The Administration of Chitosan-Tripolyphosphate-DNA Nanoparticles to Express Exogenous SREBP1a Enhances Conversion of Dietary Carbohydrates into Lipids in the Liver of Sparus aurata
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Chitosan-TPP-DNA Nanoparticles
2.3. Enzyme Activity Assays and Metabolites
2.4. Reverse Transcriptase-Coupled Quantitative Real Time PCR (RT-qPCR)
2.5. Western Blot
2.6. Obtention of Aqueous and Organic Fractions from the Liver
2.7. NMR Analysis and Liver Metabolite Identification
2.8. Fatty Acid Methyl Ester (FAME) Analysis
2.9. Statistics
3. Results
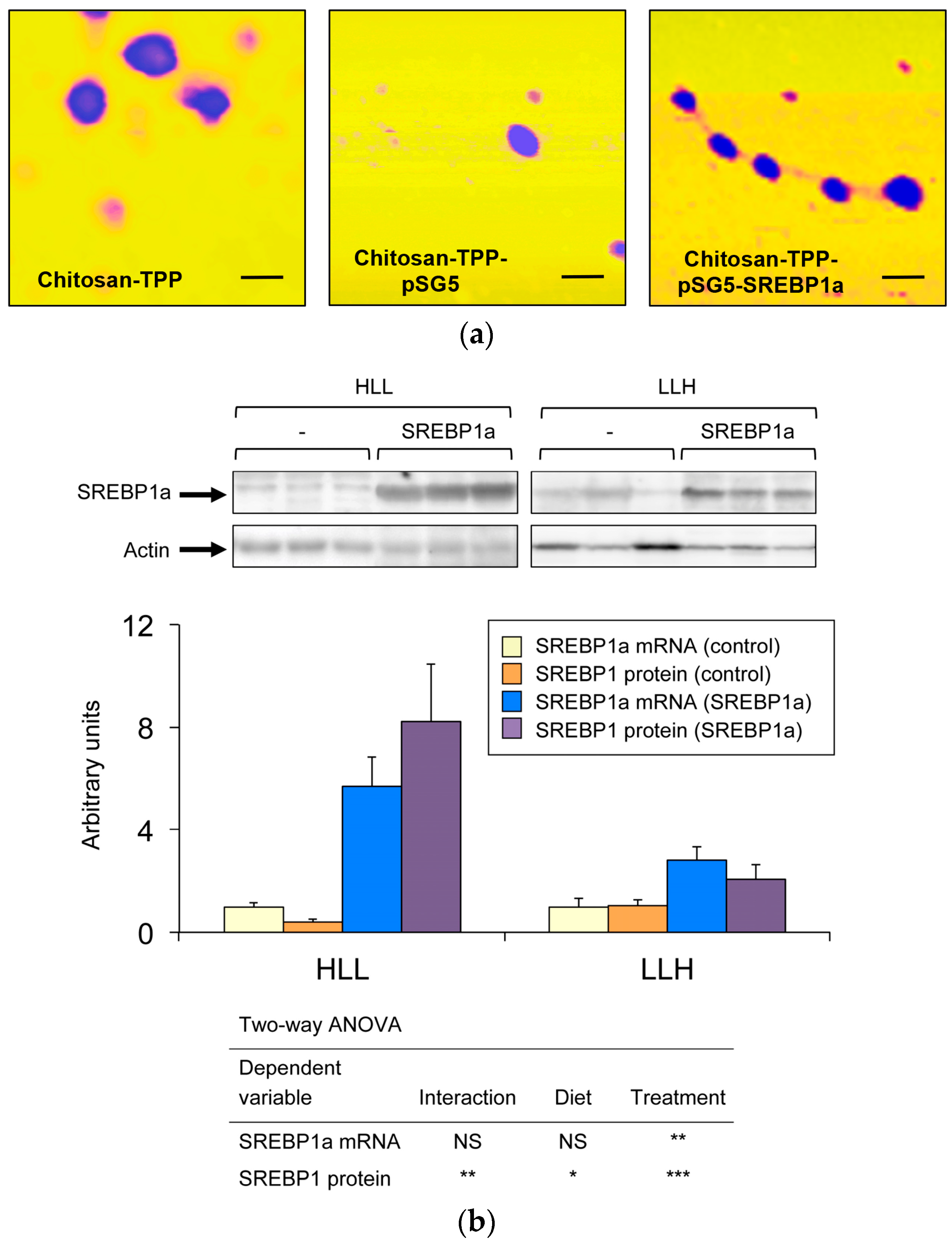
3.1. Intraperitoneal Administration of Chitosan-TPP-pSG5-SREBP1a Increases mRNA and Immunodetectable Levels of SREBP1a in the Liver
3.2. Effect of SREBP1a Overexpression on Serum and Aqueous Metabolites in the Liver
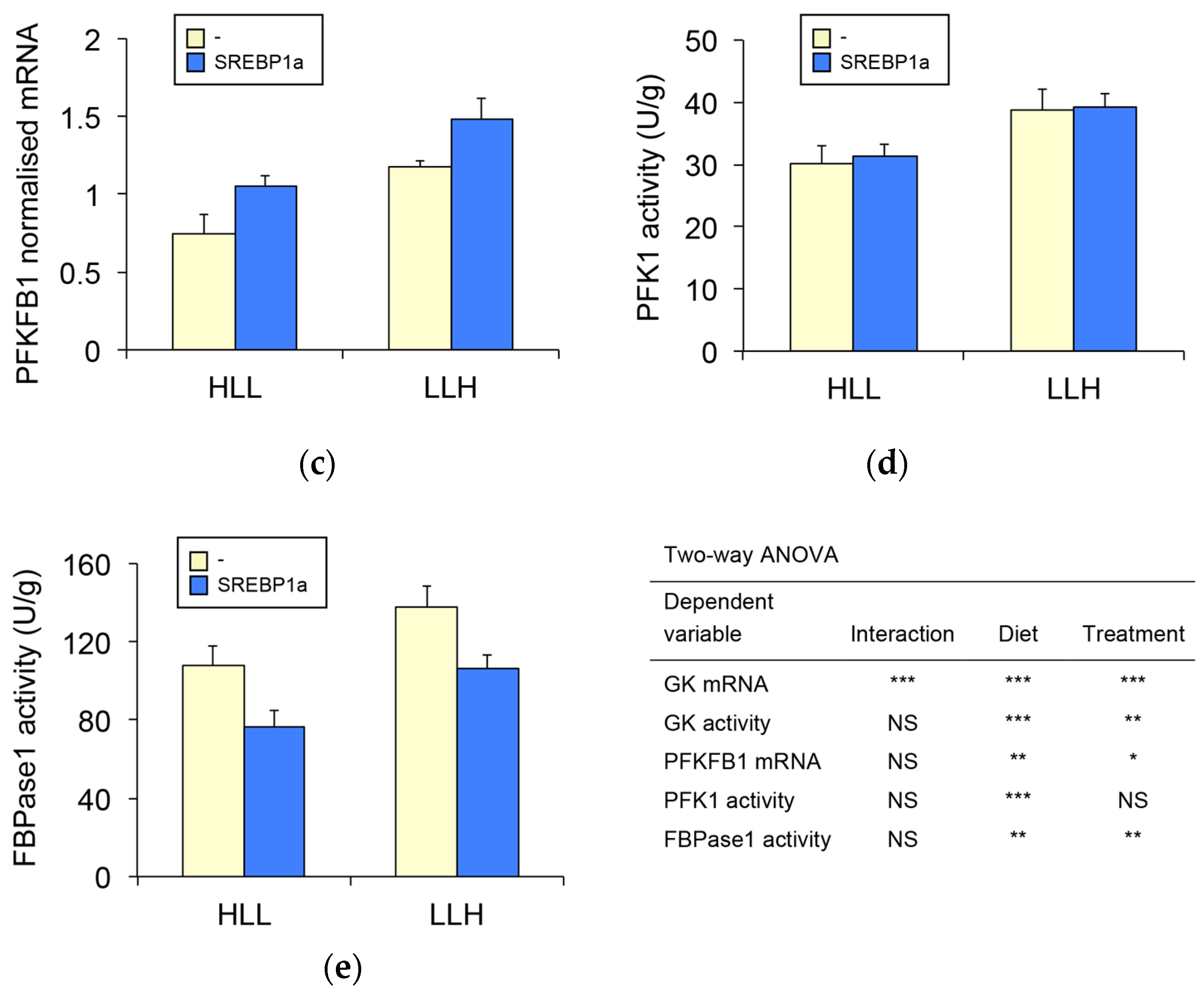
3.3. Effect of SREBP1a Overexpression on Hepatic Glycolysis-Gluconeogenesis
3.4. SREBP1a Overexpression Enhances Hepatic Biosynthesis of Fatty Acids and Cholesterol
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- DeBose-Boyd, R.A.; Ye, J. SREBPs in lipid metabolism, insulin signaling, and beyond. Trends Biochem. Sci. 2018, 43, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Bertolio, R.; Napoletano, F.; Mano, M.; Maurer-Stroh, S.; Fantuz, M.; Zannini, A.; Bicciato, S.; Sorrentino, G.; Del Sal, G. Sterol regulatory element binding protein 1 couples mechanical cues and lipid metabolism. Nat. Commun. 2019, 10, 1326. [Google Scholar] [CrossRef] [PubMed]
- Edwards, P.A.; Tabor, D.; Kast, H.R.; Venkateswaran, A. Regulation of gene expression by SREBP and SCAP. Biochim. et Biophys. Acta(BBA) - Mol. Cell Biol. Lipids 2000, 1529, 103–113. [Google Scholar] [CrossRef]
- Inoue, J.; Sato, R. New insights into the activation of sterol regulatory element-binding proteins by proteolytic processing. Biomol. Concepts 2013, 4, 417–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, M.S.; Goldstein, J.L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997, 89, 331–340. [Google Scholar] [CrossRef]
- Horton, J.D.; Shah, N.A.; Warrington, J.A.; Anderson, N.N.; Park, S.W.; Brown, M.S.; Goldstein, J.L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA 2003, 100, 12027–12032. [Google Scholar] [CrossRef] [Green Version]
- Shimano, H.; Sato, R. SREBP-regulated lipid metabolism: Convergent physiology - divergent pathophysiology. Nat. Rev. Endocrinol. 2017, 13, 710–730. [Google Scholar] [CrossRef]
- Harada, N.; Yonemoto, H.; Yoshida, M.; Yamamoto, H.; Yin, Y.; Miyamoto, A.; Hattori, A.; Wu, Q.; Nakagawa, T.; Nakano, M.; et al. Alternative splicing produces a constitutively active form of human SREBP-1. Biochem. Biophys. Res. Commun. 2008, 368, 820–826. [Google Scholar] [CrossRef]
- Felder, T.K.; Klein, K.; Patsch, W.; Oberkofler, H. A novel SREBP-1 splice variant: Tissue abundance and transactivation potency. Biochim. et Biophys. Acta(BBA) - Gene Struct. Expr. 2005, 1731, 41–47. [Google Scholar] [CrossRef]
