The Role of a Key Amino Acid Position in Species-Specific Proteinaceous dUTPase Inhibition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site-Directed Mutagenesis
2.2. Molecular Cloning, Protein Expression and Purification
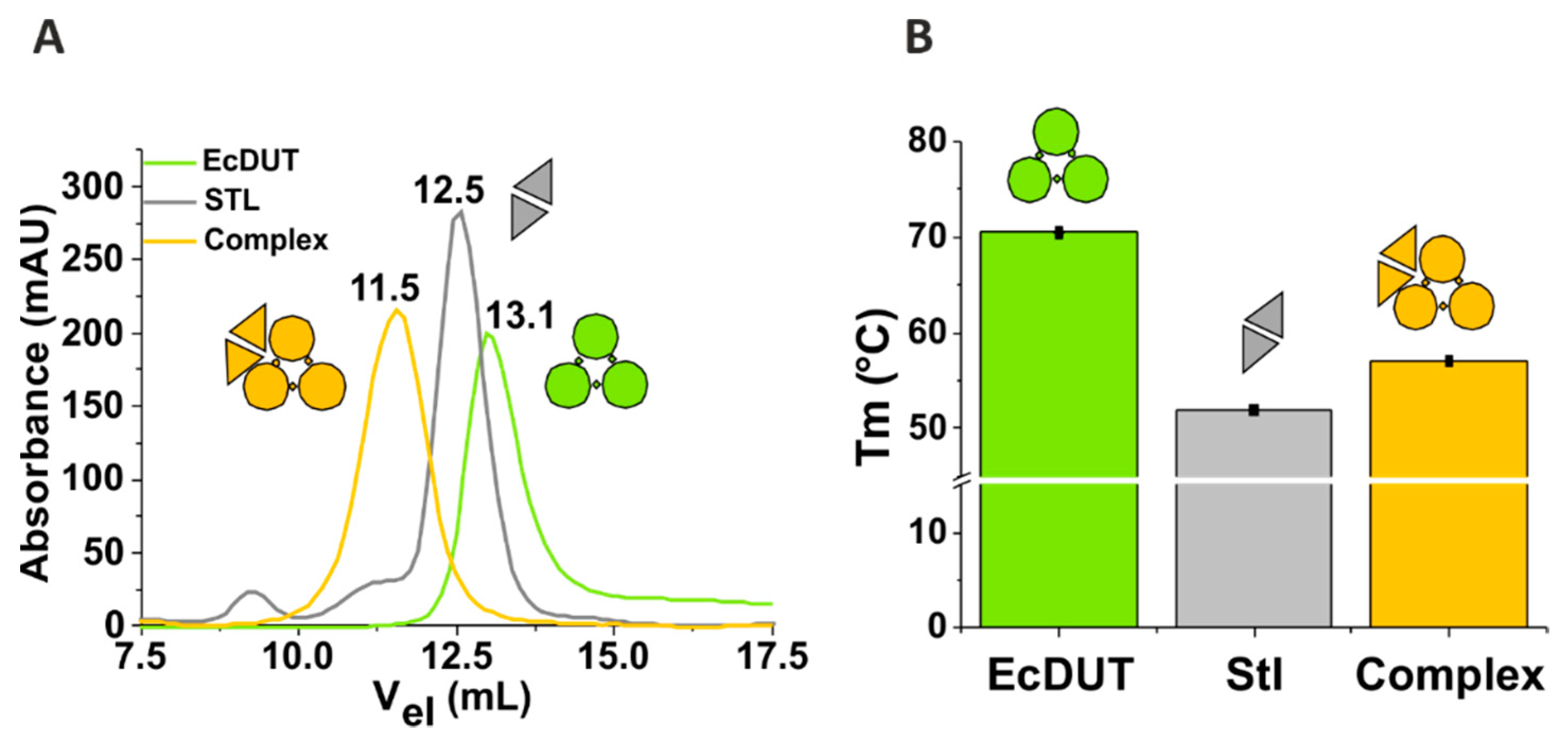
2.3. Size Exclusion Chromatography
2.4. Differential Scanning Fluorimetry (Thermofluor)
2.5. Multiple Sequence Alignment Based Mutational Screen
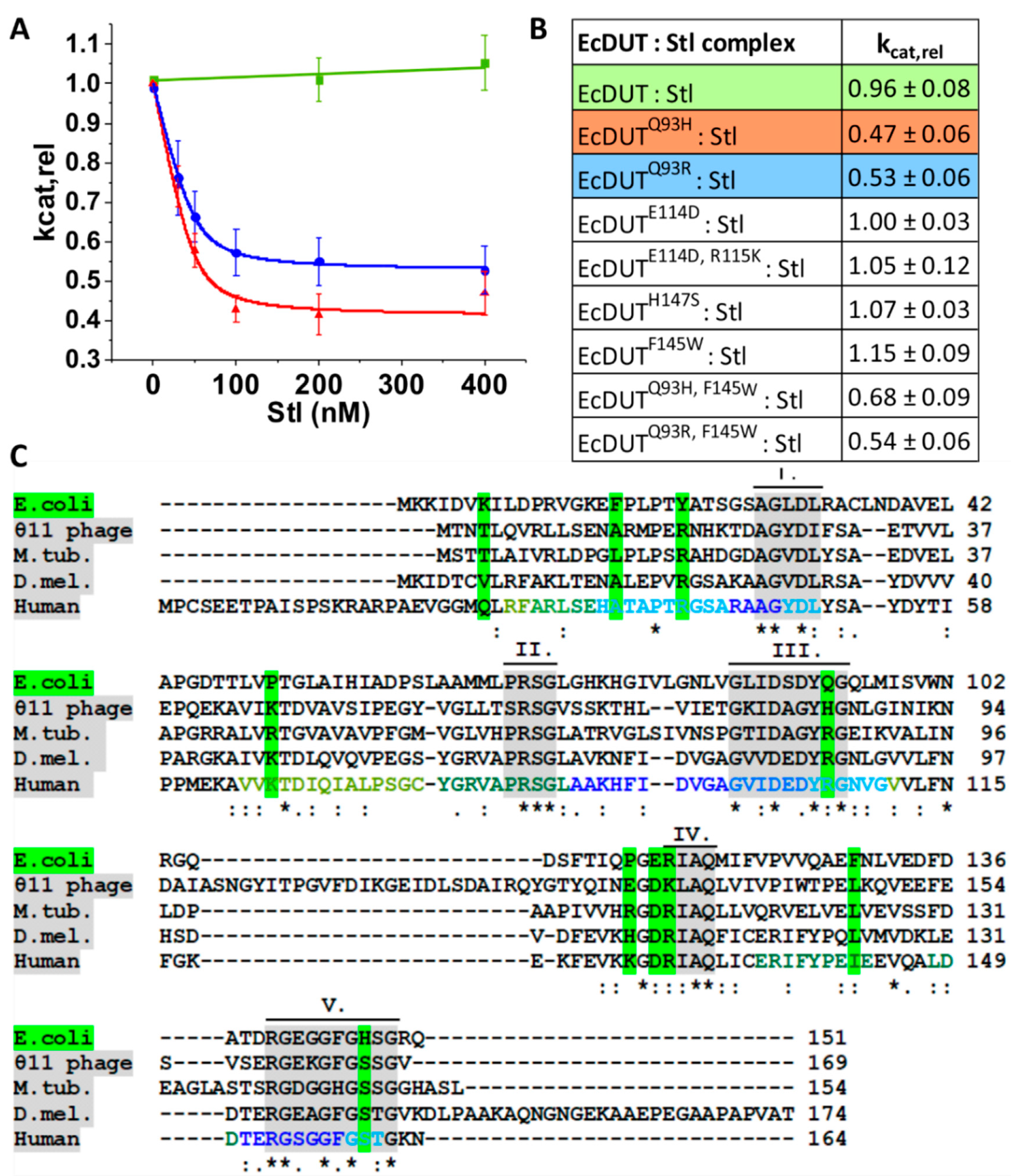
2.6. Measurement of Steady-State Enzyme Activity and Inhibition
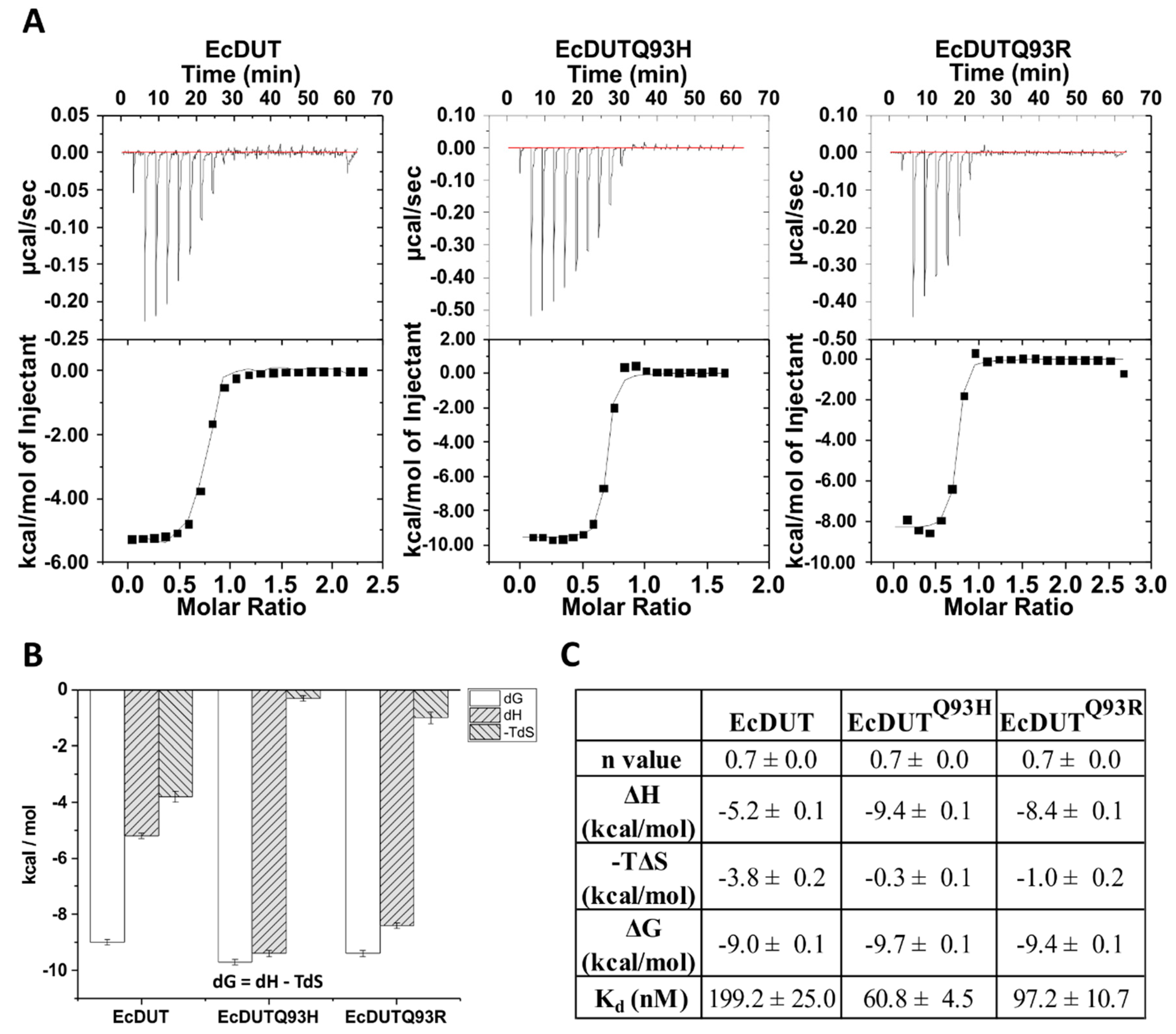
2.7. Isothermal Titration Calorimetry
2.8. Protein Crystallization and Structural Refinement
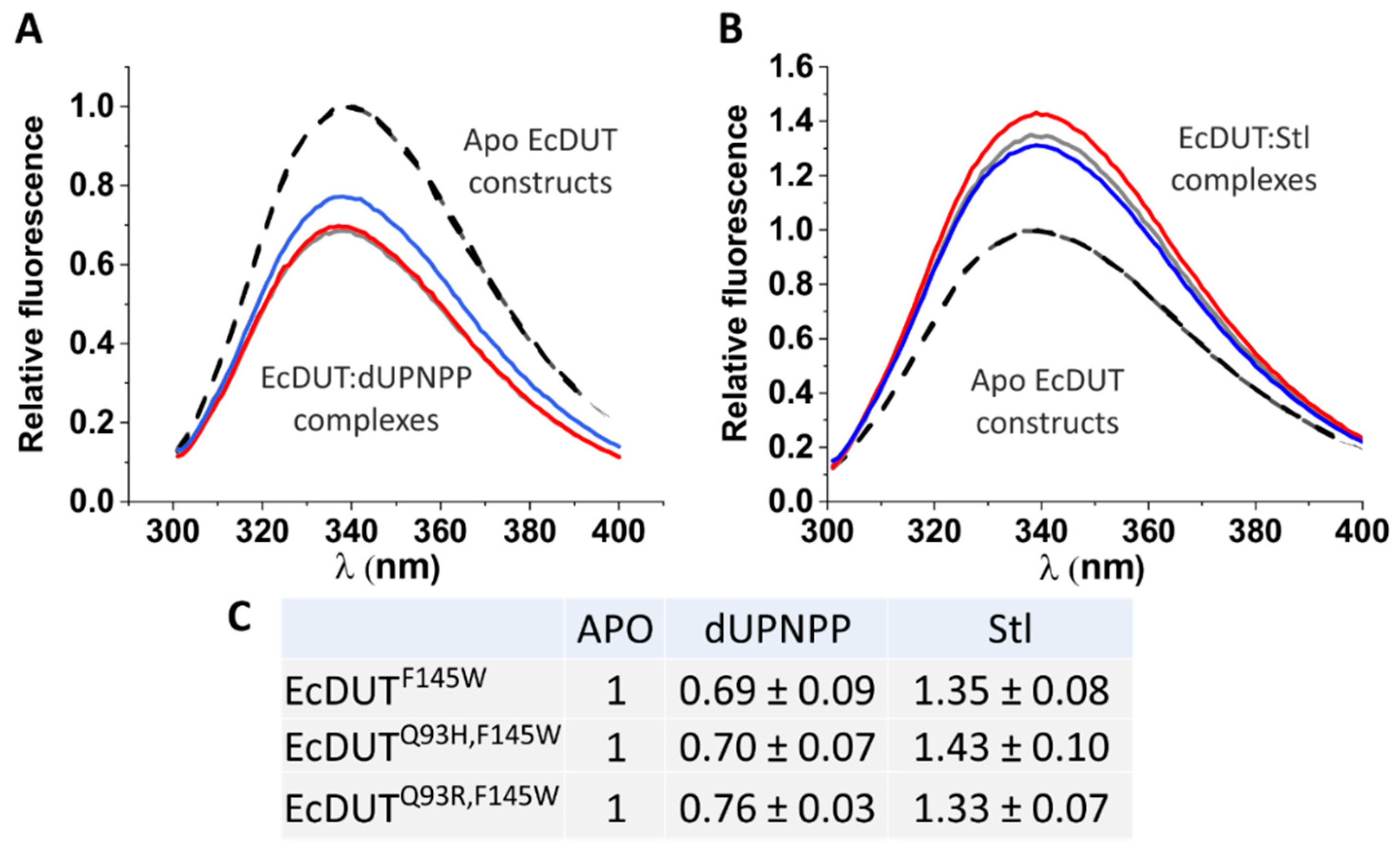
2.9. Tryptophan Fluorimetry
2.10. Acrylamide Quenching and Statistical Analysis
3. Results and discussion
3.1. Stl Binds to Escherichia coli dUTPase but Does Not Show Inhibitory Potential in Steady-State Kinetic Experiments
3.2. Searching for Key Residues in Inhibition and Identification of Such a Position
3.3. Understanding the Structural Background of Inhibitablity
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nagy, G.N.; Leveles, I.; Vertessy, B.G. Preventive DNA repair by sanitizing the cellular (deoxy)nucleoside triphosphate pool. FEBS J. 2014, 281, 4207–4223. [Google Scholar] [CrossRef] [PubMed]
- Vertessy, B.G.; Toth, J. Keeping uracil out of DNA: Physiological role, structure and catalytic mechanism of dUTPases. Acc. Chem. Res. 2009, 42, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Bjoras, M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef] [PubMed]
