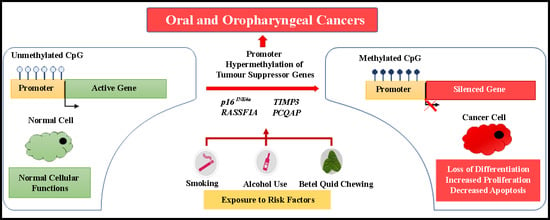
Promoter Hypermethylation of Tumor-Suppressor Genes p16INK4a, RASSF1A, TIMP3, and PCQAP/MED15 in Salivary DNA as a Quadruple Biomarker Panel for Early Detection of Oral and Oropharyngeal Cancers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Study Subjects and Data Collection
2.3. Sample Collection
2.4. DNA Extraction and Human DNA Confirmation
2.5. HPV-L1 DNA Detection
2.6. DNA Bisulfite Conversion
2.7. Target Gene Selection
2.8. Methylation Analysis of TSGs Using MS-PCR Assay
2.9. Agarose Gel Electrophoresis and Determination of Gene Methylation Levels
2.10. Statistical Analysis
3. Results
3.1. Population Characteristics of the Study Cohorts
3.2. HPV-L1 Analysis
3.3. Combined and Independent Effect Assessment of Etiologic Agents of OC and OPC
3.4. Comparative DNA Methylation Analysis of Individual TSGs
3.5. Association between Promoter Methylation of TSGs and Clinicopathological Parameters of OC and OPC
3.6. Performance of the Methylation Marker Panel in Discriminating OC and OPC from Healthy Controls
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Scully, C.; Bedi, R. Ethnicity and oral cancer. Lancet Oncol. 2000, 1, 37–42. [Google Scholar] [CrossRef]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef]
- Krishna Rao, S.V.; Mejia, G.; Roberts-Thomson, K.; Logan, R. Epidemiology of oral cancer in Asia in the past decade—An update (2000–2012). Asian Pac. J. Cancer Prev. 2013, 14, 5567–5577. [Google Scholar]
- Siriwardena, B.S.; Tilakaratne, A.; Amaratunga, E.A.; Tilakaratne, W.M. Demographic, aetiological and survival differences of oral squamous cell carcinoma in the young and the old in Sri Lanka. Oral Oncol. 2006, 42, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Somatunga, L.C.; Sinha, D.N.; Sumanasekera, P.; Galapatti, K.; Rinchen, S.; Kahandaliyanage, A.; Mehta, F.R.; Nishirani Lanka, J.D. Smokeless tobacco use in Sri Lanka. Indian J. Cancer 2012, 49, 357–363. [Google Scholar] [PubMed]
- Warnakulasuriya, S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009, 45, 309–316. [Google Scholar] [CrossRef]
- Marur, S.; D’Souza, G.; Westra, W.H.; Forastiere, A.A. HPV-associated head and neck cancer: A virus-related cancer epidemic. Lancet Oncol. 2010, 11, 781–789. [Google Scholar] [CrossRef]
- Feng, Z.; Xu, Q.; Chen, W. Epigenetic and genetic alterations-based molecular classification of head and neck cancer. Expert Rev. Mol. Diagn. 2012, 12, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.G. Hypermethylation of tumor suppressor genes in cancer. Semin. Cancer Biol. 1999, 9, 359–367. [Google Scholar] [CrossRef]
- Righini, C.A.; de Fraipont, F.; Timsit, J.F.; Faure, C.; Brambilla, E.; Reyt, E.; Favrot, M.C. Tumor-specific methylation in saliva: A promising biomarker for early detection of head and neck cancer recurrence. Clin. Cancer Res. 2007, 13, 1179–1185. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikov, D.A.; Wan, Y.; Coman, W.B.; Pandit, P.; Cooper-White, J.J.; Herman, J.G.; Punyadeera, C. DNA Methylation at the Novel CpG Sites in the Promoter of MED15/PCQAP Gene as a Biomarker for Head and Neck Cancers. Biomark. Insights 2014, 9, 53–60. [Google Scholar] [CrossRef]
- Sun, W.; Zaboli, D.; Wang, H.; Liu, Y.; Arnaoutakis, D.; Khan, T.; Khan, Z.; Koch, W.M.; Califano, J.A. Detection of TIMP3 promoter hypermethylation in salivary rinse as an independent predictor of local recurrence-free survival in head and neck cancer. Clin. Cancer Res. 2012, 18, 1082–1091. [Google Scholar] [CrossRef]
- Supic, G.; Kozomara, R.; Brankovic-Magic, M.; Jovic, N.; Magic, Z. Gene hypermethylation in tumor tissue of advanced oral squamous cell carcinoma patients. Oral Oncol. 2009, 45, 1051–1057. [Google Scholar] [CrossRef]
