Identification of SWI2/SNF2-Related 1 Chromatin Remodeling Complex (SWR1-C) Subunits in Pineapple and the Role of Pineapple SWR1 COMPLEX 6 (AcSWC6) in Biotic and Abiotic Stress Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, Growth Conditions, and Treatments
2.2. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.3. RNA Seq of Pineapple
2.4. Phylogenetic Analysis and Identification of Homologue Sequences in Pineapple
2.5. Gene Structure Analysis and Chromosomal Location
2.6. Vector Construction
2.7. Microscopy
2.8. Statistical Analysis
3. Results
3.1. Identification, Phylogenetic Analysis, and Gene Structure of SWR1-C Subunits
3.2. Chromosomal Location of SWR1-C Components
3.3. AcSWR1-C Subunits Show Differential Expression
3.4. Expression of SWR1-C Components is Regulated Under Abiotic Stress
3.5. AcSWC6 Gets Localized to the Nucleus
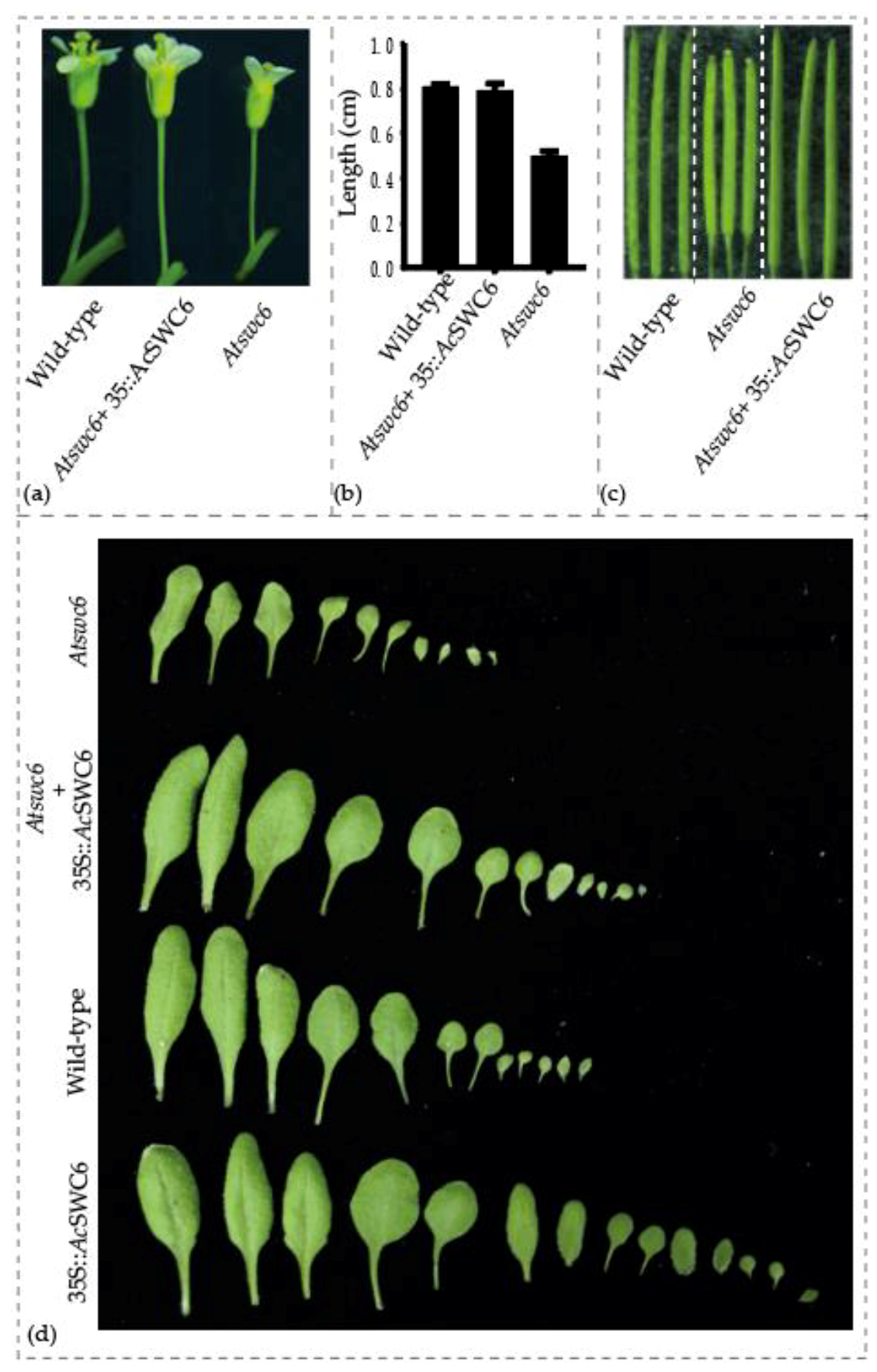
3.6. AcSWC6 Regulates the Biotic and Abiotic Stress Tolerance in Arabidopsis
3.7. SWC6 Function Is Conserved in Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SWR1 | SWI2/SNF2-RELATED 1 |
| DAB | 3,3-diaminobenzidine |
| SWC2 | SWR1 COMPLEX2 |
| SWC3 | SWR1 COMPLEX 3 |
| SWC5 | SWR1 COMPLEX 5 |
| SWC6 | SWR1 COMPLEX 6 |
| SEF | SERRATED LEAF AND EARLY FLOWERING |
| SWC7 | SWR1 COMPLEX 7 |
| ARP6 | ACTIN-RELATED PROTEIN 6 |
| YAF9 | YEAST ALL1 FUSED GENE FROM CHROMOSOME 9 |
| BDF1 | BROMODOMAIN-CONTAINING FACTOR 1 |
| ACT1 | ACTIN 1 |
| ARP4 | ACTIN-RELATED PROTEIN 4 |
| SWC4 | SWR1 COMPLEX 4 |
| RVB1 | RUVB LIKE PROTEIN 1 |
| RVB2 | RUVB LIKE PROTEIN 2 |
| UTR | Untranslated region |
| CDS | Coding sequence |
References
- Conde, D.; Perales, M.; Sreedasyam, A.; Tuskan, G.A.; Lloret, A.; Badenes, M.L.; Gonzalez-Melendi, P.; Rios, G.; Allona, I. Engineering Tree Seasonal Cycles of Growth Through Chromatin Modification. Front. Plant Sci. 2019, 10, 412. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Ono, A.; Scholten, S.; Kinoshita, T.; Zilberman, D.; Okamoto, T. DNA demethylation by ROS1a in rice vegetative cells promotes methylation in sperm. Proc. Natl. Acad. Sci. USA 2019, 116, 9652–9657. [Google Scholar] [CrossRef] [PubMed]
- Forgione, I.; Woloszynska, M.; Pacenza, M.; Chiappetta, A.; Greco, M.; Araniti, F.; Abenavoli, M.R.; Van Lijsebettens, M.; Bitonti, M.B.; Bruno, L. Hypomethylated drm1 drm2 cmt3 mutant phenotype of Arabidopsis thaliana is related to auxin pathway impairment. Plant Sci. 2019, 280, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.H.; Wu, C.H.; Ladurner, A.; Mizuguchi, G.; Wei, D.; Xiao, H.; Luk, E.; Ranjan, A.; Wu, C. N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex. J. Biol. Chem. 2009, 284, 6200–6207. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Park, C.; Lee, J.; Oh, M.; Noh, B.; Lee, I. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 2007, 134, 1931–1941. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Zambrano, A.; Crevillen, P.; Franco-Zorrilla, J.M.; Lopez, J.A.; Moreno-Romero, J.; Roszak, P.; Santos-Gonzalez, J.; Jurado, S.; Vazquez, J.; Kohler, C.; et al. Arabidopsis SWC4 Binds DNA and Recruits the SWR1 Complex to Modulate Histone H2A.Z Deposition at Key Regulatory Genes. Mol. Plant 2018, 11, 815–832. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Bartholomew, B. Regulation of ATP-dependent chromatin remodelers: Accelerators/Brakes, anchors and sensors. Biochem. Soc. Trans. 2018, 46, 1423–1430. [Google Scholar] [CrossRef] [PubMed]