- Wang, H.; Liu, F.; Millette, C.F.; Kilpatrick, D.L. Expression of a novel, sterol-insensitive form of sterol regulatory element binding protein 2 (SREBP2) in male germ cells suggests important cell- and stage-specific functions for SREBP targets during spermatogenesis. Mol. Cell. Biol. 2002, 22, 8478–8490. [Google Scholar] [CrossRef]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 2002, 109, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Pai, J.T.; Guryev, O.; Brown, M.S.; Goldstein, J.L. Differential stimulation of cholesterol and unsaturated fatty acid biosynthesis in cells expressing individual nuclear sterol regulatory element-binding proteins. J. Biol. Chem. 1998, 273, 26138–26148. [Google Scholar] [CrossRef] [PubMed]
- Shimano, H.; Horton, J.D.; Shimomura, I.; Hammer, R.E.; Brown, M.S.; Goldstein, J.L. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Investig. 1997, 99, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.D.; Bashmakov, Y.; Shimomura, I.; Shimano, H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc. Natl. Acad. Sci. USA 1998, 95, 5987–5992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foretz, M.; Guichard, C.; Ferre, P.; Foufelle, F. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc. Natl. Acad. Sci. USA 1999, 96, 12737–12742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gosmain, Y.; Lefai, E.; Ryser, S.; Roques, M.; Vidal, H. Sterol regulatory element-binding protein-1 mediates the effect of insulin on hexokinase II gene expression in human muscle cells. Diabetes 2004, 53, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, H.I.; Kim, T.H.; Im, S.S.; Park, S.K.; Lee, I.K.; Kim, K.S.; Ahn, Y.H. SREBP-1c mediates the insulin-dependent hepatic glucokinase expression. J. Biol. Chem. 2004, 279, 30823–30829. [Google Scholar] [CrossRef]
- Gosmain, Y.; Dif, N.; Berbe, V.; Loizon, E.; Rieusset, J.; Vidal, H.; Lefai, E. Regulation of SREBP-1 expression and transcriptional action on HKII and FAS genes during fasting and refeeding in rat tissues. J. Lipid Res. 2005, 46, 697–705. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.; Shimano, H.; Nakagawa, Y.; Ide, T.; Yahagi, N.; Matsuzaka, T.; Nakakuki, M.; Takahashi, A.; Suzuki, H.; Sone, H.; et al. SREBP-1 interacts with hepatocyte nuclear factor-4 alpha and interferes with PGC-1 recruitment to suppress hepatic gluconeogenic genes. J. Biol. Chem. 2004, 279, 12027–12035. [Google Scholar] [CrossRef]
- Metón, I.; Egea, M.; Anemaet, I.G.; Fernández, F.; Baanante, I.V. Sterol regulatory element binding protein-1a transactivates 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase gene promoter. Endocrinology 2006, 147, 3446–3456. [Google Scholar] [CrossRef]
- Egea, M.; Metón, I.; Córdoba, M.; Fernández, F.; Baanante, I.V. Role of Sp1 and SREBP-1a in the insulin-mediated regulation of glucokinase transcription in the liver of gilthead sea bream (Sparus aurata). Gen. Comp. Endocrinol. 2008, 155, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Polakof, S.; Panserat, S.; Soengas, J.L.; Moon, T.W. Glucose metabolism in fish: A review. J. Comp. Physiol. B 2012, 182, 1015–1045. [Google Scholar] [CrossRef] [PubMed]