- Palinkas, H.L.; Racz, G.A.; Gal, Z.; Hoffmann, O.I.; Tihanyi, G.; Rona, G.; Gocza, E.; Hiripi, L.; Vertessy, B.G. CRISPR/Cas9-Mediated Knock-Out of dUTPase in Mice Leads to Early Embryonic Lethality. Biomolecules 2019, 9, 136. [Google Scholar] [CrossRef] [PubMed]
- Gadsden, M.H.; McIntosh, E.M.; Game, J.C.; Wilson, P.J.; Haynes, R.H. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 1993, 12, 4425–4431. [Google Scholar] [CrossRef] [PubMed]
- el-Hajj, H.H.; Zhang, H.; Weiss, B. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 1988, 170, 1069–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecsi, I.; Hirmondo, R.; Brown, A.C.; Lopata, A.; Parish, T.; Vertessy, B.G.; Toth, J. The dUTPase enzyme is essential in Mycobacterium smegmatis. PLoS ONE 2012, 7, e37461. [Google Scholar] [CrossRef]
- Muha, V.; Horvath, A.; Bekesi, A.; Pukancsik, M.; Hodoscsek, B.; Merenyi, G.; Rona, G.; Batki, J.; Kiss, I.; Jankovics, F.; et al. Uracil-containing DNA in Drosophila: Stability, stage-specific accumulation, and developmental involvement. PLoS Genet. 2012, 8, e1002738. [Google Scholar] [CrossRef]
- Horvath, A.; Bekesi, A.; Muha, V.; Erdelyi, M.; Vertessy, B.G. Expanding the DNA alphabet in the fruit fly: Uracil enrichment in genomic DNA. Fly 2013, 7, 23–27. [Google Scholar] [CrossRef]
- Wilson, P.M.; Danenberg, P.V.; Johnston, P.G.; Lenz, H.J.; Ladner, R.D. Standing the test of time: Targeting thymidylate biosynthesis in cancer therapy. Nat. Rev. Clin. Oncol. 2014, 11, 282–298. [Google Scholar] [CrossRef]
- Wilson, P.M.; LaBonte, M.J.; Lenz, H.J.; Mack, P.C.; Ladner, R.D. Inhibition of dUTPase induces synthetic lethality with thymidylate synthase-targeted therapies in non-small cell lung cancer. Mol. Cancer 2012, 11, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Pearl, L.H.; Savva, R. The problem with pyrimidines. Nat. Struct. Biol. 1996, 3, 485–487. [Google Scholar] [CrossRef] [PubMed]