- Sura, W.; Kabza, M.; Karlowski, W.M.; Bieluszewski, T. Dual Role of the Histone Variant H2A.Z in Transcriptional Regulation of Stress-Response Genes. Plant Cell 2017, 29, 791–807. [Google Scholar] [CrossRef]
- Cai, H.; Zhao, L.; Wang, L.; Zhang, M.; Su, Z.; Cheng, Y.; Zhao, H.; Qin, Y. ERECTA signaling controls Arabidopsis inflorescence architecture through chromatin-mediated activation of PRE1 expression. New Phytol. 2017, 214, 1579–1596. [Google Scholar] [CrossRef]
- Berriri, S.; Gangappa, S.N.; Kumar, S.V. SWR1 Chromatin-Remodeling Complex Subunits and H2A.Z Have Non-overlapping Functions in Immunity and Gene Regulation in Arabidopsis. Mol. Plant 2016, 9, 1051–1065. [Google Scholar] [CrossRef]
- Kumar, S.V. H2A.Z at the core of transcriptional regulation in plants. Mol. Plant 2018, 11, 1112–1114. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Kim, J. Regulation of MicroRNA-Mediated Developmental Changes by the SWR1 Chromatin Remodeling Complex. Plant Physiol. 2016, 171, 1128–1143. [Google Scholar] [PubMed]
- Dai, X.; Bai, Y.; Zhao, L.; Dou, X.; Liu, Y.; Wang, L.; Li, Y.; Li, W.; Hui, Y.; Huang, X.; et al. H2A.Z Represses Gene Expression by Modulating Promoter Nucleosome Structure and Enhancer Histone Modifications in Arabidopsis. Mol. Plant 2017, 10, 1274–1292. [Google Scholar] [CrossRef] [PubMed]
- March-Diaz, R.; Reyes, J.C. The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant 2009, 2, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.H.; Alami, S.; Luk, E.; Wu, C.H.; Sen, S.; Mizuguchi, G.; Wei, D.; Wu, C. SWC2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 2005, 12, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.J.; Shen, X. Chromatin remodelling beyond transcription: The INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol. 2009, 10, 373–384. [Google Scholar] [CrossRef]
- March-Diaz, R.; Garcia-Dominguez, M.; Lozano-Juste, J.; Leon, J.; Florencio, F.J.; Reyes, J.C. Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 2008, 53, 475–487. [Google Scholar] [CrossRef]
- Cai, H.; Zhang, M.; Chai, M.; He, Q.; Huang, X.; Zhao, L.; Qin, Y. Epigenetic regulation of anthocyanin biosynthesis by an antagonistic interaction between H2A.Z and H3K4me3. New Phytol. 2019, 221, 295–308. [Google Scholar] [CrossRef]
- March-Diaz, R.; Garcia-Dominguez, M.; Florencio, F.J.; Reyes, J.C. SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 2007, 143, 893–901. [Google Scholar] [CrossRef]
- Rosa, M.; Von Harder, M.; Cigliano, R.A.; Schlogelhofer, P.; Mittelsten Scheid, O. The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell 2013, 25, 1990–2001. [Google Scholar] [CrossRef]
