Role of MCC/Eisosome in Fungal Lipid Homeostasis
Abstract
:1. Introduction
2. Ergosterol-Enriched MCC
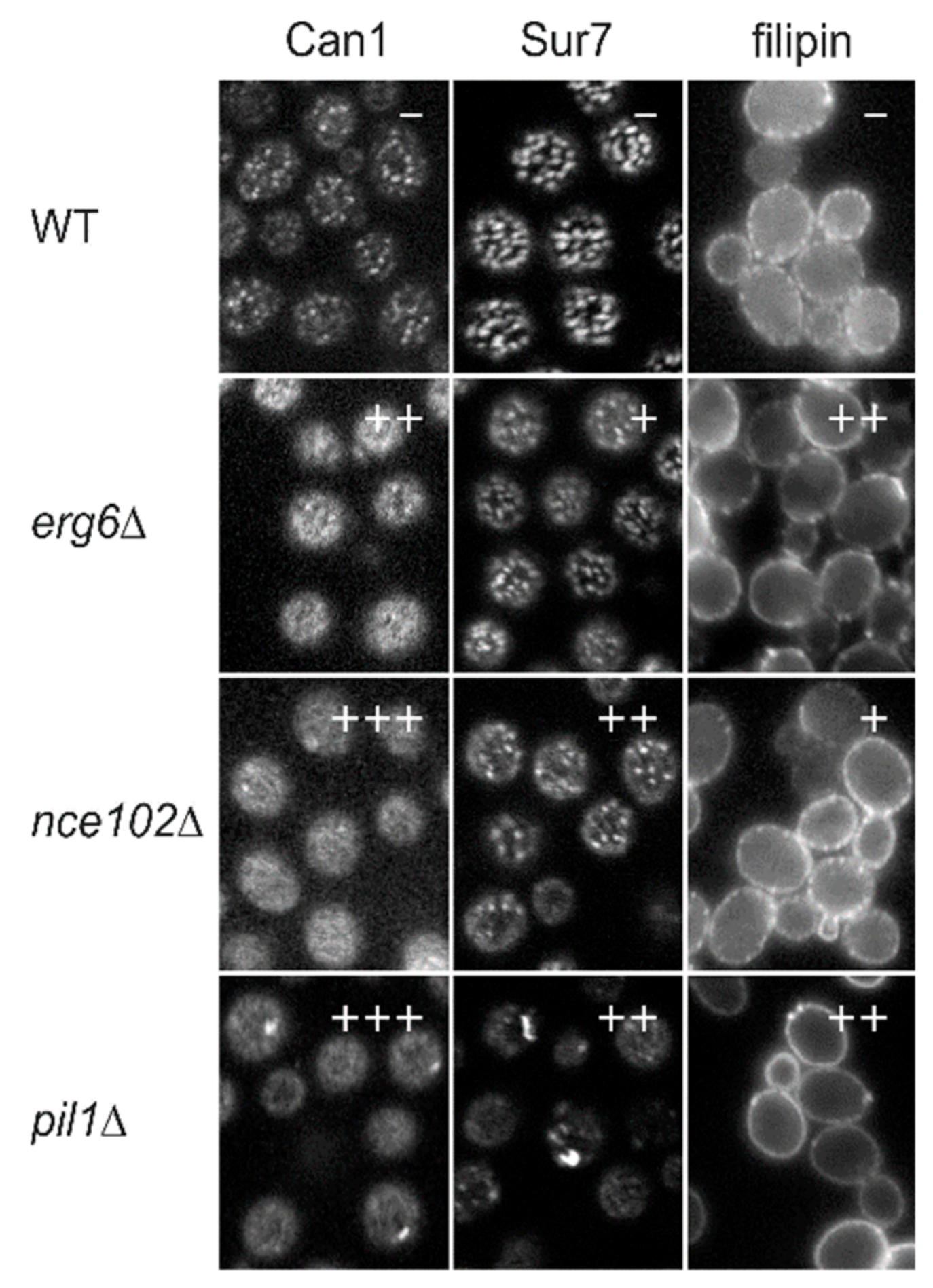
2.1. Subcellular Distribution of Ergosterol
2.2. The Link between the MCC/Eisosome and Ergosterol Homeostasis
3. Eisosome and Sphingolipids
3.1. Fungal Sphingolipids
3.2. Eisosomal Feedback Loop in Sphingolipid Biosynthesis
3.3. Further Notes on Nce102′s Function as a Sphingolipid Sensor
4. Plasma Membrane Phosphatidylinositols and MCC
5. Functional Relevance of MCC/Eisosome-Mediated Lipid Regulation
6. Concluding Remarks and Future Prospects
- Lipid composition of the MCC: As stated above, there are valid reasons to expect (sphingolipid-free) ergosterol, as well as some PI(4,5)P2, inside the MCC membrane. The question of the presence of sphingolipids in the microdomain is more complicated. The MCC membrane is detached from the cell wall, indicating a low concentration of glycosylated lipids. However, this does not exclude the presence of non-glycosylated sphingolipid precursors, namely LCBs and ceramides, within the compartment. The limited possibilities of specific labelling of lipid species in vivo accent the necessity of biochemical characterization of the microdomain lipid composition. However, all previous attempts to isolate the MCC/eisosome have failed.
- Mechanism of Nce102 action: One of the biggest gaps in the understanding of the MCC/eisosome-mediated sphingolipid biosynthesis regulation is the understanding of the molecular mechanism of Nce102 redistribution out of the MCC upon sphingolipid depletion. In this respect, S. cerevisiae represents a good model, as it contains exactly two Nce102-like proteins in the genome. In addition, Nce102 and Fhn1 expression profiles significantly differ with respect to their dependence on nutrient availability and actual lipid biosynthetic activity. For example, Fhn1 expression is induced by Upc2 following sterol biosynthesis inhibition, while Nce102 is not [54,55,58]. Whether this is just a reflection of the tight interconnection between the ergosterol and sphingolipid metabolic pathways, or whether Fhn1 plays an unknown sphingolipid-independent function in the membrane remains unknown. Even in the case of the proposed sphingolipid sensor Nce102, no direct experimental evidence has excluded the possibility that ergosterol, not sphingolipids, could interact with the Nce102 molecule. The act of sphingolipid sensing could in fact be the sensing of ergosterol-sphingolipid imbalance in the membrane.
- Cell interior and MCC/eisosome: The possible involvement of the MCC/eisosome in ergosterol metabolism opens other directions for future studies. For example, the eisosome function has not been related to the architecture of the inner cellular membranes, although the organelle morphology and membrane lipid composition are closely related. If involved in ergosterol biosynthesis and regulation, the MCC/eisosome could influence morphology of various cellular organelles. The first evidence in support of a role for eisosomes in autophagy was reported in a recent study focused on the pathogenic fungus, Beauveria bassiana [177].