- Doi, T.; Yoh, K.; Shitara, K.; Takahashi, H.; Ueno, M.; Kobayashi, S.; Morimoto, M.; Okusaka, T.; Ueno, H.; Morizane, C.; et al. First-in-human phase 1 study of novel dUTPase inhibitor TAS-114 in combination with S-1 in Japanese patients with advanced solid tumors. Investig. New Drugs 2018. [Google Scholar] [CrossRef] [PubMed]
- Hirmondo, R.; Lopata, A.; Suranyi, E.V.; Vertessy, B.G.; Toth, J. Differential control of dNTP biosynthesis and genome integrity maintenance by the dUTPase superfamily enzymes. Sci. Rep. 2017, 7, 6043. [Google Scholar] [CrossRef] [PubMed]
- Giovannetti, E.; Zucali, P.A.; Rolfo, C.; Assaraf, Y.G.; Peters, G.J. Prognostic and Predictive Roles of Thymidylate Synthase Expression in Lung Cancer: The Debate Is Still Open. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016, 34, 511–512. [Google Scholar] [CrossRef] [PubMed]
- Hagner, N.; Joerger, M. Cancer chemotherapy: Targeting folic acid synthesis. Cancer Manag. Res. 2010, 2, 293–301. [Google Scholar]
- Yao, W.; Zhu, S.; Li, P.; Zhang, S. Large tumor suppressor kinase 2 overexpression attenuates 5-FU-resistance in colorectal cancer via activating the JNK-MIEF1-mitochondrial division pathway. Cancer Cell Int. 2019, 19, 97. [Google Scholar] [CrossRef]
- Whittingham, J.L.; Leal, I.; Nguyen, C.; Kasinathan, G.; Bell, E.; Jones, A.F.; Berry, C.; Benito, A.; Turkenburg, J.P.; Dodson, E.J.; et al. dUTPase as a platform for antimalarial drug design: Structural basis for the selectivity of a class of nucleoside inhibitors. Structure 2005, 13, 329–338. [Google Scholar] [CrossRef]