- Chang, H.W.; Ling, G.S.; Wei, W.I.; Yuen, A.P. Smoking and drinking can induce p15 methylation in the upper aerodigestive tract of healthy individuals and patients with head and neck squamous cell carcinoma. Cancer 2004, 101, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, M.; Saitoh, M.; Kusano, K.; Nagayasu, H.; Kurashige, Y.; Malsantha, M.; Arakawa, T.; Takuma, T.; Chiba, I.; Kaku, T.; et al. High frequency of hypermethylation of p14, p15 and p16 in oral pre-cancerous lesions associated with betel-quid chewing in Sri Lanka. J. Oral Pathol. Med. 2008, 37, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Marsit, C.J.; McClean, M.D.; Furniss, C.S.; Kelsey, K.T. Epigenetic inactivation of the SFRP genes is associated with drinking, smoking and HPV in head and neck squamous cell carcinoma. Int. J. Cancer 2006, 119, 1761–1766. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Corn, P.G.; Baylin, S.B.; Herman, J.G. A gene hypermethylation profile of human cancer. Cancer Res. 2001, 61, 3225–3229. [Google Scholar]
- Sidransky, D.; Irizarry, R.; Califano, J.A.; Li, X.; Ren, H.; Benoit, N.; Mao, L. Serum protein MALDI profiling to distinguish upper aerodigestive tract cancer patients from control subjects. J. Natl. Cancer Inst. 2003, 95, 1711–1717. [Google Scholar] [CrossRef]
- Ahrendt, S.A.; Chow, J.T.; Xu, L.H.; Yang, S.C.; Eisenberger, C.F.; Esteller, M.; Herman, J.G.; Wu, L.; Decker, P.A.; Jen, J.; et al. Molecular detection of tumor cells in bronchoalveolar lavage fluid from patients with early stage lung cancer. J. Natl. Cancer Inst. 1999, 91, 332–339. [Google Scholar] [CrossRef]
- Cairns, P.; Esteller, M.; Herman, J.G.; Schoenberg, M.; Jeronimo, C.; Sanchez-Cespedes, M.; Chow, N.H.; Grasso, M.; Wu, L.; Westra, W.B.; et al. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin. Cancer Res. 2001, 7, 2727–2730. [Google Scholar] [PubMed]
- Evron, E.; Dooley, W.C.; Umbricht, C.B.; Rosenthal, D.; Sacchi, N.; Gabrielson, E.; Soito, A.B.; Hung, D.T.; Ljung, B.; Davidson, N.E.; et al. Detection of breast cancer cells in ductal lavage fluid by methylation-specific PCR. Lancet 2001, 357, 1335–1336. [Google Scholar] [CrossRef]
- Carvalho, A.L.; Jeronimo, C.; Kim, M.M.; Henrique, R.; Zhang, Z.; Hoque, M.O.; Chang, S.; Brait, M.; Nayak, C.S.; Jiang, W.W.; et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin. Cancer Res. 2008, 14, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Arellano, M.; Boontheung, P.; Wang, J.; Zhou, H.; Jiang, J.; Elashoff, D.; Wei, R.; Loo, J.A.; Wong, D.T. Salivary proteomics for oral cancer biomarker discovery. Clin. Cancer Res. 2008, 14, 6246–6252. [Google Scholar] [CrossRef]
- Nagata, S.; Hamada, T.; Yamada, N.; Yokoyama, S.; Kitamoto, S.; Kanmura, Y.; Nomura, M.; Kamikawa, Y.; Yonezawa, S.; Sugihara, K. Aberrant DNA methylation of tumor-related genes in oral rinse: A noninvasive method for detection of oral squamous cell carcinoma. Cancer 2012, 118, 4298–4308. [Google Scholar] [CrossRef] [PubMed]
- Pfaffe, T.; Cooper-White, J.; Beyerlein, P.; Kostner, K.; Punyadeera, C. Diagnostic potential of saliva: Current state and future applications. Clin. Chem. 2011, 57, 675–687. [Google Scholar] [CrossRef]
- Ovchinnikov, D.A.; Cooper, M.A.; Pandit, P.; Coman, W.B.; Cooper-White, J.J.; Keith, P.; Wolvetang, E.J.; Slowey, P.D.; Punyadeera, C. Tumor-suppressor Gene Promoter Hypermethylation in Saliva of Head and Neck Cancer Patients. Transl. Oncol. 2012, 5, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Shah, F.D.; Begum, R.; Vajaria, B.N.; Patel, K.R.; Patel, J.B.; Shukla, S.N.; Patel, P.S. A review on salivary genomics and proteomics biomarkers in oral cancer. Indian J. Clin. Biochem. 2011, 26, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Viet, C.T.; Schmidt, B.L. Methylation array analysis of preoperative and postoperative saliva DNA in oral cancer patients. Cancer Epidemiol. Biomarkers Prev. 2008, 17, 3603–3611. [Google Scholar] [CrossRef]