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Malinska, K.; Malinsky, J.; Opekarova, M.; Tanner, W. Visualization of protein compartmentation within the plasma membrane of living yeast cellls. Mol. Biol. Cell 2003, 14, 4427–4436. [Google Scholar] [CrossRef] [PubMed]
- Malinska, K.; Malinsky, J.; Opekarova, M.; Tanner, W. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 2004, 117, 6031–6041. [Google Scholar] [CrossRef]
- Grossmann, G.; Opekarová, M.; Malinsky, J.; Weig-Meckl, I.; Tanner, W. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 2007, 26, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, F.; Syga, L.; Moiset, G.; Spakman, D.; Schavemaker, P.E.; Punter, C.M.; Seinen, A.B.; Van Oijen, A.M.; Robinson, A.; Poolman, B. Steric exclusion and protein conformation determine the localization of plasma membrane transporters. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Busto, J.V.; Elting, A.; Haase, D.; Spira, F.; Kuhlman, J.; Schäfer-Herte, M.; Wedlich-Söldner, R. Lateral plasma membrane compartmentalization links protein function and turnover. EMBO J. 2018, 37, 1–17. [Google Scholar] [CrossRef]
- Grossmann, G.; Malinsky, J.; Stahlschmidt, W.; Loibl, M.; Weig-Meckl, I.; Frommer, W.B.; Opekarová, M.; Tanner, W. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J. Cell Biol. 2008, 183, 1075–1088. [Google Scholar] [CrossRef] [Green Version]
- Fröhlich, F.; Moreira, K.; Aguilar, P.S.; Hubner, N.C.; Mann, M.; Walter, P.; Walther, T.C. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J. Cell Biol. 2009, 185, 1227–1242. [Google Scholar] [CrossRef] [Green Version]
- Walther, T.C.; Brickner, J.H.; Aguilar, P.S.; Bernales, S.; Pantoja, C.; Walter, P. Eisosomes mark static sites of endocytosis. Nature 2006, 439, 998–1003. [Google Scholar] [CrossRef]
- Stradalova, V.; Stahlschmidt, W.; Grossmann, G.; Blazikova, M.; Rachel, R.; Tanner, W.; Malinsky, J. Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J. Cell Sci. 2009, 122, 2887–2894. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Lester, R.L.; Dickson, R.C. Pil1p and Lsp1p negatively regulate the 3-phosphoinositide-dependent protein kinase-like kinase Pkh1p and downstream signaling pathways Pkc1p and Ypk1p. J. Biol. Chem. 2004, 279, 22030–22038. [Google Scholar] [CrossRef]
- Olivera-Couto, A.; Grana, M.; Harispe, L.; Aguilar, P.S. The eisosome core is composed of BAR domain proteins. Mol. Biol. Cell 2011, 22, 2360–2372. [Google Scholar] [CrossRef] [PubMed]
- Kabeche, R.; Baldissard, S.; Hammond, J.; Howard, L.; Moseley, J.B. The filament-forming protein Pil1 assembles linear eisosomes in fission yeast. Mol. Biol. Cell 2011, 22, 4059–4067. [Google Scholar] [CrossRef] [PubMed]
- Karotki, L.; Huiskonen, J.T.; Stefan, C.J.; Ziółkowska, N.E.; Roth, R.; Surma, M.A.; Krogan, N.J.; Emr, S.D.; Heuser, J.; Grünewald, K.; et al. Eisosome proteins assemble into a membrane scaffold. J. Cell Biol. 2011, 195, 889–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabeche, R.; Roguev, A.; Krogan, N.J.; Moseley, J.B. A Pil1-Sle1-Syj1-Tax4 functional pathway links eisosomes with PI(4,5)P2 regulation. J. Cell Sci. 2014, 127, 1318–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-H.; Heuser, J.E.; Roth, R.; Goodenough, U. Eisosome Ultrastructure and Evolution in Fungi, Microalgae, and Lichens. Eukaryot. Cell 2015, 14, 1017–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blyth, J.; Makrantoni, V.; Baton, R.E.; Spanos, C.; Rappsilber, J.; Marston, A.L. Genes Important for Schizosaccharomyces pombe Genomics Screen. Genetics 2018, 208, 589–603. [Google Scholar] [CrossRef]
- Wang, H.X.; Douglas, L.M.; Veselá, P.; Rachel, R.; Malinsky, J.; Konopka, J.B. Eisosomes promote the ability of Sur7 to regulate plasma membrane organization in Candida albicans. Mol. Biol. Cell 2016, 27, 1663–1675. [Google Scholar] [CrossRef]
- Douglas, L.M.; Wang, H.X.; Keppler-Ross, S.; Dean, N.; Konopka, J.B. Sur7 Promotes Plasma Membrane Organization and Is Needed for Resistance to Stressful Conditions and to the Invasive Growth and Virulence of Candida albicans. mBio 2012, 3, 1–12. [Google Scholar] [CrossRef]
- Douglas, L.M.; Konopka, J.B. Plasma membrane architecture protects Candida albicans from killing by copper. PLoS Genet. 2019, 15, 1–26. [Google Scholar] [CrossRef]
- Carman, G.M.; Henry, S.A. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 1999, 38, 361–399. [Google Scholar] [CrossRef]
- Frohlich, F.; Christiano, R.; Olson, D.K.; Alcazar-Roman, A.; DeCamilli, P.; Walther, T.C. A role for eisosomes in maintenance of plasma membrane phosphoinositide levels. Mol. Biol. Cell 2014, 25, 2797–2806. [Google Scholar] [CrossRef] [PubMed]
- Berchtold, D.; Piccolis, M.; Chiaruttini, N.; Riezman, I.; Riezman, H.; Roux, A.; Walther, T.C.; Loewith, R. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat. Cell Biol. 2012, 14, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Grousl, T.; Opekarová, M.; Stradalova, V.; Hasek, J.; Malinsky, J. Evolutionarily conserved 5′-3′ exoribonuclease Xrn1 accumulates at plasma membrane-associated eisosomes in post-diauxic yeast. PLoS ONE 2015, 10, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Pedroso, N.; Matias, A.C.; Cyrne, L.; Antunes, F.; Borges, C.; Malhó, R.; de Almeida, R.F.M.; Herrero, E.; Marinho, H.S. Modulation of plasma membrane lipid profile and microdomains by H2O2 in Saccharomyces cerevisiae. Free Radic. Biol. Med. 2009, 46, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Beney, L.; Ritt, J.F.; Lherminier, J.; Gervais, P. Lateral reorganization of plasma membrane is involved in the yeast resistance to severe dehydration. Biochimica et Biophysica Acta Biomembranes 2010, 1798, 975–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Needham, D.; Nunn, R.S. Elastic deformation and failure of lipid bilayer membranes containing cholesterol. Biophys. J. 1990, 58, 997–1009. [Google Scholar] [CrossRef] [Green Version]
