Effect of Protein Denaturation and Enzyme Inhibitors on Proteasomal-Mediated Production of Peptides in Human Embryonic Kidney Cells
Abstract
:1. Introduction
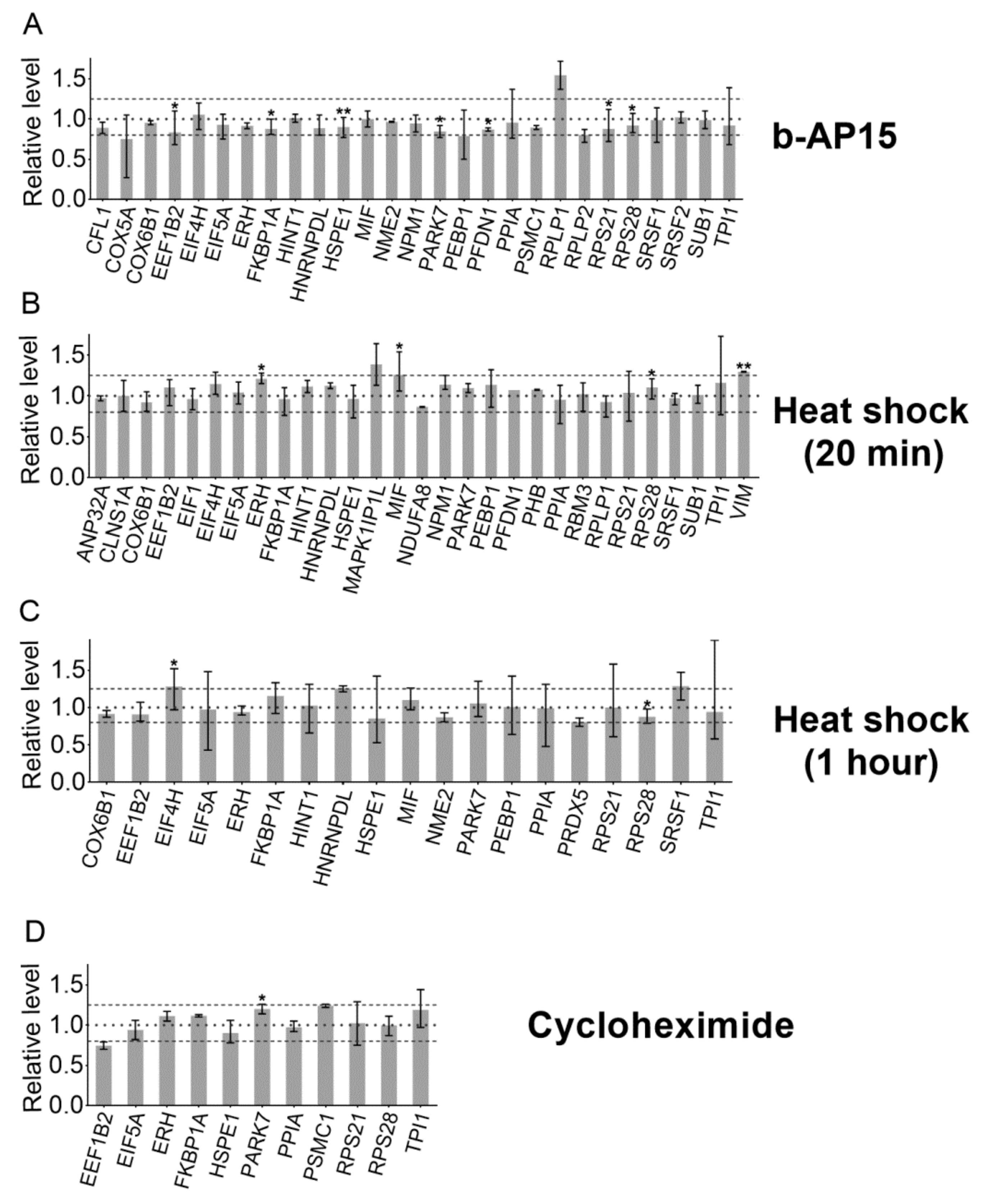
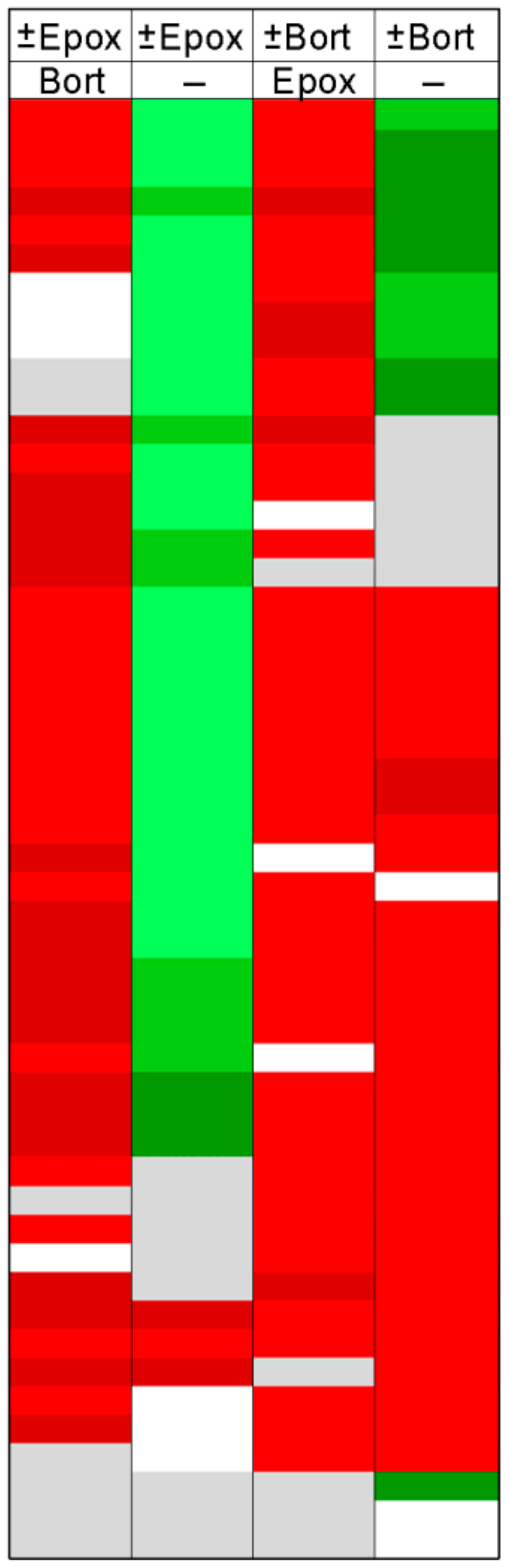
2. Results
Effect of Combinations of Proteasome Inhibitors on the Levels of Intracellular Peptides
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Methods
4.2.1. Treatment with b-AP15 and Western Blotting
4.2.2. Large-Scale Cell Culture, Induction of Heat Shock, Treatment with B-AP15, Cycloheximide and Proteasome Inhibitors, and Peptide Extraction
4.2.3. Isotopic Labeling and Mass Spectrometry
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Clynen, E.; Baggerman, G.; Veelaert, D.; Cerstiaens, A.; Van Der, H.D.; Harthoorn, L.; Derua, R.; Waelkens, E.; De Loof, A.; Schoofs, L. Peptidomics of the pars intercerebralis-corpus cardiacum complex of the migratory locust, Locusta migratoria. Eur. J. Biochem. 2001, 268, 1929–1939. [Google Scholar] [CrossRef] [PubMed]
- Schulz-Knappe, P.; Zucht, H.D.; Heine, G.; Jurgens, M.; Hess, R.; Schrader, M. Peptidomics: the comprehensive analysis of peptides in complex biological mixtures. Comb. Chem. High Throughput Screen. 2001, 4, 207–217. [Google Scholar] [CrossRef]
- Verhaert, P.; Uttenweiler-Joseph, S.; de Vries, M.; Loboda, A.; Ens, W.; Standing, K.G. Matrix-assisted laser desorption/ionization quadrupole time-of-flight mass spectrometry: An elegant tool for peptidomics. Proteomics 2001, 1, 118–131. [Google Scholar] [CrossRef]
- Che, F.Y.; Yan, L.; Li, H.; Mzhavia, N.; Devi, L.; Fricker, L.D. Identification of peptides from brain and pituitary of Cpe fat/Cpe fat mice. Proc. Natl. Acad. Sci. USA 2001, 98, 9971–9976. [Google Scholar] [CrossRef] [PubMed]
- Fricker, L.D. Analysis of mouse brain peptides using mass spectrometry-based peptidomics: Implications for novel functions ranging from non-classical neuropeptides to microproteins. Mol. Biosyst. 2010, 6, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Ferro, E.S.; Hyslop, S.; Camargo, A.C. Intracellullar peptides as putative natural regulators of protein interactions. J. Neurochem. 2004, 91, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Ferro, E.S.; Rioli, V.; Castro, L.M.; Fricker, L.D. Intracellular peptides: From discovery to function. EuPA Open Proteom. 2014, 3, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Haynes, C.M.; Yang, Y.; Blais, S.P.; Neubert, T.A.; Ron, D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol. Cell. 2010, 37, 529–540. [Google Scholar] [CrossRef]