- Herman, J.G.; Graff, J.R.; Myohanen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826. [Google Scholar] [CrossRef]
- Rand, K.; Qu, W.; Ho, T.; Clark, S.J.; Molloy, P. Conversion-specific detection of DNA methylation using real-time polymerase chain reaction (ConLight-MSP) to avoid false positives. Methods 2002, 27, 114–120. [Google Scholar] [CrossRef]
- Divine, K.K.; Liechty, K.C.; Crume, K.C.; Belinsky, S.A. Nested multigene MSP/DHPLC method for analyzing promoter hypermethylation status in clinical samples. Biotechniques 2006, 40, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Bassil, C.F.; Murphy, S.K. Methylation-specific PCR. Methods Mol. Biol. 2013, 1049, 75–82. [Google Scholar] [PubMed]
- Topkas, E.; Keith, P.; Dimeski, G.; Cooper-White, J.; Punyadeera, C. Evaluation of saliva collection devices for the analysis of proteins. Clin. Chim. Acta 2012, 413, 1066–1070. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, R.; Campbell, J.L.; Cooper-White, J.; Dimeski, G.; Punyadeera, C. The impact of saliva collection and processing methods on CRP, IgE, and Myoglobin immunoassays. Clin. Transl. Med. 2012, 1, 19. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef]
- de Roda Husman, A.M.; Walboomers, J.M.; van den Brule, A.J.; Meijer, C.J.; Snijders, P.J. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 1995, 76, 1057–1062. [Google Scholar] [CrossRef]
- Eads, C.A.; Lord, R.V.; Wickramasinghe, K.; Long, T.I.; Kurumboor, S.K.; Bernstein, L.; Peters, J.H.; DeMeester, S.R.; DeMeester, T.R.; Skinner, K.A.; et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001, 61, 3410–3418. [Google Scholar] [PubMed]
- Rettori, M.M.; de Carvalho, A.C.; Longo, A.L.; de Oliveira, C.Z.; Kowalski, L.P.; Carvalho, A.L.; Vettore, A.L. TIMP3 and CCNA1 hypermethylation in HNSCC is associated with an increased incidence of second primary tumors. J. Transl. Med. 2013, 11, 316. [Google Scholar] [CrossRef]
- Lim, Y.; Wan, Y.; Vagenas, D.; Ovchinnikov, D.A.; Perry, C.F.; Davis, M.J.; Punyadeera, C. Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers. BMC Cancer 2016, 16, 749. [Google Scholar] [CrossRef]
- Mazzara, S.; Rossi, R.L.; Grifantini, R.; Donizetti, S.; Abrignani, S.; Bombaci, M. CombiROC: An interactive web tool for selecting accurate marker combinations of omics data. Sci. Rep. 2017, 7, 45477. [Google Scholar] [CrossRef]
- Gonzalez, S.; Serrano, M. A new mechanism of inactivation of the INK4/ARF locus. Cell Cycle 2006, 5, 1382–1384. [Google Scholar] [CrossRef]
- Rosenbaum, E.; Hoque, M.O.; Cohen, Y.; Zahurak, M.; Eisenberger, M.A.; Epstein, J.I.; Partin, A.W.; Sidransky, D. Promoter hypermethylation as an independent prognostic factor for relapse in patients with prostate cancer following radical prostatectomy. Clin. Cancer Res. 2005, 11, 8321–8325. [Google Scholar] [CrossRef] [PubMed]
- Belinsky, S.A.; Klinge, D.M.; Dekker, J.D.; Smith, M.W.; Bocklage, T.J.; Gilliland, F.D.; Crowell, R.E.; Karp, D.D.; Stidley, C.A.; Picchi, M.A. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin. Cancer Res. 2005, 11, 6505–6511. [Google Scholar] [CrossRef]
- Hsu, L.S.; Lee, H.C.; Chau, G.Y.; Yin, P.H.; Chi, C.W.; Lui, W.Y. Aberrant methylation of EDNRB and p16 genes in hepatocellular carcinoma (HCC) in Taiwan. Oncol. Rep. 2006, 15, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Ogi, K.; Toyota, M.; Ohe-Toyota, M.; Tanaka, N.; Noguchi, M.; Sonoda, T.; Kohama, G.; Tokino, T. Aberrant methylation of multiple genes and clinicopathological features in oral squamous cell carcinoma. Clin. Cancer Res. 2002, 8, 3164–3171. [Google Scholar] [PubMed]
- Mino, A.; Onoda, N.; Yashiro, M.; Aya, M.; Fujiwara, I.; Kubo, N.; Sawada, T.; Ohira, M.; Kato, Y.; Hirakawa, K. Frequent p16 CpG island hypermethylation in primary remnant gastric cancer suggesting an independent carcinogenic pathway. Oncol. Rep. 2006, 15, 615–620. [Google Scholar] [CrossRef]