- Lazaro, A.; Gomez-Zambrano, A.; Lopez-Gonzalez, L.; Pineiro, M.; Jarillo, J.A. Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 2008, 59, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zhao, L.; Skaggs, M.I.; Andreuzza, S.; Tsukamoto, T.; Panoli, A.; Wallace, K.N.; Smith, S.; Siddiqi, I.; Yang, Z.; et al. ACTIN-RELATED PROTEIN6 Regulates Female Meiosis by Modulating Meiotic Gene Expression in Arabidopsis. Plant Cell 2014, 26, 1612–1628. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Tong, A.; Huo, Y.; Yan, Z.; Yang, W.; Yang, X.; Wang, X.X. SKIP controls flowering time via the alternative splicing of SEF pre-mRNA in Arabidopsis. BMC Biol. 2017, 15, 80. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; VanBuren, R.; Wai, C.M.; Tang, H. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015, 47, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Redwan, R.M.; Saidin, A.; Kumar, S.V. The draft genome of MD-2 pineapple using hybrid error correction of long reads. DNA Res. 2016, 23, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Ur Rahman, Z.; Azam, S.M.; Liu, Y.; Yan, C.; Ali, H.; Zhao, L.; Chen, P.; Yi, L.; Priyadarshani, S.; Yuan, Q. Expression profiles of Wuschel-related homeobox gene family in pineapple (Ananas comosus L.). Trop. Plant Biol. 2017, 10, 204–215. [Google Scholar] [CrossRef]
- Ming, R.; Wai, C.M.; Guyot, R. Pineapple Genome: A Reference for Monocots and CAM Photosynthesis. Trends Genet. 2016, 32, 690–696. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef]
- Nguyen, B.T.; Bujna, E.; Fekete, N.; Tran, A.T.M.; Rezessy-Szabo, J.M.; Prasad, R.; Nguyen, Q.D. Probiotic Beverage from Pineapple Juice Fermented with Lactobacillus and Bifidobacterium Strains. Front. Nutr. 2019, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Priyadarshani, S.; Hu, B.; Li, W.; Ali, H. Simple protoplast isolation system for gene expression and protein interaction studies in pineapple (Ananas comosus L.). Plant Methods 2018, 14, 95. [Google Scholar] [CrossRef]
- Ali, H.; Liu, Y.; Azam, S.M.; Rahman, Z.U.; Priyadarshani, S.; Li, W.; Huang, X.; Hu, B.; Xiong, J.; Ali, U.; et al. Genomic Survey, Characterization, and Expression Profile Analysis of the SBP Genes in Pineapple (Ananas comosus L.). Int. J. Genom. 2017, 2017, 1032846. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, Y.; Zhao, L.; Hou, Z.; Yan, M.; Hu, B.; Liu, Y.; Azam, S.M.; Zhang, Z.; Rahman, Z.U.; et al. Genome-Wide Identification and Expression Profiling of ATP-Binding Cassette (ABC) Transporter Gene Family in Pineapple (Ananas comosus (L.) Merr.) Reveal the Role of AcABCG38 in Pollen Development. Front. Plant Sci. 2017, 8, 2150. [Google Scholar] [CrossRef] [PubMed]
- Baskin, T.I.; Wilson, J.E. Inhibitors of protein kinases and phosphatases alter root morphology and disorganize cortical microtubules. Plant Physiol. 1997, 113, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A. Auxin: A regulator of cold stress response. Physiol. Plant 2013, 147, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Seifi, H.S.; Zarei, A.; Hsiang, T.; Shelp, B. Spermine is a potent plant defense activator against gray mold disease on Solanum lycopersicum, Phaseolus vulgaris and Arabidopsis thaliana. Phytopathology 2019. [Google Scholar] [CrossRef] [PubMed]