- Nyiri, K.; Vertessy, B.G. Perturbation of genome integrity to fight pathogenic microorganisms. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 3593–3612. [Google Scholar] [CrossRef]
- Szabo, J.E.; Nemeth, V.; Papp-Kadar, V.; Nyiri, K.; Leveles, I.; Bendes, A.A.; Zagyva, I.; Rona, G.; Palinkas, H.L.; Besztercei, B.; et al. Highly potent dUTPase inhibition by a bacterial repressor protein reveals a novel mechanism for gene expression control. Nucleic Acids Res. 2014, 42, 11912–11920. [Google Scholar] [CrossRef] [Green Version]
- Tormo-Mas, M.A.; Mir, I.; Shrestha, A.; Tallent, S.M.; Campoy, S.; Lasa, I.; Barbe, J.; Novick, R.P.; Christie, G.E.; Penades, J.R. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature 2010, 465, 779–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirmondo, R.; Szabo, J.E.; Nyiri, K.; Tarjanyi, S.; Dobrotka, P.; Toth, J.; Vertessy, B.G. Cross-species inhibition of dUTPase via the Staphylococcal Stl protein perturbs dNTP pool and colony formation in Mycobacterium. DNA Repair 2015, 30, 21–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyiri, K.; Mertens, H.D.T.; Tihanyi, B.; Nagy, G.N.; Kohegyi, B.; Matejka, J.; Harris, M.J.; Szabo, J.E.; Papp-Kadar, V.; Nemeth-Pongracz, V.; et al. Structural model of human dUTPase in complex with a novel proteinaceous inhibitor. Sci. Rep. 2018, 8, 4326. [Google Scholar] [CrossRef] [PubMed]
- Benedek, A.; Poloskei, I.; Ozohanics, O.; Vekey, K.; Vertessy, B.G. The Stl repressor from Staphylococcus aureus is an efficient inhibitor of the eukaryotic fruitfly dUTPase. FEBS Open Bio 2018, 8, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Nyiri, K.; Kohegyi, B.; Micsonai, A.; Kardos, J.; Vertessy, B.G. Evidence-Based Structural Model of the Staphylococcal Repressor Protein: Separation of Functions into Different Domains. PLoS ONE 2015, 10, e0139086. [Google Scholar] [CrossRef] [PubMed]