- Bretscher, M.S.; Munro, S. Cholesterol and the Golgi apparatus. Science 1993, 261, 1280–1281. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Ikonen, E. Functional rafts in cell membranes. Nature 1997, 387, 569–572. [Google Scholar] [CrossRef]
- Zinser, E.; Paltauf, F.; Daum, G. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J. Bacteriol. 1993, 175, 2853–2858. [Google Scholar] [CrossRef] [Green Version]
- Munn, A.L.; Heese-Peck, A.; Stevenson, B.J.; Pichler, H.; Riezman, H. Specific Sterols Required for the Internalization Step of Endocytosis in Yeast. Mol. Biol. Cell 1999, 10, 3943–3957. [Google Scholar] [CrossRef] [Green Version]
- Heese-Peck, A.; Pichler, H.; Zanolari, B.; Watanabe, R.; Daum, G.; Riezman, H. Multiple functions of sterols in yeast endocytosis. Mol. Biol. Cell 2002, 13, 2664–2680. [Google Scholar] [CrossRef] [PubMed]
- Schneiter, R.; Brügger, B.; Sandhoff, R.; Zellnig, G.; Leber, A.; Lampl, M.; Athenstaedt, K.; Hrastnik, C.; Eder, S.; Daum, G.; et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 1999, 146, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Munro, S. Lipid rafts: Elusive or illusive? Cell 2003, 115, 377–388. [Google Scholar] [CrossRef]
- Solanko, L.M.; Sullivan, D.P.; Sere, Y.Y.; Szomek, M.; Lunding, A.; Solanko, K.A.; Pizovic, A.; Stanchev, L.D.; Pomorski, T.G.; Menon, A.K.; et al. Ergosterol is mainly located in the cytoplasmic leaflet of the yeast plasma membrane. Traffic 2018, 19, 198–214. [Google Scholar] [CrossRef] [PubMed]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Bagnat, M.; Simons, K. Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA 2002, 99, 14183–14188. [Google Scholar] [CrossRef] [Green Version]
- Wachtler, V. Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 2003, 116, 867–874. [Google Scholar] [CrossRef]
- Nichols, C.B.; Fraser, J.A.; Heitman, J. PAK kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans. Mol. Biol. Cell 2004, 15, 4476–4489. [Google Scholar] [CrossRef]
- Grossmann, G.; Opekarova, M.; Novakova, L.; Stolz, J.; Tanner, W. Lipid Raft-Based Membrane Compartmentation of a Plant Transport Protein Expressed in Saccharomyces cerevisiae. Eukaryot. Cell 2006, 5, 945–953. [Google Scholar] [CrossRef]
- Proszynski, T.J.; Klemm, R.; Bagnat, M.; Gaus, K.; Simons, K. Plasma membrane polarization during mating in yeast cells. J. Cell Biol. 2006, 173, 861–866. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.W.; Konopka, J.B. Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot. Cell 2004, 3, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Pearson, C.L.; Xu, K.; Sharpless, K.E.; Harris, S.D. MesA, a novel fungal protein required for the stabilization of polarity axes in Aspergillus nidulans. Mol. Biol. Cell 2004, 15, 3658–3672. [Google Scholar] [CrossRef] [PubMed]
- Malinsky, J.; Opekarová, M.; Grossmann, G.; Tanner, W. Membrane Microdomains, Rafts, and Detergent-Resistant Membranes in Plants and Fungi. Annu. Rev. Plant Biol. 2013, 64, 501–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toulmay, A.; Prinz, W.A. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 2013, 202, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, T.; Fujimoto, M.; Tatematsu, T.; Cheng, J.; Orii, M.; Takatori, S.; Fujimoto, T. Niemann-Pick type C proteins promote microautophagy by expanding raft-like membrane domains in the yeast vacuole. eLife 2017, 6, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Zinser, E.; Daum, G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast 1995, 11, 493–536. [Google Scholar] [CrossRef]
- Lange, Y.; Steck, T.L. The role of intracellular cholesterol transport in cholesterol homeostasis. Trends Cell Biol. 1996, 6, 205–208. [Google Scholar] [CrossRef]
- Wang, C.W.; Miao, Y.H.; Chang, Y.S. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 2014, 206, 357–366. [Google Scholar] [CrossRef] [Green Version]
- Dupre, S.; Haguenauer-Tsapis, R. Raft partitioning of the yeast uracil permease during trafficking along the endocytic pathway. Traffic 2003, 4, 83–96. [Google Scholar] [CrossRef]
- Umebayashi, K.; Nakano, A. Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J. Cell Biol. 2003, 161, 1117–1131. [Google Scholar] [CrossRef]
- Douglas, L.M.; Wang, H.X.; Konopka, J.B. The MARVEL Domain Protein Nce102 Regulates Actin Organization and Invasive Growth of Candida albicans. mBio 2013, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Athanasopoulos, A.; Gournas, C.; Amillis, S.; Sophianopoulou, V. Characterization of AnNce102 and its role in eisosome stability and sphingolipid biosynthesis. Sci. Rep. 2015, 5, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Loibl, M.; Grossmann, G.; Stradalova, V.; Klingl, A.; Rachel, R.; Tanner, W.; Malinsky, J.; Opekarová, M. C terminus of Nce 102 determines the structure and function of microdomains in the Saccharomyces cerevisiae plasma membrane. Eukaryot. Cell 2010, 9, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.K.; Rogers, P.D.; Baerson, S.R.; Jacob, M.R.; Barker, K.S.; Cleary, J.D.; Walker, L.A.; Nagle, D.G.; Clark, A.M. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 34998–35015. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, L.J.; Balderes, D.A.; Wharton, B.; Tinkelenberg, A.H.; Rao, G.; Sturley, S.L. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 2002, 277, 32466–32472. [Google Scholar] [CrossRef] [PubMed]
- Vik, A.; Rine, J. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 6395–6405. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Tong, J.; Lee, C.W.; Ha, S.; Eom, S.H.; Im, Y.J. Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat. Commun. 2015, 6, 1–13. [Google Scholar] [CrossRef]
- Woods, K.; Höfken, T. The zinc cluster proteins Upc2 and Ecm22 promote filamentation in Saccharomyces cerevisiae by sterol biosynthesis-dependent and -independent pathways. Mol. Microbiol. 2016, 99, 512–527. [Google Scholar] [CrossRef]