- Kondo, T.; Plaza, S.; Zanet, J.; Benrabah, E.; Valenti, P.; Hashimoto, Y.; Kobayashi, S.; Payre, F.; Kageyama, Y. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 2010, 329, 336–339. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.L. Functions of the proteasome: From protein degradation and immune surveillance to cancer therapy. Biochem. Soc. Trans. 2007, 35, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The ubiquitin system for protein degradation. Annu. Rev. Biochem. 1992, 61, 761–807. [Google Scholar] [CrossRef]
- Collins, G.A.; Goldberg, A.L. The Logic of the 26S Proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadtmueller, B.M.; Hill, C.P. Proteasome Activators. Mol. Cell 2011, 41, 8–19. [Google Scholar] [CrossRef] [Green Version]
- Morozov, A.V.; Karpov, V.L. Biological consequences of structural and functional proteasome diversity. Heliyon 2018, 4, e00894. [Google Scholar] [CrossRef] [Green Version]
- Fischer, M.; Hilt, W.; Richter-Ruoff, B.; Gonen, H.; Ciechanover, A.; Wolf, D.H. The 26S proteasome of the yeast Saccharomyces cerevisiae. FEBS Lett. 1994, 355, 69–75. [Google Scholar] [CrossRef]
- Ben-Nissan, G.; Sharon, M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 2014, 4, 862–884. [Google Scholar] [CrossRef] [PubMed]
- Rock, K.L.; York, I.A.; Goldberg, A.L. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat. Immunol. 2004, 5, 670–677. [Google Scholar] [CrossRef]
- Rock, K.L.; York, I.A.; Saric, T.; Goldberg, A.L. Protein degradation and the generation of MHC class I-presented peptides. Adv. Immunol. 2002, 80, 1–70. [Google Scholar] [PubMed]
- Reits, E.; Neijssen, J.; Herberts, C.; Benckhuijsen, W.; Janssen, L.; Drijfhout, J.W.; Neefjes, J. A major role for TPPII in trimming proteasomal degradation products for MHC class I antigen presentation. Immunity 2004, 20, 495–506. [Google Scholar] [CrossRef]
- Gelman, J.S.; Sironi, J.; Castro, L.M.; Ferro, E.S.; Fricker, L.D. Peptidomic analysis of human cell lines. J. Prot. Res. 2011, 10, 1583–1592. [Google Scholar] [CrossRef]
- Fricker, L.D.; Gelman, J.S.; Castro, L.M.; Gozzo, F.C.; Ferro, E.S. Peptidomic analysis of HEK293T cells: Effect of the proteasome inhibitor epoxomicin on intracellular peptides. J. Prot. Res. 2012, 11, 1981–1990. [Google Scholar] [CrossRef] [PubMed]
- Kisselev, A.F.; Akopian, T.N.; Woo, K.M.; Goldberg, A.L. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 1999, 274, 3363–3371. [Google Scholar] [CrossRef]
- Cohen, S.; Lahav-Baratz, S.; Ciechanover, A. Two distinct ubiquitin-dependent mechanisms are involved in NF-kappaB p105 proteolysis. Biochem. Biophys. Res. Commun. 2006, 345, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Rape, M.; Jentsch, S. Productive RUPture: Activation of transcription factors by proteasomal processing. Biochim. Biophys. Acta 2004, 1695, 209–213. [Google Scholar] [CrossRef]