- Kato, K.; Hara, A.; Kuno, T.; Mori, H.; Yamashita, T.; Toida, M.; Shibata, T. Aberrant promoter hypermethylation of p16 and MGMT genes in oral squamous cell carcinomas and the surrounding normal mucosa. J. Cancer Res. Clin. Oncol. 2006, 132, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Don, K.R.; Ramani, P.; Ramshankar, V.; Sherlin, H.J.; Premkumar, P.; Natesan, A. Promoter hypermethylation patterns of P16, DAPK and MGMT in oral squamous cell carcinoma: A systematic review and meta-analysis. Indian J. Dent. Res. 2014, 25, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Sushma, P.S.; Jamil, K.; Kumar, P.U.; Satyanarayana, U.; Ramakrishna, M.; Triveni, B. PTEN and p16 genes as epigenetic biomarkers in oral squamous cell carcinoma (OSCC): A study on south Indian population. Tumour Biol. 2016, 37, 7625–7632. [Google Scholar] [CrossRef] [PubMed]
- Spandidos, D.A.; Sourvinos, G.; Tsatsanis, C.; Zafiropoulos, A. Normal RAS genes: Their onco-suppressor and pro-apoptotic functions (review). Int. J. Oncol. 2002, 21, 237–241. [Google Scholar] [CrossRef]
- Blanchard, T.G.; Lapidus, R.; Banerjee, V.; Bafford, A.C.; Czinn, S.J.; Ahmed, H.; Banerjee, A. Upregulation of RASSF1A in Colon Cancer by Suppression of Angiogenesis Signaling and Akt Activation. Cell. Physiol. Biochem. 2018, 48, 1259–1273. [Google Scholar] [CrossRef]
- Wen, G.; Wang, H.; Zhong, Z. Associations of RASSF1A, RARbeta, and CDH1 promoter hypermethylation with oral cancer risk: A PRISMA-compliant meta-analysis. Medicine 2018, 97, e9971. [Google Scholar] [CrossRef]
- Gomez, D.E.; Alonso, D.F.; Yoshiji, H.; Thorgeirsson, U.P. Tissue inhibitors of metalloproteinases: Structure, regulation and biological functions. Eur. J. Cell Biol. 1997, 74, 111–122. [Google Scholar] [PubMed]
- Rettori, M.M.; de Carvalho, A.C.; Bomfim Longo, A.L.; de Oliveira, C.Z.; Kowalski, L.P.; Carvalho, A.L.; Vettore, A.L. Prognostic significance of TIMP3 hypermethylation in post-treatment salivary rinse from head and neck squamous cell carcinoma patients. Carcinogenesis 2013, 34, 20–27. [Google Scholar] [CrossRef]
- Qi, J.H.; Ebrahem, Q.; Moore, N.; Murphy, G.; Claesson-Welsh, L.; Bond, M.; Baker, A.; Anand-Apte, B. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): Inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat. Med. 2003, 9, 407–415. [Google Scholar] [CrossRef] [PubMed]
- De Schutter, H.; Geeraerts, H.; Verbeken, E.; Nuyts, S. Promoter methylation of TIMP3 and CDH1 predicts better outcome in head and neck squamous cell carcinoma treated by radiotherapy only. Oncol. Rep. 2009, 21, 507–513. [Google Scholar] [PubMed]
- Sandhu, H.K.; Hollenbeck, N.; Wassink, T.H.; Philibert, R.A. An association study of PCQAP polymorphisms and schizophrenia. Psychiatr. Genet. 2004, 14, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Marsit, C.J.; Houseman, E.A.; Schned, A.R.; Karagas, M.R.; Kelsey, K.T. Promoter hypermethylation is associated with current smoking, age, gender and survival in bladder cancer. Carcinogenesis 2007, 28, 1745–1751. [Google Scholar] [CrossRef]
- Kaur, J.; Demokan, S.; Tripathi, S.C.; Macha, M.A.; Begum, S.; Califano, J.A.; Ralhan, R. Promoter hypermethylation in Indian primary oral squamous cell carcinoma. Int. J. Cancer 2010, 127, 2367–2373. [Google Scholar] [CrossRef]
- Hasegawa, M.; Nelson, H.H.; Peters, E.; Ringstrom, E.; Posner, M.; Kelsey, K.T. Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene 2002, 21, 4231–4236. [Google Scholar] [CrossRef] [PubMed]
- Misawa, K.; Mochizuki, D.; Imai, A.; Endo, S.; Mima, M.; Misawa, Y.; Kanazawa, T.; Carey, T.E.; Mineta, H. Prognostic value of aberrant promoter hypermethylation of tumor-related genes in early-stage head and neck cancer. Oncotarget 2016, 7, 26087–26098. [Google Scholar] [CrossRef] [PubMed]