- Foster, H.A.; Cui, M.; Naveenathayalan, A.; Unden, H.; Schwanbeck, R.; Höfken, T. The zinc cluster protein Sut1 contributes to filamentation in Saccharomyces cerevisiae. Eukaryot. Cell 2013, 12, 244–253. [Google Scholar] [CrossRef]
- Blanda, C.; Höfken, T. Regulation of mating in the budding yeast Saccharomyces cerevisiae by the zinc cluster proteins Sut1 and Sut2. Biochem. Biophys. Res. Commun. 2013, 438, 66–70. [Google Scholar] [CrossRef] [Green Version]
- Shively, C.A.; Eckwahl, M.J.; Dobry, C.J.; Mellacheruvu, D.; Nesvizhskii, A.; Kumar, A. Genetic Networks Inducing Invasive Growth in Saccharomyces cerevisiae Identified Through Systematic Genome-Wide Overexpression. Genetics 2013, 193, 1297–1310. [Google Scholar] [CrossRef] [PubMed]
- Kubler, E.; Dohlman, H.G.; Lisanti, M.P. Identification of triton X-100 insoluble membrane domains in the yeast Saccharomyces cerevisiae—Lipid requirements for targeting of heterotrimeric G-protein subunits. J. Biol. Chem. 1996, 271, 32975–32980. [Google Scholar] [CrossRef] [PubMed]
- Bagnat, M.; Keranen, S.; Shevchenko, A.; Simons, K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 3254–3259. [Google Scholar] [CrossRef] [PubMed]
- Souza, C.M.; Pichler, H. Lipid requirements for endocytosis in yeast. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Patton, J.L.; Lester, R.L. The phosphoinositol sphingolipids of Saccharomyces cerevisiae are highly localized in the plasma membrane. J. Bacteriol. 1991, 173, 3101–3108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lester, R.L.; Wells, G.B.; Oxford, G.; Dickson, R.C. Mutant strains of Saccharomyces cerevisiae lacking sphingolipids synthesize novel inositol glycerophospholipids that mimic sphingolipid structures. J. Biol. Chem. 1993, 268, 845–856. [Google Scholar] [PubMed]
- Lee, M.C.S.; Hamamoto, S.; Schekman, R. Ceramide Biosynthesis Is Required for the Formation of the Oligomeric H+-ATPase Pma1p in the Yeast Endoplasmic Reticulum. J. Biol. Chem. 2002, 277, 22395–22401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Chang, A. Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, Pma1. Proc. Natl. Acad. Sci. USA 2002, 99, 12853–12858. [Google Scholar] [CrossRef] [Green Version]
- Lauwers, E.; Grossmann, G.; André, B. Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: Essential role in transport activity and normal control by ubiquitination. Mol. Biol. Cell 2007, 18, 3068–3080. [Google Scholar] [CrossRef]
- Serrano, R.; Kielland-Brandt, M.C.; Fink, G.R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature 1986, 319, 689–693. [Google Scholar] [CrossRef]
- Serrano, R. Structure and function of proton translocafing ATPase in plasma membranes of plants and fungi. Biochem. Biophys. Acta 1988, 947, 1–28. [Google Scholar] [PubMed]
- Stanbrough, M.; Magasanik, B. Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. J. Bacterial. 1995, 177, 94–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, N.; Jenkins, G.; Hannun, Y.A.; Heitman, J.; Obeid, L.M. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J. Biol. Chem. 2000, 275, 17229–17232. [Google Scholar] [CrossRef] [PubMed]
- Cowart, L.A.; Okamoto, Y.; Pinto, F.R.; Gandy, J.L.; Almeida, J.S.; Hannun, Y.A. Roles for sphingolipid biosynthesis in mediation of specific programs of the heat stress response determined through gene expression profiling. J. Biol. Chem. 2003, 278, 30328–30338. [Google Scholar] [CrossRef] [PubMed]
- Dickson, R.C.; Nagiec, E.E.; Skrzypek, M.; Tillman, P.; Wells, G.B.; Lester, R.L. Sphingolipids are potential heat stress signals in Saccharomyces. J. Biol. Chem. 1997, 272, 30196–30200. [Google Scholar] [CrossRef]
- Jenkins, G.M. The emerging role for sphingolipids in the eukaryotic heat shock response. Cell. Mol. Life Sci. 2003, 60, 701–710. [Google Scholar] [CrossRef]
- Wells, G.B.; Dickson, R.C.; Lester, R.L. Heat-induced Elevation of Ceramide in Saccharomyces cerevisiae via de Novo Synthesis. J. Biol. Chem. 1998, 273, 7235–7243. [Google Scholar] [CrossRef] [Green Version]
- Levin, D.E. Cell Wall Integrity Signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005, 69, 262–291. [Google Scholar] [CrossRef]
- Liu, M.; Huang, C.; Polu, S.R.; Schneiter, R.; Chang, A. Regulation of sphingolipid synthesis through Orm1 and Orm2 in yeast. J. Cell Sci. 2012, 125, 2428–2435. [Google Scholar] [CrossRef]
- Piña, F.; Yagisawa, F.; Obara, K.; Gregerson, J.D.; Kihara, A.; Niwa, M. Sphingolipids activate the endoplasmic reticulum stress surveillance pathway. J. Cell Biol. 2018, 217, 495–505. [Google Scholar] [CrossRef]
- Friant, S. Increased protein kinase or decreased PP2A activity bypasses sphingoid base requirement in endocytosis. EMBO J. 2000, 19, 2834–2844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanolari, B.; Friant, S.; Funato, K.; Suetterlin, C.; Stevenson, B.J.; Riezman, H. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 2000, 19, 2824–2833. [Google Scholar] [CrossRef] [PubMed]
- Dickson, R.C. Roles for Sphingolipids in Saccharomyces cerevisiae. In Sphingolipids as Signaling and Regulatory Molecules; Chalfant, C., DelPoeta, M., Eds.; Springer: Berlin, Germany, 2010; Volume 688, pp. 217–231. [Google Scholar]
- Frisz, J.F.; Lou, K.; Klitzing, H.A.; Hanafin, W.P.; Lizunov, V.; Wilson, R.L.; Carpenter, K.J.; Kim, R.; Hutcheon, I.D.; Zimmerberg, J.; et al. Direct chemical evidence for sphingolipid domains in the plasma membranes of fibroblasts. Proc. Natl. Acad. Sci. USA 2013, 110, E613–E622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frisz, J.F.; Klitzing, H.A.; Lous, K.; Hutcheon, I.D.; Weber, P.K.; Zimmerberg, J.; Kraft, M.L. Sphingolipid domains in the plasma membranes of fibroblasts are not enriched with cholesterol. J. Biol. Chem. 2013, 288, 16855–16861. [Google Scholar] [CrossRef] [PubMed]