- Caseras, A.; Metón, I.; Fernández, F.; Baanante, I.V. Glucokinase gene expression is nutritionally regulated in liver of gilthead sea bream (Sparus aurata). Biochim. et Biophys. Acta(BBA) - Gene Struct. Expr. 2000, 1493, 135–141. [Google Scholar] [CrossRef]
- Panserat, S.; Médale, F.; Brèque, J.; Plagnes-Juan, E.; Kaushik, S. Lack of significant long-term effect of dietary carbohydrates on hepatic glucose-6-phosphatase expression in rainbow trout (Oncorhynchus mykiss). J. Nutr. Biochem. 2000, 11, 22–29. [Google Scholar] [CrossRef]
- Panserat, S.; Capilla, E.; Gutierrez, J.; Frappart, P.O.; Vachot, C.; Plagnes-Juan, E.; Aguirre, P.; Brèque, J.; Kaushik, S. Glucokinase is highly induced and glucose-6-phosphatase poorly repressed in liver of rainbow trout (Oncorhynchus mykiss) by a single meal with glucose. Comp. Biochem. Physiol. Part B. Biochem. Mol. Biol. 2001, 128, 275–283. [Google Scholar] [CrossRef]
- Caseras, A.; Metón, I.; Vives, C.; Egea, M.; Fernández, F.; Baanante, I.V. Nutritional regulation of glucose-6-phosphatase gene expression in liver of the gilthead sea bream (Sparus aurata). Br. J. Nutr. 2002, 88, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Metón, I.; Mediavilla, D.; Caseras, A.; Cantó, E.; Fernández, F.; Baanante, I.V. Effect of diet composition and ration size on key enzyme activities of glycolysis-gluconeogenesis, the pentose phosphate pathway and amino acid metabolism in liver of gilthead sea bream (Sparus aurata). Br. J. Nutr. 1999, 82, 223–232. [Google Scholar] [CrossRef]
- Metón, I.; Caseras, A.; Fernández, F.; Baanante, I.V. 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase gene expression is regulated by diet composition and ration size in liver of gilthead sea bream, Sparus aurata. Biochim. et Biophys. Acta(BBA) -Gene Struct. Expr. 2000, 1491, 220–228. [Google Scholar] [CrossRef]
- Fernández, F.; Miquel, A.G.; Cordoba, M.; Varas, M.; Metón, I.; Caseras, A.; Baanante, I.V. Effects of diets with distinct protein-to-carbohydrate ratios on nutrient digestibility, growth performance, body composition and liver intermediary enzyme activities in gilthead sea bream (Sparus aurata, L.) fingerlings. J. Exp. Mar. Biol. Ecol. 2007, 343, 1–10. [Google Scholar] [CrossRef]
- Rashidpour, A.; Silva-Marrero, J.I.; Seguí, L.; Baanante, I.V.; Metón, I. Metformin counteracts glucose-dependent lipogenesis and impairs transdeamination in the liver of gilthead sea bream (Sparus aurata). Am. J. Physiol. Integr. Comp. Physiol. 2019, 316, R265–R273. [Google Scholar] [CrossRef]
- Silva-Marrero, J.I.; Sáez, A.; Caballero-Solares, A.; Viegas, I.; Almajano, M.P.; Fernández, F.; Baanante, I.V.; Metón, I. A transcriptomic approach to study the effect of long-term starvation and diet composition on the expression of mitochondrial oxidative phosphorylation genes in gilthead sea bream (Sparus aurata). BMC Genom. 2017, 18, 768. [Google Scholar] [CrossRef] [PubMed]