- Barabas, O.; Pongracz, V.; Kovari, J.; Wilmanns, M.; Vertessy, B.G. Structural insights into the catalytic mechanism of phosphate ester hydrolysis by dUTPase. J. Biol. Chem. 2004, 279, 42907–42915. [Google Scholar] [CrossRef] [PubMed]
- Maiques, E.; Quiles-Puchalt, N.; Donderis, J.; Ciges-Tomas, J.R.; Alite, C.; Bowring, J.Z.; Humphrey, S.; Penades, J.R.; Marina, A. Another look at the mechanism involving trimeric dUTPases in Staphylococcus aureus pathogenicity island induction involves novel players in the party. Nucleic Acids Res. 2016, 44, 5457–5469. [Google Scholar] [CrossRef]
- Leveles, I.; Nemeth, V.; Szabo, J.E.; Harmat, V.; Nyiri, K.; Bendes, A.A.; Papp-Kadar, V.; Zagyva, I.; Rona, G.; Ozohanics, O.; et al. Structure and enzymatic mechanism of a moonlighting dUTPase. Acta Cryst. D Biol. Cryst. 2013, 69, 2298–2308. [Google Scholar] [CrossRef] [Green Version]
- Varga, B.; Barabas, O.; Kovari, J.; Toth, J.; Hunyadi-Gulyas, E.; Klement, E.; Medzihradszky, K.F.; Tolgyesi, F.; Fidy, J.; Vertessy, B.G. Active site closure facilitates juxtaposition of reactant atoms for initiation of catalysis by human dUTPase. FEBS Lett. 2007, 581, 4783–4788. [Google Scholar] [CrossRef] [Green Version]
- Varga, B.; Barabas, O.; Takacs, E.; Nagy, N.; Nagy, P.; Vertessy, B.G. Active site of mycobacterial dUTPase: Structural characteristics and a built-in sensor. Biochem. Biophys. Res. Commun. 2008, 373, 8–13. [Google Scholar] [CrossRef]