- Aresta-Branco, F.; Cordeiro, A.M.; Marinho, H.S.; Cyrne, L.; Antunes, F.; de Almeida, R.F.M. Gel Domains in the Plasma Membrane of Saccharomyces cerevisiae: Highly Ordered, Ergosterol-Free, And Sphingolipid-Enriched Lipid Rafts. J. Biol. Chem. 2011, 286, 5043–5054. [Google Scholar] [CrossRef]
- Vecer, J.; Vesela, P.; Malinsky, J.; Herman, P. Sphingolipid levels crucially modulate lateral microdomain organization of plasma membrane in living yeast. FEBS Lett. 2014, 588, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Herman, P.; Vecer, J.; Opekarova, M.; Vesela, P.; Jancikova, I.; Zahumensky, J.; Malinsky, J. Depolarization affects the lateral microdomain structure of yeast plasma membrane. FEBS J. 2015, 282, 419–434. [Google Scholar] [CrossRef]
- Malinsky, J.; Tanner, W.; Opekarova, M. Transmembrane voltage: Potential to induce lateral microdomains. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 806–811. [Google Scholar] [CrossRef]
- Spira, F.; Mueller, N.S.; Beck, G.; Von Olshausen, P.; Beig, J.; Wedlich-Söldner, R. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat. Cell Biol. 2012, 14, 640–648. [Google Scholar] [CrossRef]
- Breslow, D.K.; Collins, S.R.; Bodenmiller, B.; Aebersold, R.; Simons, K.; Shevchenko, A.; Ejsing, C.S.; Weissman, J.S. Orm family proteins mediate sphingolipid homeostasis. Nature 2010, 463, 1048–1053. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Lone, M.A.; Schneiter, R.; Chang, A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. USA 2010, 107, 5851–5856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hjelmqvist, L.; Tuson, M.; Marfany, G.; Herrero, E.; Balcells, S.; Gonzalez-Duarte, R. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [PubMed]
- Funato, K.; Vallee, B.; Riezman, H. Biosynthesis and Trafficking of Sphingolipids in the Yeast Saccharomyces cerevisiae. Biochemistry 2002, 41, 15105–15113. [Google Scholar] [CrossRef] [PubMed]
- Gable, K.; Han, G.; Monaghan, E.; Bacikova, D.; Natarajan, M.; Williams, R.; Dunn, T.M. Mutations in the yeast LCB1 and LCB2 genes, including those corresponding to the hereditary sensory neuropathy type I mutations, dominantly inactivate serine palmitoyltransferase. J. Biol. Chem. 2002, 277, 10194–10200. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Gupta, S.D.; Gable, K.; Bacikova, D.; Sengupta, N.; Somashekarappa, N.; Proia, R.L.; Harmon, J.M.; Dunn, T.M. The ORMs interact with transmembrane domain 1 of Lcb1 and regulate serine palmitoyltransferase oligomerization, activity and localization. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2019, 1864, 245–259. [Google Scholar] [CrossRef] [PubMed]
- Casamayor, A.; Torrance, P.D.; Kobayashi, T.; Thorner, J.; Alessi, D.R. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 1999, 9, S184–S186. [Google Scholar] [CrossRef]
- Roelants, F.M.; Breslow, D.K.; Muir, A.; Weissman, J.S.; Thorner, J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2011, 108, 19222–19227. [Google Scholar] [CrossRef]
- Gururaj, C.; Federman, R.; Chang, A. Orm proteins integrate multiple signals to maintain sphingolipid homeostasis. J. Biol. Chem. 2013, 288, 20453–20463. [Google Scholar] [CrossRef]
- Kimberlin, A.N.; Han, G.; Chen, M.; Cahoon, R.E.; Luttgeharm, K.D.; Stone, J.M.; Markham, J.E.; Dunn, T.M.; Cahoon, E.B. ORM Expression Alters Sphingolipid Homeostasis and Differentially Affects Ceramide Synthase Activity. Plant Physiol. 2016, 172, 00965. [Google Scholar] [CrossRef]
- Roelants, F.M.; Torrance, P.D.; Bezman, N.; Thorner, J. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell 2002, 13, 3005–3028. [Google Scholar] [CrossRef]
- Roelants, F.M.; Torrance, P.D.; Thorner, J. Differential roles of PDK1- and PDK2- phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology 2004, 150, 3289–3304. [Google Scholar] [CrossRef] [PubMed]
- Kamada, Y.; Fujioka, Y.; Suzuki, N.N.; Inagaki, F.; Wullschleger, S.; Loewith, R.; Hall, M.N.; Ohsumi, Y. Tor2 Directly Phosphorylates the AGC Kinase Ypk2 To Regulate Actin Polarization. Mol. Cell. Biol. 2005, 25, 7239–7248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Miao, Y.; Yamane, Y.; Zhang, C.; Shokat, K.M.; Takematsu, H.; Kozutsumi, Y.; Drubin, D.G. Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol. Biol. Cell 2012, 23, 2388–2398. [Google Scholar] [CrossRef] [PubMed]
- Leskoske, K.L.; Roelants, F.M.; Marshall, M.N.M.; Hill, J.M.; Thorner, J. The stress-sensing TORC2 complex activates yeast AGC-family protein kinase ypk1 at multiple novel sites. Genetics 2017, 207, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.C.; Aguilar, P.S.; Fröhlich, F.; Chu, F.; Moreira, K.; Burlingame, A.L.; Walter, P. Pkh-kinases control eisosome assembly and organization. EMBO J. 2007, 26, 4946–4955. [Google Scholar] [CrossRef] [Green Version]
- Berchtold, D.; Walther, T.C. TORC2 Plasma Membrane Localization Is Essential for Cell Viability and Restricted to a Distinct Domain. Mol. Biol. Cell 2009, 20, 1565–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, G.; Gruhler, A.; Liu, Y.; Jensen, O.N.; Dickson, R.C. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J. Biol. Chem. 2008, 283, 10433–10444. [Google Scholar] [CrossRef]
- Baxter, B.K.; DiDone, L.; Ogu, D.; Schor, S.; Krysan, D.J. Identification, in Vitro Activity and Mode of Action of Phosphoinositide-Dependent-1 Kinase Inhibitors as Antifungal Molecules. ACS Chem. Biol. 2011, 6, 502–510. [Google Scholar] [CrossRef]
- Audhya, A.; Loewith, R.; Parsons, A.B.; Gao, L.; Tabuchi, M.; Zhou, H.; Boone, C.; Hall, M.N.; Emr, S.D. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 2004, 23, 3747–3757. [Google Scholar] [CrossRef] [Green Version]
- Niles, B.J.; Mogri, H.; Hill, A.; Vlahakis, A.; Powers, T. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc. Natl. Acad. Sci. USA 2012, 109, 1536–1541. [Google Scholar] [CrossRef] [Green Version]