- González, J.D.; Silva-Marrero, J.I.; Metón, I.; Caballero-Solares, A.; Viegas, I.; Fernández, F.; Miñarro, M.; Fàbregas, A.; Ticó, J.R.; Jones, J.G.; et al. Chitosan-mediated shRNA knockdown of cytosolic alanine aminotransferase improves hepatic carbohydrate metabolism. Mar. Biotechnol. 2016, 18, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [PubMed]
- Ravanbakhsh, S.; Liu, P.; Bjordahl, T.C.; Mandal, R.; Grant, J.R.; Wilson, M.; Eisner, R.; Sinelnikov, I.; Hu, X.; Luchinat, C.; et al. Accurate, fully-automated NMR spectral profiling for metabolomics. PLoS ONE 2015, 10, e0124219. [Google Scholar] [CrossRef] [PubMed]
- Villasante, J.; Girbal, M.; Metón, I.; Almajano, M. Effects of pecan nut (Carya illinoiensis) and roselle flower (Hibiscus sabdariffa) as antioxidant and antimicrobial agents for sardines (Sardina pilchardus). Molecules 2019, 24, 85. [Google Scholar] [CrossRef]
- Ragelle, H.; Riva, R.; Vandermeulen, G.; Naeye, B.; Pourcelle, V.; Le Duff, C.S.; D’Haese, C.; Nysten, B.; Braeckmans, K.; De Smedt, S.C.; et al. Chitosan nanoparticles for siRNA delivery: Optimizing formulation to increase stability and efficiency. J. Control. Release 2014, 176, 54–63. [Google Scholar] [CrossRef]
- Sáez, M.; Vizcaíno, A.; Alarcón, F.; Martínez, T. Comparison of lacZ reporter gene expression in gilthead sea bream (Sparus aurata) following oral or intramuscular administration of plasmid DNA in chitosan nanoparticles. Aquaculture 2017, 474, 1–10. [Google Scholar] [CrossRef]
- Gaspar, C.; Silva-Marrero, J.I.; Fàbregas, A.; Miñarro, M.; Ticó, J.R.; Baanante, I.V.; Metón, I. Administration of chitosan-tripolyphosphate-DNA nanoparticles to knockdown glutamate dehydrogenase expression impairs transdeamination and gluconeogenesis in the liver. J. Biotechnol. 2018, 286, 5–13. [Google Scholar] [CrossRef]
- Wu, S.; Pan, H.; Tan, S.; Ding, C.; Huang, G.; Liu, G.; Guo, J.; Su, Z. In vitro inhibition of lipid accumulation induced by oleic acid and in vivo pharmacokinetics of chitosan microspheres (CTMS) and chitosan-capsaicin microspheres (CCMS). Food Nutr. Res. 2017, 61, 1331658. [Google Scholar] [CrossRef]
- Liu, X.; Zhi, X.; Liu, Y.; Wu, B.; Sun, Z.; Shen, J. Effect of chitosan, O -carboxymethyl chitosan, and N -[(2-hydroxy-3- N, N -dimethylhexadecyl ammonium)propyl] chitosan chloride on overweight and insulin resistance in a murine diet-induced obesity. J. Agric. Food Chem. 2012, 60, 3471–3476. [Google Scholar] [CrossRef] [PubMed]
- Houddane, A.; Bultot, L.; Novellasdemunt, L.; Johanns, M.; Gueuning, M.-A.; Vertommen, D.; Coulie, P.G.; Bartrons, R.; Hue, L.; Rider, M.H. Role of Akt/PKB and PFKFB isoenzymes in the control of glycolysis, cell proliferation and protein synthesis in mitogen-stimulated thymocytes. Cell. Signal. 2017, 34, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Okar, D.A.; Wu, C.; Lange, A.J. Regulation of the regulatory enzyme, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Adv. Enzyme Regul. 2004, 44, 123–154. [Google Scholar] [CrossRef] [PubMed]