- Liu, H.; Naismith, J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008, 8, 91. [Google Scholar] [CrossRef] [PubMed]
- Benedek, A.; Horvath, A.; Hirmondo, R.; Ozohanics, O.; Bekesi, A.; Modos, K.; Revesz, A.; Vekey, K.; Nagy, G.N.; Vertessy, B.G. Potential steps in the evolution of a fused trimeric all-beta dUTPase involve a catalytically competent fused dimeric intermediate. FEBS J. 2016, 283, 3268–3286. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef]
- Winn, M.D.; Isupov, M.N.; Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D, Biol. Crystallogr. 2001, 57, 122–133. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System; Version 2.1.0; Schrödinger, LLC: New York, NY, USA.
- Lakowicz, J. Principles of Fluorescence Spectroscopy, 2nd ed.; Springer: New York, NY, USA, 1999; pp. 237–251; 291–306. [Google Scholar]
- Toth, J.; Varga, B.; Kovacs, M.; Malnasi-Csizmadia, A.; Vertessy, B.G. Kinetic mechanism of human dUTPase, an essential nucleotide pyrophosphatase enzyme. J. Biol. Chem. 2007, 282, 33572–33582. [Google Scholar] [CrossRef] [PubMed]
- Lopata, A.; Leveles, I.; Bendes, A.A.; Viskolcz, B.; Vertessy, B.G.; Jojart, B.; Toth, J. A Hidden Active Site in the Potential Drug Target Mycobacterium tuberculosis dUTPase Is Accessible through Small Amplitude Protein Conformational Changes. J. Biol. Chem. 2016, 291, 26320–26331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matulis, D.; Kranz, J.K.; Salemme, F.R.; Todd, M.J. Thermodynamic stability of carbonic anhydrase: Measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry 2005, 44, 5258–5266. [Google Scholar] [CrossRef] [PubMed]