- Dasgupta, S.; Fishman, M.A.; Mahallati, H.; Castro, L.M.; Tashima, A.K.; Ferro, E.S.; Fricker, L.D. Reduced Levels of Proteasome Products in a Mouse Striatal Cell Model of Huntington’s Disease. PLoS ONE 2015, 10, e0145333. [Google Scholar] [CrossRef]
- Dasgupta, S.; Castro, L.M.; Dulman, R.; Yang, C.; Schmidt, M.; Ferro, E.S.; Fricker, L.D. Proteasome inhibitors alter levels of intracellular peptides in HEK293T and SH-SY5Y cells. PLoS ONE 2014, 9, e103604. [Google Scholar] [CrossRef]
- Gelman, J.S.; Sironi, J.; Berezniuk, I.; Dasgupta, S.; Castro, L.M.; Gozzo, F.C.; Ferro, E.S.; Fricker, L.D. Alterations of the intracellular peptidome in response to the proteasome inhibitor bortezomib. PLoS ONE 2013, 8, e53263. [Google Scholar] [CrossRef]
- Harris, J.L.; Alper, P.B.; Li, J.; Rechsteiner, M.; Backes, B.J. Substrate specificity of the human proteasome. Chem. Biol. 2001, 8, 1131–1141. [Google Scholar] [CrossRef] [Green Version]
- Wenzel, T.; Eckerskorn, C.; Lottspeich, F.; Baumeister, W. Existence of a molecular ruler in proteasomes suggested by analysis of degradation products. FEBS Lett. 1994, 349, 205–209. [Google Scholar] [CrossRef] [Green Version]
- Fricker, L.D. Limitations of Mass Spectrometry-Based Peptidomic Approaches. J. Am. Soc. Mass Spectrom. 2015, 26, 1981–1991. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, S.; Yang, C.; Castro, L.M.; Tashima, A.K.; Ferro, E.S.; Moir, R.D.; Willis, I.M.; Fricker, L.D. Analysis of the Yeast Peptidome and Comparison with the Human Peptidome. PLoS ONE 2016, 11, e0163312. [Google Scholar] [CrossRef]
- Thibaudeau, T.A.; Smith, D.M. A Practical Review of Proteasome Pharmacology. Pharmacol. Rev. 2019, 71, 170–197. [Google Scholar] [CrossRef] [Green Version]
- Tian, Z.; D’Arcy, P.; Wang, X.; Ray, A.; Tai, Y.T.; Hu, Y.; Carrasco, R.D.; Richardson, P.; Linder, S.; Chauhan, D.; Anderson, K.C. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood 2014, 123, 706–716. [Google Scholar] [CrossRef] [PubMed]
- D’Arcy, P.; Brnjic, S.; Olofsson, M.H.; Fryknas, M.; Lindsten, K.; De Cesare, M.; Perego, P.; Sadeghi, B.; Hassan, M.; Larsson, R.; Linder, S. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 2011, 17, 1636–1640. [Google Scholar] [CrossRef]
- Yewdell, J.W.; Nicchitta, C.V. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 2006, 27, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Yewdell, J.W.; Anton, L.C.; Bennink, J.R. Defective ribosomal products (DRiPs): A major source of antigenic peptides for MHC class I molecules? J. Immunol. 1996, 157, 1823–1826. [Google Scholar]