- Mediavilla, D.; Metón, I.; Baanante, I.V. Purification and kinetic characterization of 6-phosphofructo-1-kinase from the liver of gilthead sea bream (Sparus aurata). J. Biochem. 2008, 144, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Rome, S.; Lecomte, V.; Meugnier, E.; Rieusset, J.; Debard, C.; Euthine, V.; Vidal, H.; Lefai, E. Microarray analyses of SREBP-1a and SREBP-1c target genes identify new regulatory pathways in muscle. Physiol. Genom. 2008, 34, 327–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.K.; Kyriakou, T.; Weston, A.J.; O’Dell, S.D. Functional single-nucleotide polymorphism in acetyl-CoA carboxylase ACACB gene promoter. DNA Cell Biol. 2010, 29, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Shikama, A.; Shinozaki, H.; Takeuchi, Y.; Matsuzaka, T.; Aita, Y.; Murayama, T.; Sawada, Y.; Piao, X.; Toya, N.; Oya, Y.; et al. Identification of human ELOVL5 enhancer regions controlled by SREBP. Biochem. Biophys. Res. Commun. 2015, 465, 857–863. [Google Scholar] [CrossRef]
- Carmona-Antoñanzas, G.; Tocher, D.R.; Martinez-Rubio, L.; Leaver, M.J. Conservation of lipid metabolic gene transcriptional regulatory networks in fish and mammals. Gene 2014, 534, 1–9. [Google Scholar] [CrossRef]
- Dong, X.; Tan, P.; Cai, Z.; Xu, H.; Li, J.; Ren, W.; Xu, H.; Zuo, R.; Zhou, J.; Mai, K.; et al. Regulation of FADS2 transcription by SREBP-1 and PPAR-α influences LC-PUFA biosynthesis in fish. Sci. Rep. 2017, 7, 40024. [Google Scholar] [CrossRef]
- Du, X.; Cai, C.; Yao, J.; Zhou, Y.; Yu, H.; Shen, W. Histone modifications in FASN modulated by sterol regulatory element-binding protein 1c and carbohydrate responsive-element binding protein under insulin stimulation are related to NAFLD. Biochem. Biophys. Res. Commun. 2017, 483, 409–417. [Google Scholar] [CrossRef]
- Kim, C.-W.; Addy, C.; Kusunoki, J.; Anderson, N.N.; Deja, S.; Fu, X.; Burgess, S.C.; Li, C.; Ruddy, M.; Chakravarthy, M.; et al. Acetyl CoA carboxylase inhibition reduces hepatic steatosis but elevates plasma triglycerides in mice and humans: A bedside to bench investigation. Cell Metab. 2017, 26, 394–406.e6. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Viscarra, J.; Kim, S.-J.; Sul, H.S. Transcriptional regulation of hepatic lipogenesis. Nat. Rev. Mol. Cell Biol. 2015, 16, 678–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lansard, M.; Panserat, S.; Plagnes-Juan, E.; Seiliez, I.; Skiba-Cassy, S. Integration of insulin and amino acid signals that regulate hepatic metabolism-related gene expression in rainbow trout: Role of TOR. Amino Acids 2010, 39, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Skiba-Cassy, S.; Panserat, S.; Larquier, M.; Dias, K.; Surget, A.; Plagnes-Juan, E.; Kaushik, S.; Seiliez, I. Apparent low ability of liver and muscle to adapt to variation of dietary carbohydrate:protein ratio in rainbow trout (Oncorhynchus mykiss). Br. J. Nutr. 2013, 109, 1359–1372. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Mokrani, A.; Chisomo-Kasiya, H.; Ji, K.; Ge, X.; Ren, M.; Liu, B.; Xi, B.; Sun, A. Dietary leucine affects glucose metabolism and lipogenesis involved in TOR/PI3K/Akt signaling pathway for juvenile blunt snout bream Megalobrama amblycephala. Fish Physiol. Biochem. 2019, 45, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.E.; Bobik, E.G.; Dorado, J.; Kroeger, P.E.; Chuang, L.T.; Thurmond, J.M.; Parker-Barnes, J.M.; Das, T.; Huang, Y.S.; Mukerji, P. Cloning of a human cDNA encoding a novel enzyme involved in the elongation of long-chain polyunsaturated fatty acids. Biochem. J. 2000, 350, 765–770. [Google Scholar] [CrossRef]
- Geay, F.; Tinti, E.; Mellery, J.; Michaux, C.; Larondelle, Y.; Perpète, E.; Kestemont, P. Cloning and functional characterization of Δ6 fatty acid desaturase (FADS2) in Eurasian perch (Perca fluviatilis). Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2016, 191, 112–125. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Kothapalli, K.S.D.; Brenna, J.T. Desaturase and elongase-limiting endogenous long-chain polyunsaturated fatty acid biosynthesis. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 103–110. [Google Scholar] [CrossRef]
- Castro, L.F.C.; Tocher, D.R.; Monroig, O. Long-chain polyunsaturated fatty acid biosynthesis in chordates: Insights into the evolution of Fads and Elovl gene repertoire. Prog. Lipid Res. 2016, 62, 25–40. [Google Scholar] [CrossRef]



| HLL | LLH | |
|---|---|---|
| Formulation (%) | ||
| Fish meal 1 | 81.6 | 54.3 |
| Fish oil 2 | 0.8 | 6.0 |
| Starch 3 | 15.0 | 37.1 |
| Vitamin mixture 4 | 0.2 | 0.2 |
| Mineral mixture 5 | 0.9 | 0.9 |
| Carrageenan 6 | 1.5 | 1.5 |
| Proximate composition (%) | ||
| Protein | 58.0 | 38.6 |
| Carbohydrates 7 | 15.0 | 37.1 |
| Fat | 9.9 | 12.1 |
| Ash | 15.4 | 10.5 |
| Gross energy (kJ/g) 8 | 20.1 | 20.0 |
| Gene | Forward Sequences (5’ to 3’) | Reverse Sequences (5’ to 3’) | GenBank Accession |
|---|---|---|---|
| SREBP1a | CCTCCTGCCTCCGAGTTTCC | GAAGGAAGGCTAGAATACCCC | U09103 |
| GK | TGTGTCAGCTCTCAACTCGACC | AGGATCTGCTCTACCATGTGGAT | AF169368 |
| PFKFB1 | TCGTGATGGTGGGACTGCCG | CTCGGCGTTGTCGGCTCTGAAG | U84724 |
| ACC1 | CCCAACTTCTTCTACCACAG | GAACTGGAACTCTACTACAC | JX073712 |
| ACC2 | TGACATGAGTCCTGTGCTGG | GCCTCAGTTCGTATGATGGT | JX073714 |
| FASN | TGGGTCAGGGGGAGTTGG | TTGTGTGGAGAAGAACGTGCTGT | JQ277708 |
| ELOVL5 | GGGATGGCTACTGCTCGACA | CAGGAGAGTGAGGCCCAGAT | AY660879 |
| FADS2 | CACTATGCTGGAGAGGATGCC | TATTTCGGTCCTGGCTGGGC | AY055749 |
| G6PD | TGATGATCCAACAGTTCCTA | GCTCGTTCCTGACACACTGA | JX073711 |
| HMGCR | ACTGATGGCTGCTCTGGCTG | GGGACTGAGGGATGACGCAC | MN047456 |
| 18s | TTACGCCCATGTTGTCCTGAG | AGGATTCTGCATGATGGTCACC | AM490061 |
| β-actin | CTGGCATCACACCTTCTACAACGAG | GCGGGGGTGTTGAAGGTCTC | X89920 |
| EF1α | CCCGCCTCTGTTGCCTTCG | CAGCAGTGTGGTTCCGTTAGC | AF184170 |
| HLL | LLH | Two-way ANOVA | |||||
|---|---|---|---|---|---|---|---|
| Metabolite (mM) | - | SREBP1a | - | SREBP1a | Interaction | Diet | Treatment |
| Lactate + Thr | 0.96 ± 0.16 | 1.13 ± 0.19 | 0.78 ± 0.16 | 1.20 ± 0.35 | NS | NS | NS |
