellular amastigotes. Antimicrob. Agents Chemother. 2005, 49, 1988–1993. [Google Scholar] [CrossRef]
- Khanra, S.; Sarraf, N.R.; Das, A.K.; Roy, S.; Manna, M. Miltefosine Resistant Field Isolate from Indian Kala-Azar Patient Shows Similar Phenotype in Experimental Infection. Sci. Rep. 2017, 7, 10330. [Google Scholar] [CrossRef] [PubMed]
- Leprohon, P.; Légaré, D.; Girard, I.; Papadopoulou, B.; Ouellette, M. Modulation of Leishmania ABC protein gene expression through life stages and among drug-resistant parasites. Eukaryot. Cell 2006, 5, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Obonaga, R.; Fernández, O.L.; Valderrama, L.; Rubiano, L.C.; Castro, M.D.M.; Barrera, M.C.; Gomez, M.A.; Saravia, N.G. Treatment failure and miltefosine susceptibility in dermal leishmaniasis caused by leishmania subgenus viannia species. Antimicrob. Agents Chemother. 2014, 58, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Teklemariam, S.; Hiwot, A.G.; Frommel, D.; Mike, T.L.; Ganlov, G.; Bryceson, A. Aminosidine and its combination with sodium stibogluconate in the treatment of diffuse cutaneous leishmaniasis caused by leishmania aethiopica. Trans. R. Soc. Trop. Med. Hyg. 1994, 88, 334–339. [Google Scholar] [CrossRef]







| Donor | Acceptor | Bond Type | Persistence (>40%) |
|---|---|---|---|
| ARG254 | GLU180 | SB | 100.00 |
| LYS131 | ASP129 | SB | 98.45 |
| ARG152 | GLU53 | SB | 85.66 |
| LYS37 | ASP147 | SB | 82.46 |
| ARG128-Main | ASP192-Side | HB | 60.49 |
| LEU137-Main | GLY88-Main | HB | 48.95 |
| ASN134-Side | ASP129-Main | HB | 40.61 |
| Rank | IUPAC Name | Chemical Name | MW | LogP | HBA (Lipinski) | HBD (Lipinski) | Violations (Lipinski) | Docking Score (kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| 1 | (2S,3S,4R,5R,6R)-5-amino-2-(aminomethyl)-6-[(2R,3S,4R,5S)-5-[(1R,2R,3S,5R,6S)-3,5-diamino-2-[(2S,3R,4R,5S,6R)-3-amino-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-6-hydroxycyclohexyl]oxy-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl]oxyoxane-3,4-diol | Paromomycin sulfate | 615.6 | −8.67 | 19 | 18 | 2 | −11.46 |
| 2 | (2R,3S,4R,5R,6R)-5-amino-2-(aminomethyl)-6-[(1R,2R,3S,4R,6S)-4,6-diamino-2-[(2S,3R,4S,5R)-4-[(2R,3R,4R,5S,6S)-3-amino-6-(aminomethyl)-4,5-dihydroxyoxan-2-yl]oxy-3-hydroxy-5-(hydroxymethyl)oxolan-2-yl]oxy-3-hydroxycyclohexyl]oxyoxane-3,4-diol | Neomycin sulfate stereoisomer A | 614.6 | −8.96 | 19 | 19 | 2 | −10.93 |
| 3 | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-3-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-[[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxymethyl]oxan-2-yl]oxychromen-4-one | Rutin | 610.5 | −1.16 | 16 | 10 | 2 | −10.84 |
| 4 | 1,4-dihydroxy-5,8-bis[2-(2-hydroxyethylamino)ethylamino]anthracene-9,10-dione | Mitoxantrone | 444.5 | 0.07 | 10 | 8 | 1 | −9.14 |
| 5 | (2S,3R,4S,5S,6R)-4-amino-2-[(1S,2S,3R,4S,6R)-4,6-diamino-3-[(2R,3R,5S,6R)-3-amino-6-(aminomethyl)-5-hydroxyoxan-2-yl]oxy-2-hydroxycyclohexyl]oxy-6-(hydroxymethyl)oxane-3,5-diol | Tobramycin sulfate | 467.5 | −6.86 | 14 | 15 | 2 | −9.06 |
| 6 | 5-hydroxy-2-(3-hydroxy-4-methoxyphenyl)-7-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-[[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxymethyl]oxan-2-yl]oxychromen-4-one | Diosmin | 608.5 | −0.4 | 15 | 8 | 2 | −9.05 |
| 7 | (2S)-4-amino-N-[(1R,2S,3S,4R,5S)-5-amino-2-[(2S,3R,4S,5S,6R)-4-amino-3,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-trihydroxyoxan-2-yl]oxy-3-hydroxycyclohexyl]-2-hydroxybutanamide | Amikacin stereoisomer | 585.6 | −8.43 | 18 | 17 | 2 | −9.04 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vacas, A.; Fernández-Rubio, C.; Algarabel, M.; Peña-Guerrero, J.; Larrea, E.; Rocha Formiga, F.; García-Sosa, A.T.; Nguewa, P.A. The Novel Serine/Threonine Protein Kinase LmjF.22.0810 from Leishmania major May Be Involved in the Resistance to Drugs such as Paromomycin. Biomolecules 2019, 9, 723. https://doi.org/10.3390/biom9110723
Vacas A, Fernández-Rubio C, Algarabel M, Peña-Guerrero J, Larrea E, Rocha Formiga F, García-Sosa AT, Nguewa PA. The Novel Serine/Threonine Protein Kinase LmjF.22.0810 from Leishmania major May Be Involved in the Resistance to Drugs such as Paromomycin. Biomolecules. 2019; 9(11):723. https://doi.org/10.3390/biom9110723
Chicago/Turabian StyleVacas, Andrés, Celia Fernández-Rubio, Miriam Algarabel, José Peña-Guerrero, Esther Larrea, Fabio Rocha Formiga, Alfonso T. García-Sosa, and Paul A. Nguewa. 2019. "The Novel Serine/Threonine Protein Kinase LmjF.22.0810 from Leishmania major May Be Involved in the Resistance to Drugs such as Paromomycin" Biomolecules 9, no. 11: 723. https://doi.org/10.3390/biom9110723
APA StyleVacas, A., Fernández-Rubio, C., Algarabel, M., Peña-Guerrero, J., Larrea, E., Rocha Formiga, F., García-Sosa, A. T., & Nguewa, P. A. (2019). The Novel Serine/Threonine Protein Kinase LmjF.22.0810 from Leishmania major May Be Involved in the Resistance to Drugs such as Paromomycin. Biomolecules, 9(11), 723. https://doi.org/10.3390/biom9110723