- García-Marqués, S.; Randez-Gil, F.; Dupont, S.; Garre, E.; Prieto, J.A. Sng1 associates with Nce102 to regulate the yeast Pkh–Ypk signalling module in response to sphingolipid status. BBA Mol. Cell Res. 2016, 1863, 1319–1333. [Google Scholar] [CrossRef] [PubMed]
- García-López, M.C.; Mirón-García, M.C.; Garrido-Godino, A.I.; Mingorance, C.; Navarro, F. Overexpression of SNG1 causes 6-azauracil resistance in Saccharomyces cerevisiae. Curr. Genet. 2010, 56, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Cabiscol, E.; Herrero, E.; Ros, J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. Biochemistry 2000, 275, 27393–27398. [Google Scholar] [CrossRef] [PubMed]
- Breitenbach, M.; Laun, P.; Dickinson, J.R.; Klocker, A.; Rinnerthaler, M.; Dawes, I.W.; Aung-htut, M.T.; Breitenbach-koller, L.; Caballero, A.; Nyström, T.; et al. The Role of Mitochondria in the Aging Processes of Yeast. In Aging Research in Yeast; Breitenbach, M., Jazwinski, S.M., Laun, P., Eds.; Springer: New York, NY, USA, 2012; Volume 57, pp. 55–78. [Google Scholar]
- Niles, B.J.; Joslin, A.C.; Fresques, T.; Powers, T. TOR Complex 2-Ypk1 signaling maintains sphingolipid homeostasis by sensing and regulating ROS accumulation. Cell Rep. 2014, 6, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Mulet, J.M.; Martin, D.E.; Loewith, R.; Hall, M.N. Mutual antagonism of target of rapamycin and calcineurin signaling. J. Biol. Chem. 2006, 281, 33000–33007. [Google Scholar] [CrossRef]
- Tabuchi, M.; Audhya, A.; Parsons, A.B.; Boone, C.; Emr, S.D. The Phosphatidylinositol 4,5-Biphosphate and TORC2 Binding Proteins Slm1 and Slm2 Function in Sphingolipid Regulation. Mol. Cell. Biol. 2006, 26, 5861–5875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cyert, M.S. Calcineurin signaling in Saccharomyces cerevisiae: How yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 2003, 311, 1143–1150. [Google Scholar] [CrossRef]
- Deutscher, D.; Meilijson, I.; Kupiec, M.; Ruppin, E. Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nat. Genet. 2006, 38, 993–998. [Google Scholar] [CrossRef]
- Zahumensky, J.; Malinsky, J. Intracellular engagement of Nce102-like proteins. Unpublished.
- Jenkins, G.M.; Richards, A.; Wahl, T.; Mao, C.; Obeid, L.; Hannun, Y. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 32566–32572. [Google Scholar] [CrossRef]
- Skrzypek, M.S.; Nagiec, M.M.; Lester, R.L.; Dickson, R.C. Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J. Bacteriol. 1999, 181, 1134–1140. [Google Scholar] [PubMed]
- Vaskovicova, K.; Awadova, T.; Vesela, P.; Balazova, M.; Opekarova, M.; Malinsky, J. mRNA decay is regulated via sequestration of the conserved 5′-3′ exoribonuclease Xrn1 at eisosome in yeast. Eur. J. Cell Biol. 2017, 96, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Gournas, C.; Gkionis, S.; Carquin, M.; Twyffels, L.; Tyteca, D.; André, B. Conformation-dependent partitioning of yeast nutrient transporters into starvation-protective membrane domains. Proc. Natl. Acad. Sci. USA 2018, 115, E3145–E3154. [Google Scholar] [CrossRef] [PubMed]
- Payrastre, B.; Missy, K.; Giuriato, S.; Bodin, S.; Plantavid, M.; Gratacap, M.P. Phosphoinositides: Key players in cell signalling, in time and space. Cell. Signal. 2001, 13, 377–387. [Google Scholar] [CrossRef]
- Balla, T. Phosphoinositides: Tiny Lipids with Giant Impact on Cell Regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef] [PubMed]
- Posor, Y.; Eichhorn-Grünig, M.; Haucke, V. Phosphoinositides in endocytosis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2015, 1851, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Fruman, D.A.; Meyers, R.E.; Cantley, L.C. Phosphoinositide kinases. Ann. Rev. Biochem. 1998, 67, 481–507. [Google Scholar] [CrossRef]
- Martin, T.F.J. Phosphoinositide lipids as signaling molecules: Common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu. Rev. Cell Dev. Biol. 1998, 14, 231–264. [Google Scholar] [CrossRef]
- Scharenberg, A.M.; El-Hillal, O.; Fruman, D.A.; Beitz, L.O.; Li, Z.M.; Lin, S.Q.; Gout, I.; Cantley, L.C.; Rawlings, D.J.; Kinet, J.P. Phosphatidylinositol-3,4,5-trisphosphate (PtdIns-3,4,5-P-3) Tec kinase-dependent calcium signaling pathway: A target for SHIP-mediated inhibitory signals. EMBO J. 1998, 17, 1961–1972. [Google Scholar] [CrossRef]
- Simonsen, A.; Wurmser, A.E.; Emr, S.D.; Stenmark, H. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 2001, 13, 485–492. [Google Scholar] [CrossRef]
- Takenawa, T.; Itoh, T. Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2001, 1533, 190–206. [Google Scholar] [CrossRef]
- Di Paolo, G.; De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Delage, E.; Puyaubert, J.; Zachowski, A.; Ruelland, E. Signal transduction pathways involving phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: Convergences and divergences among eukaryotic kingdoms. Prog. Lipid Res. 2013, 52, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Ohya, Y.; Goebl, M.; Nakano, A.; Anraku, Y. A novel gene, STT4, encodes a phosphatidylinositol 4-kinase in the PKC1 protein kinase pathway of Saccharomyces cerevisiae. J. Biol. Chem. 1994, 269, 1166–1172. [Google Scholar] [PubMed]
- Cutler, N.S.; Heitman, J.; Cardenas, M.E. STT4 Is an Essential Phosphatidylinositol 4-Kinase That Is a Target of Wortmannin in Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 27671–27677. [Google Scholar] [CrossRef] [Green Version]
- Boronenkov, I.V.; Anderson, R.A. The sequence of phosphatidylinositol-4-phosphate 5-kinase defines a novel family of lipid kinases. J. Biol. Chem. 1995, 270, 2881–2884. [Google Scholar] [CrossRef]
- Homma, K.; Terui, S.; Minemura, M.; Qadota, H.; Anraku, Y.; Kanaho, Y.; Ohya, Y. Phosphatidylinositol-4-phosphate 5-Kinase Localized on the Plasma Membrane Is Essential for Yeast Cell Morphogenesis. J. Biol. Chem. 1998, 273, 15779–15786. [Google Scholar] [CrossRef] [Green Version]
- McLaughlin, S.; Wang, J.; Gambhir, A.; Murray, D. PIP2 and Proteins: Interactions, Organization, and Information Flow. Annu. Rev. Biophys. Biomol. Struct. 2002, 31, 151–175. [Google Scholar] [CrossRef]