| Alanine | 3.21 ± 0.67 | 4.12 ± 0.72 | 2.87 ± 0.29 | 4.35 ± 0.31 | NS | NS | * |
| Glucose | 3.22 ± 0.44 | 4.44 ± 0.98 | 1.87 ± 0.31 | 3.27 ± 1.12 | NS | NS | NS |
| Acetate | 0.22 ± 0.02 | 0.21 ± 0.03 | 0.27 ± 0.02 | 0.28 ± 0.03 | NS | * | NS |
| Isoleucine | 0.04 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.00 | 0.04 ± 0.01 | NS | NS | NS |
| Valine | 0.08 ± 0.01 | 0.08 ± 0.02 | 0.07 ± 0.00 | 0.07 ± 0.01 | NS | NS | NS |
| Creatinine | 0.09 ± 0.02 | 0.09 ± 0.01 | 0.29 ± 0.05 | 0.23 ± 0.05 | *** | NS | NS |
| Choline compounds | 0.44 ± 0.03 | 0.43 ± 0.08 | 0.36 ± 0.03 | 0.39 ± 0.03 | NS | NS | NS |
| Succinate | 0.37 ± 0.05 | 0.45 ± 0.09 | 0.24 ± 0.04 | 0.38 ± 0.09 | NS | NS | NS |
| Sarcosine | 0.07 ± 0.01 | 0.07 ± 0.01 | 0.05 ± 0.01 | 0.05 ± 0.01 | NS | NS | NS |
| Taurine | 6.46 ± 0.45 | 5.51 ± 0.75 | 5.77 ± 0.26 | 4.66 ± 0.40 | NS | NS | NS |
| Glycine | 0.86 ± 0.09 | 1.34 ± 0.20 | 1.90 ± 0.25 | 2.75 ± 0.38 | NS | *** | * |
| Formate | 1.31 ± 0.11 | 0.94 ± 0.14 | 1.66 ± 0.18 | 1.32 ± 0.10 | NS | * | * |
| Glycogen | 1.28 ± 0.30 | 1.75 ± 0.16 | 1.84 ± 0.61 | 2.05 ± 0.63 | NS | NS | NS |
| Lac/Ala | 0.31 ± 0.03 | 0.28 ± 0.03 | 0.27 ± 0.03 | 0.26 ± 0.06 | NS | NS | NS |
| HLL | LLH | Two-way ANOVA | |||||
|---|---|---|---|---|---|---|---|
| Fatty Acid | - | SREBP1a | - | SREBP1a | Interaction | Diet | Treatment |
| 14:0 | 3.59 ± 0.31 | 3.10 ± 0.15 | 2.85 ± 0.12 | 2.76 ± 0.44 | NS | NS | NS |
| 15:0 | 0.28 ± 0.05 | 0.30 ± 0.02 | 0.11 ± 0.00 | 0.32 ± 0.01 | * | * | ** |
| 16:0 | 22.26 ± 0.76 | 21.32 ± 0.54 | 22.09 ± 1.11 | 23.73 ± 0.96 | NS | NS | NS |
| 17:0 | 0.32 ± 0.03 | 0.41 ± 0.01 | 0.33 ± 0.01 | 0.35 ± 0.01 | NS | NS | * |
| 18:0 | 7.14 ± 1.05 | 7.65 ± 0.37 | 8.69 ± 0.30 | 6.93 ± 0.37 | NS | NS | NS |
| 20:0 | 0.10 ± 0.02 | 0.12 ± 0.00 | 0.11 ± 0.01 | 0.10 ± 0.01 | NS | NS | NS |
| 21:0 | 0.89 ± 0.14 | 1.21 ± 0.14 | 1.21 ± 0.47 | 1.27 ± 0.14 | NS | NS | NS |
| 22:0 | 0.15 ± 0.02 | 0.16 ± 0.02 | 0.13 ± 0.02 | 0.16 ± 0.00 | NS | NS | NS |
| 23:0 | 0.01 ± 0.01 | 0.03 ± 0.01 | 0.00 ± 0.00 | 0.01 ± 0.01 | NS | * | NS |
| 24:0 | 0.13 ± 0.01 | 0.15 ± 0.02 | 0.18 ± 0.06 | 0.18 ± 0.02 | NS | NS | NS |
| 14:1n-5 | 0.09 ± 0.02 | 0.11 ± 0.01 | 0.00 ± 0.00 | 0.11 ± 0.04 | NS | NS | * |
| 15:1n-5 | 0.07 ± 0.02 | 0.09 ± 0.01 | 0.00 ± 0.00 | 0.08 ± 0.02 | NS | * | * |
| 16:1n-7 | 4.42 ± 0.26 | 4.53 ± 0.35 | 4.25 ± 0.14 | 3.96 ± 0.20 | NS | NS | NS |
| 17:1n-7 | 0.28 ± 0.02 | 0.32 ± 0.01 | 0.29 ± 0.05 | 0.33 ± 0.01 | NS | NS | NS |
| 18:1n-9c | 26.48 ± 0.95 | 27.15 ± 1.83 | 26.10 ± 0.40 | 23.69 ± 1.53 | NS | NS | NS |
| 18:1n-9t | 0.11 ± 0.01 | 0.12 ± 0.01 | 0.10 ± 0.02 | 0.10 ± 0.01 | NS | NS | NS |
| 20:1n-9 | 1.29 ± 0.14 | 1.40 ± 0.01 | 1.70 ± 0.03 | 1.40 ± 0.08 | NS | NS | NS |
| 22:1n-9 | 0.74 ± 0.08 | 0.80 ± 0.13 | 0.96 ± 0.07 | 1.05 ± 0.08 | NS | * | NS |
| 24:1n-9 | 0.30 ± 0.04 | 0.39 ± 0.04 | 0.31 ± 0.06 | 0.24 ± 0.02 | NS | NS | NS |
| 18:2n-6c | 2.12 ± 0.38 | 2.37 ± 0.59 | 2.09 ± 0.05 | 2.43 ± 0.07 | NS | NS | NS |
| 18:2n-6t | 0.01 ± 0.01 | 0.02 ± 0.00 | 0.02 ± 0.02 | 0.03 ± 0.01 | NS | NS | NS |