- Misawa, K.; Mochizuki, D.; Imai, A.; Misawa, Y.; Endo, S.; Mima, M.; Kawasaki, H.; Carey, T.E.; Kanazawa, T. Epigenetic silencing of SALL3 is an independent predictor of poor survival in head and neck cancer. Clin. Epigenet. 2017, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- von Zeidler, S.V.; Miracca, E.C.; Nagai, M.A.; Birman, E.G. Hypermethylation of the p16 gene in normal oral mucosa of smokers. Int. J. Mol. Med. 2004, 14, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.M.; Sun, D.I.; Benoit, N.E.; Kuzmin, I.; Lerman, M.I.; Sidransky, D. Epigenetic inactivation of RASSF1A in head and neck cancer. Clin. Cancer Res. 2003, 9, 3635–3640. [Google Scholar]
- Ai, L.; Stephenson, K.K.; Ling, W.; Zuo, C.; Mukunyadzi, P.; Suen, J.Y.; Hanna, E.; Fan, C.Y. The p16 (CDKN2a/INK4a) tumor-suppressor gene in head and neck squamous cell carcinoma: A promoter methylation and protein expression study in 100 cases. Mod. Pathol. 2003, 16, 944–950. [Google Scholar] [CrossRef]
- Lee, K.W.; Pausova, Z. Cigarette smoking and DNA methylation. Front. Genet. 2013, 4, 132. [Google Scholar] [CrossRef]
- Vertino, P.M.; Yen, R.W.; Gao, J.; Baylin, S.B. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol. Cell. Biol. 1996, 16, 4555–4565. [Google Scholar] [CrossRef] [PubMed]
- Belinsky, S.A.; Nikula, K.J.; Baylin, S.B.; Issa, J.P. Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc. Natl. Acad. Sci. USA 1996, 93, 4045–4050. [Google Scholar] [CrossRef]
- Sharan, R.N.; Mehrotra, R.; Choudhury, Y.; Asotra, K. Association of betel nut with carcinogenesis: Revisit with a clinical perspective. PLoS ONE 2012, 7, e42759. [Google Scholar] [CrossRef] [PubMed]
- Ariyawardana, A.; Athukorala, A.D.; Arulanandam, A. Effect of betel chewing, tobacco smoking and alcohol consumption on oral submucous fibrosis: A case-control study in Sri Lanka. J. Oral Pathol. Med. 2006, 35, 197–201. [Google Scholar] [CrossRef]
- Amarasena, N.; Ekanayaka, A.N.; Herath, L.; Miyazaki, H. Association between smoking, betel chewing and gingival bleeding in rural Sri Lanka. J. Clin. Periodontol. 2003, 30, 403–408. [Google Scholar] [CrossRef]
- Tran, T.N.; Liu, Y.; Takagi, M.; Yamaguchi, A.; Fujii, H. Frequent promoter hypermethylation of RASSF1A and p16INK4a and infrequent allelic loss other than 9p21 in betel-associated oral carcinoma in a Vietnamese non-smoking/non-drinking female population. J. Oral Pathol. Med. 2005, 34, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.M.; Ritchie, J.M.; Summersgill, K.F.; Klussmann, J.P.; Lee, J.H.; Wang, D.; Haugen, T.H.; Turek, L.P. Age, sexual behavior and human papillomavirus infection in oral cavity and oropharyngeal cancers. Int. J. Cancer 2004, 108, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Gunasekera, S.K.; Perera, K.A.; Fernando, C.; Udagama, P.V. A shifting paradigm in the aetiology of oral and pharyngeal cancer in Sri Lanka: A case-control study providing serologic evidence for the role of oncogenic HPV types 16 and 18. Infect. Agents Cancer 2015, 10, 12. [Google Scholar] [CrossRef]
- Kostareli, E.; Holzinger, D.; Bogatyrova, O.; Hielscher, T.; Wichmann, G.; Keck, M.; Lahrmann, B.; Grabe, N.; Flechtenmacher, C.; Schmidt, C.R.; et al. HPV-related methylation signature predicts survival in oropharyngeal squamous cell carcinomas. J. Clin. Investig. 2013, 123, 2488–2501. [Google Scholar] [CrossRef]
- Toyooka, S.; Maruyama, R.; Toyooka, K.O.; McLerran, D.; Feng, Z.; Fukuyama, Y.; Virmani, A.K.; Zochbauer-Muller, S.; Tsukuda, K.; Sugio, K.; et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int. J. Cancer 2003, 103, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Demokan, S.; Chang, X.; Chuang, A.; Mydlarz, W.K.; Kaur, J.; Huang, P.; Khan, Z.; Khan, T.; Ostrow, K.L.; Brait, M.; et al. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int. J. Cancer 2010, 127, 2351–2359. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Ganguly, P.; Ganguly, N. Modulation of DNA methylation by human papillomavirus E6 and E7 oncoproteins in cervical cancer. Oncol. Lett. 2018, 15, 11–22. [Google Scholar] [CrossRef]