- Stefan, C.J.; Audhya, A.; Emr, S.D. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell 2002, 13, 542–557. [Google Scholar] [CrossRef]
- Riggi, M.; Niewola-Staszkowska, K.; Chiaruttini, N.; Colom, A.; Kusmider, B.; Mercier, V.; Soleimanpour, S.; Stahl, M.; Matile, S.; Roux, A.; et al. Decrease in plasma membrane tension triggers PtdIns(4,5)P2 phase separation to inactivate TORC2. Nat. Cell Biol. 2018, 20, 1043–1051. [Google Scholar] [CrossRef]
- Tomioku, K.n.; Shigekuni, M.; Hayashi, H.; Yoshida, A.; Futagami, T.; Tamaki, H.; Tanabe, K.; Fujita, A. Nanoscale domain formation of phosphatidylinositol 4-phosphate in the plasma and vacuolar membranes of living yeast cells. Eur. J. Cell Biol. 2018, 97, 269–278. [Google Scholar] [CrossRef]
- Vernay, A.; Schaub, S.; Guillas, I.; Bassilana, M.; Arkowitz, R.A. A steep phosphoinositide bis-phosphate gradient forms during fungal filamentous growth. J. Cell Biol. 2012, 198, 711–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hairfield, M.L.; Westwater, C.; Dolan, J.W. Phosphatidylinositol-4-phosphate 5-kinase activity is stimulated during temperature-induced morphogenesis in Candida albicans. Microbiology 2002, 148, 1737–1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozelle, A.L.; Machesky, L.M.; Yamamoto, M.; Driessens, M.H.E.; Insall, R.H.; Roth, M.G.; Luby-Phelps, K.; Marriott, G.; Hall, A.; Yin, H.L. Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr. Biol. 2000, 10, 311–320. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.J.; Guzman-Hernandez, M.L.; Balla, T. A Highly Dynamic ER-Derived Phosphatidylinositol-Synthesizing Organelle Supplies Phosphoinositides to Cellular Membranes. Dev. Cell 2011, 21, 813–824. [Google Scholar] [CrossRef] [Green Version]
- Ziółkowska, N.E.; Karotki, L.; Rehman, M.; Huiskonen, J.T.; Walther, T.C. Eisosome-driven plasma membrane organization is mediated by BAR domains. Nat. Struct. Mol. Biol. 2011, 18, 854–856. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, K.; Gadila, S.K.G.; Tenay, B.; McDermott, H.; Alcox, B.; Kim, K. TORC2 and eisosomes are spatially interdependent, requiring optimal level of phosphatidylinositol 4, 5-bisphosphate for their integrity. J. Biosci. 2015, 40, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.R.; Boxberger, J.; Colvin, R.; Lee, S.J.; Zahn, G.; Loor, F.; Kim, K. Pil1, an eisosome organizer, plays an important role in the recruitment of synaptojanins and amphiphysins to facilitate receptor-mediated endocytosis in yeast. Eur. J. Cell Biol. 2011, 90, 825–833. [Google Scholar] [CrossRef]
- Badrane, H.; Nguyen, M.H.; Blankenship, J.R.; Cheng, S.; Hao, B.; Mitchell, A.P.; Clancy, C.J. Rapid Redistribution of Phosphatidylinositol-(4,5)-Bisphosphate and Septins during the Candida albicans Response to Caspofungin. Antimicrob. Agents Chemother. 2012, 56, 4614–4624. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, F.J.; Douglas, L.M.; Rosebrock, A.; Konopka, J.B. The Sur7 Protein Regulates Plasma Membrane Organization and Prevents Intracellular Cell Wall Growth in Candida albicans. Mol. Biol. Cell 2008, 19, 5214–5225. [Google Scholar] [CrossRef]
- Kabeche, R.; Madrid, M.; Cansado, J.; Moseley, J.B. Eisosomes regulate phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) cortical clusters and mitogen-activated protein (MAP) kinase signaling upon osmotic stress. J. Biol. Chem. 2015, 290, 25960–25973. [Google Scholar] [CrossRef] [PubMed]
- Scheek, S.; Brown, M.S.; Goldstein, J.L. Sphingomyelin depletion in cultured cells blocks proteolysis of sterol regulatory element binding proteins at site 1. Proc. Natl. Acad. Sci. USA 1997, 94, 11179–11183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beeler, T.; Bacikova, D.; Gable, K.; Hopkins, L.; Johnson, C.; Slife, H.; Dunn, T. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2 Delta mutant. J. Biol. Chem. 1998, 273, 30688–30694. [Google Scholar] [CrossRef] [PubMed]
- Baudry, K.; Swain, E.; Rahier, A.; Germann, M.; Batta, A.; Rondet, S.; Mandala, S.; Henry, K.; Tint, G.S.; Edlind, T.; et al. The effect of the erg26-1 mutation on the regulation of lipid metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 12702–12711. [Google Scholar] [CrossRef] [PubMed]
- Eisenkolb, M.; Zenzmaier, C.; Leitner, E.; Schneiter, R. A Specific Structural Requirement for Ergosterol in Long-chain Fatty Acid Synthesis Mutants Important for Maintaining Raft Domains in Yeast. Mol. Biol. Cell 2002, 13, 4414–4428. [Google Scholar] [CrossRef] [PubMed]
- Swain, E.; Baudry, K.; Stukey, J.; McDonough, V.; Germann, M.; Nickels, J.T. Sterol-dependent regulation of sphingolipid metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 26177–26184. [Google Scholar] [CrossRef] [PubMed]
- Sano, T.; Kihara, A.; Kurotsu, F.; Iwaki, S.; Igarashi, Y. Regulation of the sphingoid long-chain base kinase Lcb4p by ergosterol and heme - Studies in phytosphingosine-resistant mutants. J. Biol. Chem. 2005, 280, 36674–36682. [Google Scholar] [CrossRef]
- Valachovic, M.; Wilcox, L.J.; Sturley, S.L.; Bard, M. A mutation in sphingolipid synthesis suppresses defects in yeast ergosterol metabolism. Lipids 2004, 39, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Valachovic, M.; Bareither, B.M.; Bhuiyan, M.S.A.; Eckstein, J.; Barbuch, R.; Balderes, D.; Wilcox, L.; Sturley, S.L.; Dickson, R.C.; Bard, M. Cumulative Mutations Affecting Sterol Biosynthesis in the Yeast Saccharomyces cerevisiae Result in Synthetic Lethality That Is Suppressed by Alterations in Sphingolipid Profiles. Genetics 2006, 173, 1893–1908. [Google Scholar] [CrossRef] [Green Version]
- Brice, S.E.; Alford, C.W.; Cowart, L.A. Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae. J. Biol. Chem. 2009, 284, 7588–7596. [Google Scholar] [CrossRef]