- Zang, Y.; Thomas, S.M.; Chan, E.T.; Kirk, C.J.; Freilino, M.L.; DeLancey, H.M.; Grandis, J.R.; Li, C.; Johnson, D.E. The next generation proteasome inhibitors carfilzomib and oprozomib activate prosurvival autophagy via induction of the unfolded protein response and ATF4. Autophagy 2012, 8, 1873–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, N.N.; Ng, A.H.; Measday, V.; Mayor, T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat. Cell. Biol. 2011, 13, 1344–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz, R.; Streller, F.; Scheel, A.H.; Ruschoff, J.; Reinert, M.C.; Dobbelstein, M.; Marchenko, N.D.; Moll, U.M. HER2/ErbB2 activates HSF1 and thereby controls HSP90 clients including MIF in HER2-overexpressing breast cancer. Cell Death Dis. 2014, 5, e980. [Google Scholar] [CrossRef] [PubMed]
- Lev, A.; Princiotta, M.F.; Zanker, D.; Takeda, K.; Gibbs, J.S.; Kumagai, C.; Waffarn, E.; Dolan, B.P.; Burgevin, A.; Van Endert, P.; et al. Compartmentalized MHC class I antigen processing enhances immunosurveillance by circumventing the law of mass action. Proc. Natl. Acad. Sci. USA 2010, 107, 6964–6969. [Google Scholar] [CrossRef] [Green Version]
- Berti, D.A.; Morano, C.; Russo, L.C.; Castro, L.M.; Cunha, F.M.; Zhang, X.; Sironi, J.; Klitzke, C.F.; Ferro, E.S.; Fricker, L.D. Analysis of intracellular substrates and products of thimet oligopeptidase (EC 3.4.24.15) in human embryonic kidney 293 cells. J. Biol. Chem. 2009, 284, 14105–14116. [Google Scholar] [CrossRef]
- Wardman, J.H.; Zhang, X.; Gagnon, S.; Castro, L.M.; Zhu, X.; Steiner, D.F.; Day, R.; Fricker, L.D. Analysis of peptides in prohormone convertase 1/3 null mouse brain using quantitative peptidomics. J. Neurochem. 2010, 114, 215–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, H.; Nanno, D.; Che, F.Y.; Zhu, X.; Salton, S.R.; Steiner, D.F.; Fricker, L.D.; Devi, L.A. Neuropeptide processing profile in mice lacking prohormone convertase-1. Biochemistry 2005, 44, 4939–4948. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Che, F.Y.; Peng, B.; Steiner, D.F.; Pintar, J.E.; Fricker, L.D. The role of prohormone convertase-2 in hypothalamic neuropeptide processing: A quantitative neuropeptidomic study. J. Neurochem. 2006, 98, 1763–1777. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Che, F.Y.; Berezniuk, I.; Sonmez, K.; Toll, L.; Fricker, L.D. Peptidomics of Cpe<fat/fat> mouse brain regions: Implications for neuropeptide processing. J. Neurochem. 2008, 107, 1596–1613. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Berezniuk, I.; Che, F.Y.; Parikh, R.; Biswas, R.; Pan, H.; Fricker, L.D. Altered neuropeptide processing in prefrontal cortex of Cpe mice: Implications for neuropeptide discovery. J. Neurochem. 2006, 96, 1169–1181. [Google Scholar] [CrossRef]
- Che, F.Y.; Biswas, R.; Fricker, L.D. Relative quantitation of peptides in wild type and Cpe<fat/fat> mouse pituitary using stable isotopic tags and mass spectrometry. J. Mass Spectrom. 2005, 40, 227–237. [Google Scholar] [CrossRef]