- De Freitas Cordeiro-Silva, M.; Stur, E.; Agostini, L.P.; de Podesta, J.R.; de Oliveira, J.C.; Soares, M.S.; Mendonca, E.F.; Gouvea, S.A.; Von Zeidler, S.V.; Louro, I.D. Promoter hypermethylation in primary squamous cell carcinoma of the oral cavity and oropharynx: A study of a Brazilian cohort. Mol. Biol. Rep. 2012, 39, 10111–10119. [Google Scholar] [CrossRef]
- Leygo, C.; Williams, M.; Jin, H.C.; Chan, M.W.Y.; Chu, W.K.; Grusch, M.; Cheng, Y.Y. DNA Methylation as a Noninvasive Epigenetic Biomarker for the Detection of Cancer. Dis. Markers 2017, 2017, 3726595. [Google Scholar] [CrossRef]
- Arantes, L.M.; de Carvalho, A.C.; Melendez, M.E.; Centrone, C.C.; Gois-Filho, J.F.; Toporcov, T.N.; Caly, D.N.; Tajara, E.H.; Goloni-Bertollo, E.M.; Carvalho, A.L.; et al. Validation of methylation markers for diagnosis of oral cavity cancer. Eur. J. Cancer 2015, 51, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Al-Kaabi, A.; van Bockel, L.W.; Pothen, A.J.; Willems, S.M. p16INK4A and p14ARF gene promoter hypermethylation as prognostic biomarker in oral and oropharyngeal squamous cell carcinoma: A review. Dis. Markers 2014, 2014, 260549. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Bahadur, S.; Thakar, A.; Matta, A.; Macha, M.; Ralhan, R.; Gupta, S.D. Significance of promoter hypermethylation of p16 gene for margin assessment in carcinoma tongue. Head Neck 2009, 31, 1423–1430. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, X.W.; Dong, G.; Zhou, J.; Liu, Y.; Gao, Y.; Liu, X.Y.; Gu, L.; Sun, Z.; Deng, D. P16 Methylation as an Early Predictor for Cancer Development from Oral Epithelial Dysplasia: A Double-blind Multicentre Prospective Study. EBioMedicine 2015, 2, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.L.; Henrique, R.; Jeronimo, C.; Nayak, C.S.; Reddy, A.N.; Hoque, M.O.; Chang, S.; Brait, M.; Jiang, W.W.; Kim, M.M.; et al. Detection of promoter hypermethylation in salivary rinses as a biomarker for head and neck squamous cell carcinoma surveillance. Clin. Cancer Res. 2011, 17, 4782–4789. [Google Scholar] [CrossRef]
- Liang, X.H.; Lewis, J.; Foote, R.; Smith, D.; Kademani, D. Prevalence and significance of human papillomavirus in oral tongue cancer: The Mayo Clinic experience. J. Oral Maxillofac. Surg. 2008, 66, 1875–1880. [Google Scholar] [CrossRef] [PubMed]
- Machado, J.; Reis, P.P.; Zhang, T.; Simpson, C.; Xu, W.; Perez-Ordonez, B.; Goldstein, D.P.; Brown, D.H.; Gilbert, R.W.; Gullane, P.J.; et al. Low prevalence of human papillomavirus in oral cavity carcinomas. Head Neck Oncol. 2010, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Claus, R.; Wilop, S.; Hielscher, T.; Sonnet, M.; Dahl, E.; Galm, O.; Jost, E.; Plass, C. A systematic comparison of quantitative high-resolution DNA methylation analysis and methylation-specific PCR. Epigenetics 2012, 7, 772–780. [Google Scholar] [CrossRef]
- Shaw, R.J.; Hall, G.L.; Lowe, D.; Liloglou, T.; Field, J.K.; Sloan, P.; Risk, J.M. The role of pyrosequencing in head and neck cancer epigenetics: Correlation of quantitative methylation data with gene expression. Arch. Otolaryngol. Head Neck Surg. 2008, 134, 251–256. [Google Scholar] [CrossRef]
- Roessler, J.; Lehmann, U. Quantitative DNA Methylation Analysis by Pyrosequencing(R). Methods Mol. Biol. 2015, 1315, 175–188. [Google Scholar]
- Delaney, C.; Garg, S.K.; Yung, R. Analysis of DNA Methylation by Pyrosequencing. Methods Mol. Biol. 2015, 1343, 249–264. [Google Scholar] [PubMed]
- Mikeska, T.; Bock, C.; El-Maarri, O.; Hubner, A.; Ehrentraut, D.; Schramm, J.; Felsberg, J.; Kahl, P.; Buttner, R.; Pietsch, T.; et al. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J. Mol. Diagn. 2007, 9, 368–381. [Google Scholar] [CrossRef] [PubMed]