- Pecsi, I.; Leveles, I.; Harmat, V.; Vertessy, B.G.; Toth, J. Aromatic stacking between nucleobase and enzyme promotes phosphate ester hydrolysis in dUTPase. Nucleic Acids Res 2010, 38, 7179–7186. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Q93H _FW | 5’ GATCGATTCTGACTATCATGGCCAGTTGATGATTTCC 3’ |
| Q93H_REV | 5’ GGAAATCATCAACTGGCCATGATAGTCAGAATCGATC 3’ |
| Q93R_fw | 5’ GACTATCGGGGCCAGTTGATGATTTCCGTGTGG 3’ |
| Q93R_rev | 5’ CTGGCCCCGATAGTCAGAATCGATCAATCCTACCAGG 3’ |
| E114D_fw | 5’ CATTCAACCTGGCGATCGCATCGCCCAG 3’ |
| E114D_rev | 5’ CTGGGCGATGCGATCGCCAGGTTGAATG 3’ |
| R115K_fw | 5’ CCTGGCGAAAAAATCGCCCAGATGATTTTTGTTCCGGTAGTACAGGCTGAATTTAATCTGGTGG 3’ |
| R115K_rev | 5’ CTGGGCGATTTTTTCGCCAGGTTGAATGGTGAAGCTGTCCTGACCACGG 3’ |
| ER114-5DK_fw | 5’ CAACCTGGCGATAAAATCGCCCAGATGATTTTTGTTCCGGTAGTACAGGCTGAATTTAATCTGGTGG 3’ |
| ER114-5DK_rev | 5’ CATCTGGGCGATTTTATCGCCAGGTTGAATGGTGAAGCTGTCCTGACCACGG 3’ |
| F145W_fw | 5’ GAAGGCGGCTGGGGTCACTCTGGTCGTCAGTAACACATACGGATCCGGC 3’ |
| F145W_rev | 5’ AGAGTGACCCCAGCCGCCTTCACCGCGGTCGGTGGCGTC 3’ |
| H147S_fw | 5’ CCGCGGTGAAGGCGGCTTTGGTAGCTCTGGTCGTCAGTAACAC 3’ |
| H147S_rev | 5’ GTGTTACTGACGACCAGAGCTACCAAAGCCGCCTTCACCGCGG 3’ |
| Data Collection | Parameters |
|---|---|
| Space group | P 21 21 21 |
| Unit-cell parameters (A°) | a = 63.20, b = 66.50, c = 95.30 |
| Unit-cell parameters (angles) | α = β = γ = 90° |
| Resolution range (A°) | 45.811-1.8 |
| Total No. of reflections | 113402 |
| No. of unique reflections | 36491 |
| Completeness (%) | 99.2 |
| <I/σ(I)> | 1.64 (at 1.82 Å) |
| Rmeas | 0.043 |
| Refinement | |
| No. of dUTPase subunits in asymmetric unit | 3 |
| No. of protein atoms | 3257 |
| No. of ligand atoms | 84 |
| No. of waters | 162 |