| 18:3n-3 | 0.11 ± 0.03 | 0.10 ± 0.03 | 0.11 ± 0.03 | 0.10 ± 0.01 | NS | NS | NS |
| 18:3n-6 | 0.36 ± 0.05 | 0.42 ± 0.09 | 0.32 ± 0.04 | 0.39 ± 0.03 | NS | NS | NS |
| 20:2n-6 | 0.13 ± 0.02 | 0.17 ± 0.01 | 0.14 ± 0.02 | 0.19 ± 0.01 | NS | NS | * |
| 20:3n-3 | 0.17 ± 0.02 | 0.24 ± 0.01 | 0.25 ± 0.04 | 0.36 ± 0.04 | NS | * | * |
| 20:4n-6 | 1.02 ± 0.07 | 0.96 ± 0.10 | 0.94 ± 0.02 | 1.38 ± 0.19 | NS | NS | NS |
| 20:5n-3 | 4.77 ± 0.14 | 4.27 ± 0.49 | 3.64 ± 0.22 | 4.98 ± 0.53 | NS | NS | NS |
| 22:2n-6 | 0.01 ± 0.00 | 0.03 ± 0.00 | 0.00 ± 0.00 | 0.02 ± 0.01 | NS | NS | * |
| 22:6n-3 | 14.46 ± 1.18 | 14.87 ± 1.84 | 12.46 ± 0.68 | 15.31 ± 1.16 | NS | NS | NS |
| Saturated | 35.48 ± 2.86 | 34.45 ± 0.85 | 35.76 ± 0.93 | 35.81 ± 0.74 | NS | NS | NS |
| Monounsaturated | 34.50 ± 0.52 | 34.91 ± 2.12 | 33.71 ± 0.54 | 30.95 ± 1.79 | NS | NS | NS |
| PUFAs | 23.01 ± 1.38 | 23.47 ± 3.11 | 19.99 ± 0.80 | 25.20 ± 1.83 | NS | NS | NS |
| n-9 | 29.90 ± 0.52 | 29.86 ± 2.00 | 29.17 ± 0.46 | 26.47 ± 1.61 | NS | NS | NS |
| n-6 | 3.49 ± 0.40 | 3.97 ± 0.79 | 3.53 ± 0.04 | 4.45 ± 0.18 | NS | NS | NS |
| n-3 | 19.52 ± 1.23 | 19.49 ± 2.34 | 16.46 ± 0.82 | 20.75 ± 1.68 | NS | NS | NS |
| n-6/n-3 | 0.18 ± 0.02 | 0.20 ± 0.02 | 0.22 ± 0.01 | 0.22 ± 0.01 | NS | NS | NS |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva-Marrero, J.I.; Villasante, J.; Rashidpour, A.; Palma, M.; Fàbregas, A.; Almajano, M.P.; Viegas, I.; Jones, J.G.; Miñarro, M.; Ticó, J.R.; et al. The Administration of Chitosan-Tripolyphosphate-DNA Nanoparticles to Express Exogenous SREBP1a Enhances Conversion of Dietary Carbohydrates into Lipids in the Liver of Sparus aurata. Biomolecules 2019, 9, 297. https://doi.org/10.3390/biom9080297
Silva-Marrero JI, Villasante J, Rashidpour A, Palma M, Fàbregas A, Almajano MP, Viegas I, Jones JG, Miñarro M, Ticó JR, et al. The Administration of Chitosan-Tripolyphosphate-DNA Nanoparticles to Express Exogenous SREBP1a Enhances Conversion of Dietary Carbohydrates into Lipids in the Liver of Sparus aurata. Biomolecules. 2019; 9(8):297. https://doi.org/10.3390/biom9080297
Chicago/Turabian StyleSilva-Marrero, Jonás I., Juliana Villasante, Ania Rashidpour, Mariana Palma, Anna Fàbregas, María Pilar Almajano, Ivan Viegas, John G. Jones, Montserrat Miñarro, Josep R. Ticó, and et al. 2019. "The Administration of Chitosan-Tripolyphosphate-DNA Nanoparticles to Express Exogenous SREBP1a Enhances Conversion of Dietary Carbohydrates into Lipids in the Liver of Sparus aurata" Biomolecules 9, no. 8: 297. https://doi.org/10.3390/biom9080297
APA StyleSilva-Marrero, J. I., Villasante, J., Rashidpour, A., Palma, M., Fàbregas, A., Almajano, M. P., Viegas, I., Jones, J. G., Miñarro, M., Ticó, J. R., Baanante, I. V., & Metón, I. (2019). The Administration of Chitosan-Tripolyphosphate-DNA Nanoparticles to Express Exogenous SREBP1a Enhances Conversion of Dietary Carbohydrates into Lipids in the Liver of Sparus aurata. Biomolecules, 9(8), 297. https://doi.org/10.3390/biom9080297