- Siegel, E.M.; Riggs, B.M.; Delmas, A.L.; Koch, A.; Hakam, A.; Brown, K.D. Quantitative DNA methylation analysis of candidate genes in cervical cancer. PLoS ONE 2015, 10, e0122495. [Google Scholar] [CrossRef] [PubMed]




| Gene | Primer Sense (5′–3′) | Primer Antisense (5′–3′) | PCR Product Size (bp) |
|---|---|---|---|
| β-globin | PC03: ACACAACTGTGTTCACTAGC | KM38: TGGTCTCCTTAAACCTGTCTTG | 167 |
| HPV-L1 | GP5+: TTTGTTACTGTGGTAGATACTAC | GP6+: GAAAAATAAACTGTAAATCATATT | 150 |
| p16INK4a (MI) | GAGGAAGAAAGAGGAGGGGTTG | ACAAACCCTCTACCCACCTAAATC | 274 |
| p16INK4a (M) | GAGGGTGGGGCGGATCGC | GACCCCGAACCGCGACCG | 143 |
| p16INK4a (U) | TTATTAGAGGGTGGGGTGGATTGT | CAACCCCAAACCACAACCATAA | 145 |
| RASSF1A (MI) | GGAGGGAAGGAAGGGTAAGG | CAACTCAATAAACTCAAACTCCC | 260 |
| RASSF1A (M) | GGGGGTTTTGCGAGAGCGC | CCCGATTAAACCCGTACTTCG | 203 |
| RASSF1A (U) | GGTTTTGTGAGAGTGTGTTTAG | ACACTAACAAACACAAACCAAAC | 172 |
| TIMP3 (M) | GCGTCGGAGGTTAAGGTTGTT | CTCTCCAAAATTACCGTACGCG | 116 |
| TIMP3 (U) | TGTGTTGGAGGTTAAGGTTGTTTT | ACTCTCCAAAATTACCATACACACC | 122 |
| PCQAP 5′ (M) | GTTTTGTGATTGAGGYGGCGGC | AAAAATCCCACAATCCAACCC | 167 |
| PCQAP 5′ (U) | GTTTTGTGATTGAGGYGGTGGT | AAAAATCCCACAATCCAACCC | 167 |
| OC (N = 54) | OPC (N = 34) | Healthy Controls (N = 60) | |
|---|---|---|---|
| Demographic characteristics | |||
| Mean age | 62 | 62 | 60 |
| <50 | 7 (13.0) | 3 (8.8) | 1 (1.6) |
| 50–59 | 11 (20.3) | 13 (38.2) | 29 (48.3) |
| >60 | 36 (66.7) | 18 (53.0) | 30 (50) |
| Gender | |||
| Male | 49 (90.7) | 32 (94.1) | 55 (91.6) |
| Female | 5 (9.3) | 2 (5.9) | 5 (8.4) |
| Smoking | |||
| Cigarettes/day smoked | |||
| Non-smokers | 11 (20.3) | 3 (8.8) | 39 (65.0) |
| 1 to 5 | 14 (26.0) | 11 (32.4) | 8 (13.3) |
| >5 | 29 (53.7) | 20 (58.8) | 13 (21.6) |
| Alcohol use | |||
| Number of years in use | |||
| Non-drinkers | 14 (25.9) | 3 (8.8) | 31 (51.6) |
| 1–25 | 14 (25.9) | 12 (35.3) | 10 (16.6) |
| >25 | 26 (48.2) | 19 (55.9) | 19 (31.6) |
| Betel quid chewing | |||
| Number of years in use | |||
| Non-consumers | 12 (22.2) | 8 (23.5) | 41 (68.3) |
| 1–25 | 16 (29.6) | 9 (26.5) | 7 (11.6) |
| >25 | 26 (48.2) | 17 (50.0) | 12 (20) |
| HPV infection | |||
| HPV Positive | 5 (9.2) | 3 (8.8) | 1 (1.6) |
| HPV Negative | 49 (90.8) | 31(91.2) | 59 (98.4) |
| Tumor characteristics | |||
| Anatomic site | |||
| Lips | 2 (3.7) | ||
| Tongue (Front 2/3) | 26 (48.1) | ||
| Hard palate | 1 (1.9) | ||
| Buccal mucosa | 21 (38.9) | ||
| Mouth floor | 3 (5.5) | ||
| Retromolar | 1 (1.9) | ||
| Tongue (Back 1/3) | 0 (0) | ||
| Soft palate | 7 (20.6) | ||
| Tonsillar pillar | 12 (35.3) | ||
| Back wall of the throat | 15 (44.1) | ||
| Tumor grade | |||
| Well differentiated (1) | 24 (44.4) | 17 (50.0) | |
| Moderately differentiated (2) | 7 (13.0) | 5 (14.8) | |
| Poorly differentiated (3) | 0 (0) | 2 (5.9) | |
| Undifferentiated (4) | 19 (35.2) | 6 (17.6) | |
| Unknown | 4 (7.4) | 4 (11.7) | |
| Tumor stage | |||
| Early stage (I, II) | 8 (14.8) | 2 (5.9) | |
| Advanced stage (III, IV) | 23 (42.6) | 13 (38.2) | |
| Unknown | 23 (42.6) | 19 (55.9) |
| Cancer Type | Predictor Variable/Risk Factor | Crude OR (95% CI) | p-Value a | Adjusted ORR (95% CI) | p-Value b |
|---|---|---|---|---|---|
| OC | Smoking | 7.3 (2.8–18.6) | <0.0001 *** | 7.8 (2.1–28.4) | <0.05 * |
| Alcohol use | 3.1 (1.2–7.3) | <0.05 * | 0.6 (0.1–2.0) | 0.385 | |
| Betel quid chewing | 7.1 (3.0–19.2) | <0.0001 *** | 5.7 (2.2–14.3) | <0.05 * | |
| HPV infection | 6.0 (0.6–140.9) | 0.07 | 6.7 (0.6–123.8) | <0.05 * | |
| OPC | Smoking | 19.2 (4.7–89.8) | <0.0001 *** | 20.8 (2.4–178.2) | <0.05 * |
| Alcohol use | 11.0 (2.7–51.0) | <0.0001 *** | 0.8 (0.1–6.7) | 0.854 | |
| Betel quid chewing | 7.0 (2.4–20.7) | <0.0001 *** | 3.9 (1.2–12.4) | <0.05 * | |
| HPV infection | 5.7 (0.4–148.9) | 0.133 | 19.6 (1.0–146.4) | <0.05 * |