- Teixeira, C.M.M.; Correa, C.N.; Iwai, L.K.; Ferro, E.S.; Castro, L.M. Characterization of Intracellular Peptides from Zebrafish (Danio rerio) Brain. Zebrafish 2019, 16, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.H.; Goldberg, A.L.; Qiu, X.B. New insights into the role of the ubiquitin-proteasome pathway in the regulation of apoptosis. Chang Gung. Med. J. 2007, 30, 469–479. [Google Scholar]
- Bijur, G.N.; Davis, R.E.; Jope, R.S. Rapid activation of heat shock factor-1 DNA binding by H2O2 and modulation by glutathione in human neuroblastoma and Alzheimer’s disease cybrid cells. Brain Res. Mol. Brain Res. 1999, 71, 69–77. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Reuven, N.; Shaul, Y. Ubiquitin-independent p53 proteasomal degradation. Cell Death Differ. 2010, 17, 103–108. [Google Scholar] [CrossRef]
- Ngoc, L.V.; Wauquier, C.; Soin, R.; Bousbata, S.; Twyffels, L.; Kruys, V.; Gueydan, C. Rapid proteasomal degradation of posttranscriptional regulators of the TIS11/tristetraprolin family is induced by an intrinsically unstructured region independently of ubiquitination. Mol. Cell. Biol. 2014, 34, 4315–4328. [Google Scholar] [CrossRef]
- Schubert, U.; Anton, L.C.; Gibbs, J.; Norbury, C.C.; Yewdell, J.W.; Bennink, J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000, 404, 770–774. [Google Scholar] [CrossRef]
- Reits, E.A.; Vos, J.C.; Gromme, M.; Neefjes, J. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature 2000, 404, 774–778. [Google Scholar] [CrossRef] [PubMed]
- Dolan, B.P.; Sharma, A.A.; Gibbs, J.S.; Cunningham, T.J.; Bennink, J.R.; Yewdell, J.W. MHC class I antigen processing distinguishes endogenous antigens based on their translation from cellular vs. viral mRNA. Proc. Natl. Acad. Sci. USA 2012, 109, 7025–7030. [Google Scholar] [CrossRef] [Green Version]
- Dolan, B.P.; Li, L.; Veltri, C.A.; Ireland, C.M.; Bennink, J.R.; Yewdell, J.W. Distinct pathways generate peptides from defective ribosomal products for CD8+ T cell immunosurveillance. J. Immunol. 2011, 186, 2065–2072. [Google Scholar] [CrossRef]
- Princiotta, M.F.; Finzi, D.; Qian, S.B.; Gibbs, J.; Schuchmann, S.; Buttgereit, F.; Bennink, J.R.; Yewdell, J.W. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity 2003, 18, 343–354. [Google Scholar] [CrossRef]
- Meng, L.; Mohan, R.; Kwok, B.H.; Elofsson, M.; Sin, N.; Crews, C.M. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA 1999, 96, 10403–10408. [Google Scholar] [CrossRef]