| No. of Mg2+ ions | 3 |
| Rcryst/Rfree‡ | 0.1806/0.2209 |
| Average B factors (Å2) (all atoms) | 33.0 |
| Wilson B factor (Å2) | 27.4 |
| Protein atoms | 3257 |
| Ligand atoms | 84 |
| Water | 162 |
| Mg2+ ions | 3 |
| R.m.s. deviations from ideal values | |
| Bond lengths (A°) | 0.007 |
| Bond angles (°) | 1.01 |
| Ramachandan plot analysis, residues in (%) | |
| Favoured region | 96.97 |
| Allowed region | 3.03 |
| Disallowed region | 0.00 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benedek, A.; Temesváry-Kis, F.; Khatanbaatar, T.; Leveles, I.; Surányi, É.V.; Szabó, J.E.; Wunderlich, L.; Vértessy, B.G. The Role of a Key Amino Acid Position in Species-Specific Proteinaceous dUTPase Inhibition. Biomolecules 2019, 9, 221. https://doi.org/10.3390/biom9060221
Benedek A, Temesváry-Kis F, Khatanbaatar T, Leveles I, Surányi ÉV, Szabó JE, Wunderlich L, Vértessy BG. The Role of a Key Amino Acid Position in Species-Specific Proteinaceous dUTPase Inhibition. Biomolecules. 2019; 9(6):221. https://doi.org/10.3390/biom9060221
Chicago/Turabian StyleBenedek, András, Fanni Temesváry-Kis, Tamjidmaa Khatanbaatar, Ibolya Leveles, Éva Viola Surányi, Judit Eszter Szabó, Lívius Wunderlich, and Beáta G. Vértessy. 2019. "The Role of a Key Amino Acid Position in Species-Specific Proteinaceous dUTPase Inhibition" Biomolecules 9, no. 6: 221. https://doi.org/10.3390/biom9060221
APA StyleBenedek, A., Temesváry-Kis, F., Khatanbaatar, T., Leveles, I., Surányi, É. V., Szabó, J. E., Wunderlich, L., & Vértessy, B. G. (2019). The Role of a Key Amino Acid Position in Species-Specific Proteinaceous dUTPase Inhibition. Biomolecules, 9(6), 221. https://doi.org/10.3390/biom9060221