- Carman, G.M.; Han, G.-S. Phosphatidic Acid Phosphatase, a Key Enzyme in the Regulation of Lipid Synthesis. J. Biol. Chem. 2009, 284, 2593–2597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, X.L.; Souza, C.M.; Pichler, H.; Schaad, O.; Kajiwara, K.; Wakabayashi, H.; Ivanova, T.; Castillon, G.A.; Piccolis, M.; Abe, F.; et al. Functional Interactions between Sphingolipids and Sterols in Biological Membranes Regulating Cell Physiology. Mol. Biol. Cell 2009, 20, 2083–2095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raychaudhuri, S.; Im, Y.J.; Hurley, J.H.; Prinz, W.A. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J. Cell Biol. 2006, 173, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Schulz, T.A.; Choi, M.G.; Raychaudhuri, S.; Mears, J.A.; Ghirlando, R.; Hinshaw, J.E.; Prinz, W.A. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J. Cell Biol. 2009, 187, 889–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Saint-Jean, M.; Delfosse, V.; Douguet, D.; Chicanne, G.; Payrastre, B.; Bourguet, W.; Antonny, B.; Drin, G. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 2011, 195, 965–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefan, C.J.; Manford, A.G.; Baird, D.; Yamada-Hanff, J.; Mao, Y.; Emr, S.D. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 2011, 144, 389–401. [Google Scholar] [CrossRef]
- Guo, S.; Stolz, L.E.; Lemrow, S.M.; York, J.D. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 1999, 274, 12990–12995. [Google Scholar] [CrossRef]
- Hughes, W.E.; Woscholski, R.; Cooke, F.T.; Patrick, R.S.; Dove, S.K.; McDonald, N.Q.; Parker, P.J. SAC1 encodes a regulated lipid phosphoinositide phosphatase, defects in which can be suppressed by the homologous Inp52p and Inp53p phosphatases. J. Biol. Chem. 2000, 275, 801–808. [Google Scholar] [CrossRef]
- Stradalova, V.; Blazikova, M.; Grossmann, G.; Opekarova, M.; Tanner, W.; Malinsky, J. Distribution of cortical endoplasmic reticulum determines positioning of endocytic events in yeast plasma membrane. PLoS ONE 2012, 7, e35132. [Google Scholar] [CrossRef]
- Roelants, F.M.; Chauhan, N.; Muir, A.; Davis, J.C.; Menon, A.K.; Levine, T.P.; Thorner, J. TOR complex 2-regulated protein kinase Ypk1 controls sterol distribution by inhibiting StARkin domain-containing proteins located at plasma membrane-endoplasmic reticulum contact sites. Mol. Biol. Cell 2018, 29, 2128–2136. [Google Scholar] [CrossRef]
- Kabeche, R.; Howard, L.; Moseley, J.B. Eisosomes provide membrane reservoirs for rapid expansion of the yeast plasma membrane. J. Cell Sci. 2015, 128, 4057–4062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riggi, M.; Bourgoint, C.; Macchione, M.; Matile, S.; Loewith, R.; Roux, A. TORC2 controls endocytosis through plasma membrane tension. J. Cell Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Li, L.F.; Naseem, S.; Sharma, S.; Konopka, J.B. Flavodoxin-Like Proteins Protect Candida albicans from Oxidative Stress and Promote Virulence. PLoS Pathog. 2015, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Desmyter, L.; Verstraelen, J.; Dewaele, S.; Libert, C.; Contreras, R.; Chen, C. Nonclassical export pathway: Overexpression of NCE102 reduces protein and DNA damage and prolongs lifespan in an SGS1 deficient Saccharomyces cerevisiae. Biogerontology 2007, 8, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.B.; Tang, L.; Ying, S.H.; Feng, M.G. Two eisosome proteins play opposite roles in autophagic control and sustain cell integrity, function and pathogenicity in Beauveria bassiana. Environ. Microbiol. 2017, 19, 2037–2052. [Google Scholar] [CrossRef] [PubMed]

| MCC/Eisosome Components | |
| Lsp1/Pil1 | - BAR (Bin/Amphiphysin/Rvs) domain proteins—bind PI(4,5)P2 at the plasma membrane - Core eisosome constituents, essential for eisosome formation (Pil1) - Differentially phosphorylated by Pkh1/2 in response to LCB levels |
| Nce102-like -proteins | - Tetraspan proteins -Anchor nutrient transporters to MCC - Change their plasma membrane distribution in response to sphingolipid content (Nce102) |
| nutrient -transporters | - APC (amino acid polyamine organocation) transporters localizing to MCC - Plasma membrane trafficking of most of these was shown to be ergosterol-dependent |
| Slm1/2 | - BAR and pleckstrin homology domain proteins—bind PI(4,5)P2 at the plasma membrane - Travel between MCC and TORC2 (Tor Complex 2) in response to membrane stress - Activate TORC2 |
| Kinases | |
| Pkh1/2 | - Sphingolipid-dependent kinases - Inhibited by Nce102 at eisosomes |
| TORC2 | - Tor Complex 2, composed of six known proteins (including Avo1-3) - Associated to plasma membrane, likely via pleckstrin homology domain protein Avo1 - Activated during membrane stress by Slm1/2 |
| Ypk1 | - Inhibitor of Orm2, i.e., indirect activator of SPT (serine palmitoyltransferase) - Dually activated by Pkh1/2 and TORC2 |
| Phosphatases | |
| Inp51 | - PI(4,5)P2 phosphatase - Recruited to eisosome via interaction with Pil1 |
| Sac1 | - PI(4)P phosphatase - Forms a higher-order complex with SPT |
| Others | |
| Orm1/2 | - Inhibitors of SPT - Inactivated by Ypk1-mediated phosphorylation (Orm2) |
| SPT | - Serine palmitoyltransferase complex, composed of Lcb1, Lcb2 and Tsc3 - Catalyses the first and rate-limiting step of sphingolipid biosynthesis |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zahumensky, J.; Malinsky, J. Role of MCC/Eisosome in Fungal Lipid Homeostasis. Biomolecules 2019, 9, 305. https://doi.org/10.3390/biom9080305
Zahumensky J, Malinsky J. Role of MCC/Eisosome in Fungal Lipid Homeostasis. Biomolecules. 2019; 9(8):305. https://doi.org/10.3390/biom9080305
Chicago/Turabian StyleZahumensky, Jakub, and Jan Malinsky. 2019. "Role of MCC/Eisosome in Fungal Lipid Homeostasis" Biomolecules 9, no. 8: 305. https://doi.org/10.3390/biom9080305
APA StyleZahumensky, J., & Malinsky, J. (2019). Role of MCC/Eisosome in Fungal Lipid Homeostasis. Biomolecules, 9(8), 305. https://doi.org/10.3390/biom9080305