- Harshbarger, W.; Miller, C.; Diedrich, C.; Sacchettini, J. Crystal structure of the human 20 S proteasome in complex with carfilzomib. Structure 2015, 23, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Huber, E.M.; Basler, M.; Schwab, R.; Heinemeyer, W.; Kirk, C.J.; Groettrup, M.; Groll, M. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 2012, 148, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Murata, S.; Takahama, Y.; Kasahara, M.; Tanaka, K. The immunoproteasome and thymoproteasome: Functions, evolution and human disease. Nat. Immunol. 2018, 19, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Osmulski, P.A.; Hochstrasser, M.; Gaczynska, M. A tetrahedral transition state at the active sites of the 20 S proteasome is coupled to opening of the alpha-ring channel. Structure 2009, 17, 1137–1147. [Google Scholar] [CrossRef]
- Cai, F.; Frey, J.U.; Sanna, P.P.; Behnisch, T. Protein degradation by the proteasome is required for synaptic tagging and the heterosynaptic stabilization of hippocampal late-phase long-term potentiation. Neuroscience 2010, 169, 1520–1526. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, K.V.; Margolis, S.S. A mammalian nervous-system-specific plasma membrane proteasome complex that modulates neuronal function. Nat. Struct. Mol. Biol. 2017, 24, 419–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Araujo, C.B.; Heimann, A.S.; Remer, R.A.; Russo, L.C.; Colquhoun, A.; Forti, F.L.; Ferro, E.S. Intracellular Peptides in Cell Biology and Pharmacology. Biomolecules 2019, 9, 150. [Google Scholar] [CrossRef] [PubMed]
- Morano, C.; Zhang, X.; Fricker, L.D. Multiple isotopic labels for quantitative mass spectrometry. Anal. Chem. 2008, 80, 9298–9309. [Google Scholar] [CrossRef] [PubMed]
- Wardman, J.; Fricker, L.D. Quantitative peptidomics of mice lacking Peptide-processing enzymes. Methods Mol. Biol. 2011, 768, 307–323. [Google Scholar]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dasgupta, S.; Fishman, M.A.; Castro, L.M.; Tashima, A.K.; Ferro, E.S.; Fricker, L.D. Effect of Protein Denaturation and Enzyme Inhibitors on Proteasomal-Mediated Production of Peptides in Human Embryonic Kidney Cells. Biomolecules 2019, 9, 207. https://doi.org/10.3390/biom9060207
Dasgupta S, Fishman MA, Castro LM, Tashima AK, Ferro ES, Fricker LD. Effect of Protein Denaturation and Enzyme Inhibitors on Proteasomal-Mediated Production of Peptides in Human Embryonic Kidney Cells. Biomolecules. 2019; 9(6):207. https://doi.org/10.3390/biom9060207
Chicago/Turabian StyleDasgupta, Sayani, Michael A. Fishman, Leandro M. Castro, Alexandre K. Tashima, Emer S. Ferro, and Lloyd D. Fricker. 2019. "Effect of Protein Denaturation and Enzyme Inhibitors on Proteasomal-Mediated Production of Peptides in Human Embryonic Kidney Cells" Biomolecules 9, no. 6: 207. https://doi.org/10.3390/biom9060207
APA StyleDasgupta, S., Fishman, M. A., Castro, L. M., Tashima, A. K., Ferro, E. S., & Fricker, L. D. (2019). Effect of Protein Denaturation and Enzyme Inhibitors on Proteasomal-Mediated Production of Peptides in Human Embryonic Kidney Cells. Biomolecules, 9(6), 207. https://doi.org/10.3390/biom9060207