| Variable | Category | N | p16INK4a | RASSF1A | TIMP3 | PCQAP/MED15 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Meth | p-Value b | Meth | p-Value b | Meth | p-Value b | Meth | p-Value b | ||||
| Age | ≥62 a | 28 | 13 | 0.455 | 12 | 0.244 | 18 | 0.414 | 21 | 0.550 | |
| <62 a | 60 | 58 | 54 | 52 | 52 | ||||||
| Gender | Male | 80 | 63 | 0.053 | 61 | 0.133 | 69 | 0.152 | 66 | 0.164 | |
| Female | 8 | 5 | 4 | 5 | 5 | ||||||
| Smoking | Consumers | 74 | 57 | <0.05 * | 58 | <0.05 * | 61 | 0.199 | 62 | 0.240 | |
| Non-consumers | 14 | 11 | 7 | 13 | 9 | ||||||
| Alcohol use | Consumers | 71 | 56 | <0.05 * | 56 | <0.05 * | 61 | 0.665 | 60 | 0.060 | |
| Non-consumers | 17 | 12 | 9 | 13 | 11 | ||||||
| Betel quid Chewing | Consumers | 68 | 51 | <0.05 * | 52 | <0.05 * | 61 | <0.001 ** | 57 | <0.05 * | |
| Non-consumers | 20 | 17 | 13 | 13 | 14 | ||||||
| HPV-L1 | HPV-positive | 8 | 6 | <0.001 ** | 6 | 0.061 | 8 | <0.001 ** | 7 | <0.001 ** | |
| HPV-negative | 80 | 62 | 54 | 66 | 64 | ||||||
| Tumor grade | OC | Grade 3, 4 | 19 | 14 | <0.05 * | 13 | <0.05 * | 15 | 0.311 | 16 | 0.289 |
| Grade 1, 2 | 31 | 25 | 24 | 28 | 27 | ||||||
| OPC | Grade 3, 4 | 8 | 6 | <0.05 * | 5 | <0.05 * | 4 | <0.05 * | 2 | 0.146 | |
| Grade 1, 2 | 22 | 17 | 17 | 21 | 21 | ||||||
| Tumor stage | OC | Stage (III, IV) | 23 | 18 | <0.001 ** | 17 | <0.05 * | 17 | 0.233 | 19 | 0.712 |
| Stage (I, II) | 8 | 7 | 6 | 7 | 6 | ||||||
| OPC | Stage (III, IV) | 13 | 8 | 0.525 | 10 | 0.930 | 10 | 0.076 | 11 | 0.964 | |
| Stage (I, II) | 2 | 2 | 2 | 2 | 1 | ||||||
| AUC | ACC | Error Rate (1 − ACC) | SE% | SP% | PPV% | NPV% | |
|---|---|---|---|---|---|---|---|
| (a) Performance in discriminating OC from healthy controls | |||||||
| Whole cohort | 0.92 | 0.92 | 0.08 | 91.7 | 92.3 | 95.7 | 85.7 |
| 10-fold CV | 0.85 | 0.87 | 0.14 | 83.3 | 92.3 | ||
| Permutated models | 0.71 | 0.71 | 0.29 | 69.7 | 76.0 | ||
| (b) Performance in discriminating OPC from healthy controls | |||||||
| Whole cohort | 0.97 | 0.96 | 0.04 | 99.8 | 92.1 | 93.7 | 98.6 |
| 10-fold CV | 0.82 | 0.89 | 0.11 | 86.7 | 92.3 | ||
| Permutated models | 0.60 | 0.66 | 0.33 | 65.1 | 68.7 | ||
| (c) Performance in discriminating OC from OPC | |||||||
| Whole cohort | 0.65 | 0.72 | 0.28 | 91.7 | 40.0 | 61.2 | 53.3 |
| 10-fold CV | 0.60 | 0.56 | 0.44 | 33.3 | 93.3 | ||
| Permutated models | 0.70 | 0.70 | 0.30 | 68.8 | 73.9 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liyanage, C.; Wathupola, A.; Muraleetharan, S.; Perera, K.; Punyadeera, C.; Udagama, P. Promoter Hypermethylation of Tumor-Suppressor Genes p16INK4a, RASSF1A, TIMP3, and PCQAP/MED15 in Salivary DNA as a Quadruple Biomarker Panel for Early Detection of Oral and Oropharyngeal Cancers. Biomolecules 2019, 9, 148. https://doi.org/10.3390/biom9040148
Liyanage C, Wathupola A, Muraleetharan S, Perera K, Punyadeera C, Udagama P. Promoter Hypermethylation of Tumor-Suppressor Genes p16INK4a, RASSF1A, TIMP3, and PCQAP/MED15 in Salivary DNA as a Quadruple Biomarker Panel for Early Detection of Oral and Oropharyngeal Cancers. Biomolecules. 2019; 9(4):148. https://doi.org/10.3390/biom9040148
Chicago/Turabian StyleLiyanage, Chamikara, Asanga Wathupola, Sanjayan Muraleetharan, Kanthi Perera, Chamindie Punyadeera, and Preethi Udagama. 2019. "Promoter Hypermethylation of Tumor-Suppressor Genes p16INK4a, RASSF1A, TIMP3, and PCQAP/MED15 in Salivary DNA as a Quadruple Biomarker Panel for Early Detection of Oral and Oropharyngeal Cancers" Biomolecules 9, no. 4: 148. https://doi.org/10.3390/biom9040148
APA StyleLiyanage, C., Wathupola, A., Muraleetharan, S., Perera, K., Punyadeera, C., & Udagama, P. (2019). Promoter Hypermethylation of Tumor-Suppressor Genes p16INK4a, RASSF1A, TIMP3, and PCQAP/MED15 in Salivary DNA as a Quadruple Biomarker Panel for Early Detection of Oral and Oropharyngeal Cancers. Biomolecules, 9(4), 148. https://doi.org/10.3390/biom9040148