- Raad, M.; Glare, T.R.; Brochero, H.L.; Muller, C.; Rostas, M. Transcriptional Reprogramming of Arabidopsis thaliana Defence Pathways by the Entomopathogen Beauveria bassiana Correlates with Resistance Against a Fungal Pathogen but Not Against Insects. Front. Microbiol. 2019, 10, 615. [Google Scholar] [CrossRef]
- Daudi, A.; O’Brien, J.A. Detection of Hydrogen Peroxide by DAB Staining in Arabidopsis Leaves. Plant Cell 2012, 2, e263. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, H.; Hu, B.; Zhao, L.; Shi, D.; She, Z.; Huang, X.; Priyadarshani, S.; Niu, X.; Qin, Y. Differential Expression Analysis of Reference Genes in Pineapple (Ananas comosus L.) during Reproductive Development and Response to Abiotic Stress, Hormonal Stimuli. Trop. Plant Biol. 2019, 12, 67–77. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yan, M.; Hu, B.; Priyadarshani, S.; Hou, Z.; Ojolo, S.P.; Xiong, J.; Zhao, H.; Qin, Y. Characterization and the Expression Analysis of Nitrate Transporter (NRT) Gene Family in Pineapple. Trop. Plant Biol. 2018, 11, 177–191. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Potok, M.E.; Wang, Y.; Xu, L.; Zhong, Z.; Liu, W.; Feng, S.; Naranbaatar, B.; Rayatpisheh, S.; Wang, Z.; Wohlschlegel, J.A.; et al. Arabidopsis SWR1-associated protein methyl-CpG-binding domain 9 is required for histone H2A.Z deposition. Nat. Commun. 2019, 10, 3352. [Google Scholar] [CrossRef] [PubMed]
- Henderson, I.R.; Jacobsen, S.E. Epigenetic inheritance in plants. Nature 2007, 447, 418–424. [Google Scholar] [CrossRef]
- Sarnowska, E.; Gratkowska, D.M.; Sacharowski, S.P.; Cwiek, P.; Tohge, T.; Fernie, A.R.; Siedlecki, J.A.; Koncz, C.; Sarnowski, T.J. The Role of SWI/SNF Chromatin Remodeling Complexes in Hormone Crosstalk. Trends Plant Sci. 2016, 21, 594–608. [Google Scholar] [CrossRef] [PubMed]
- Deal, R.B.; Kandasamy, M.K.; McKinney, E.C.; Meagher, R.B. The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 2005, 17, 2633–2646. [Google Scholar] [CrossRef]
- Deal, R.B.; Topp, C.N.; McKinney, E.C.; Meagher, R.B. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 2007, 19, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.Q.; Ranjan, A.; Stengel, F.; Wei, D.; Aebersold, R.; Wu, C.; Leschziner, A.E. Molecular architecture of the ATP-dependent chromatin-remodeling complex SWR1. Cell 2013, 154, 1220–1231. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Hyun, Y.; Kang, M.J.; In Yun, H.; Yun, J.Y.; Lister, C.; Dean, C.; Amasino, R.M.; Noh, B.; Noh, Y.S.; et al. Resetting and regulation of Flowering Locus C expression during Arabidopsis reproductive development. Plant J. Cell Mol. Biol. 2009, 57, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Ojolo, S.P.; Cao, S.; Priyadarshani, S.; Li, W.; Yan, M.; Aslam, M.; Zhao, H.; Qin, Y. Regulation of Plant Growth and Development: A Review from a Chromatin Remodeling Perspective. Front. Plant Sci. 2018, 9, 1232. [Google Scholar] [CrossRef] [PubMed]
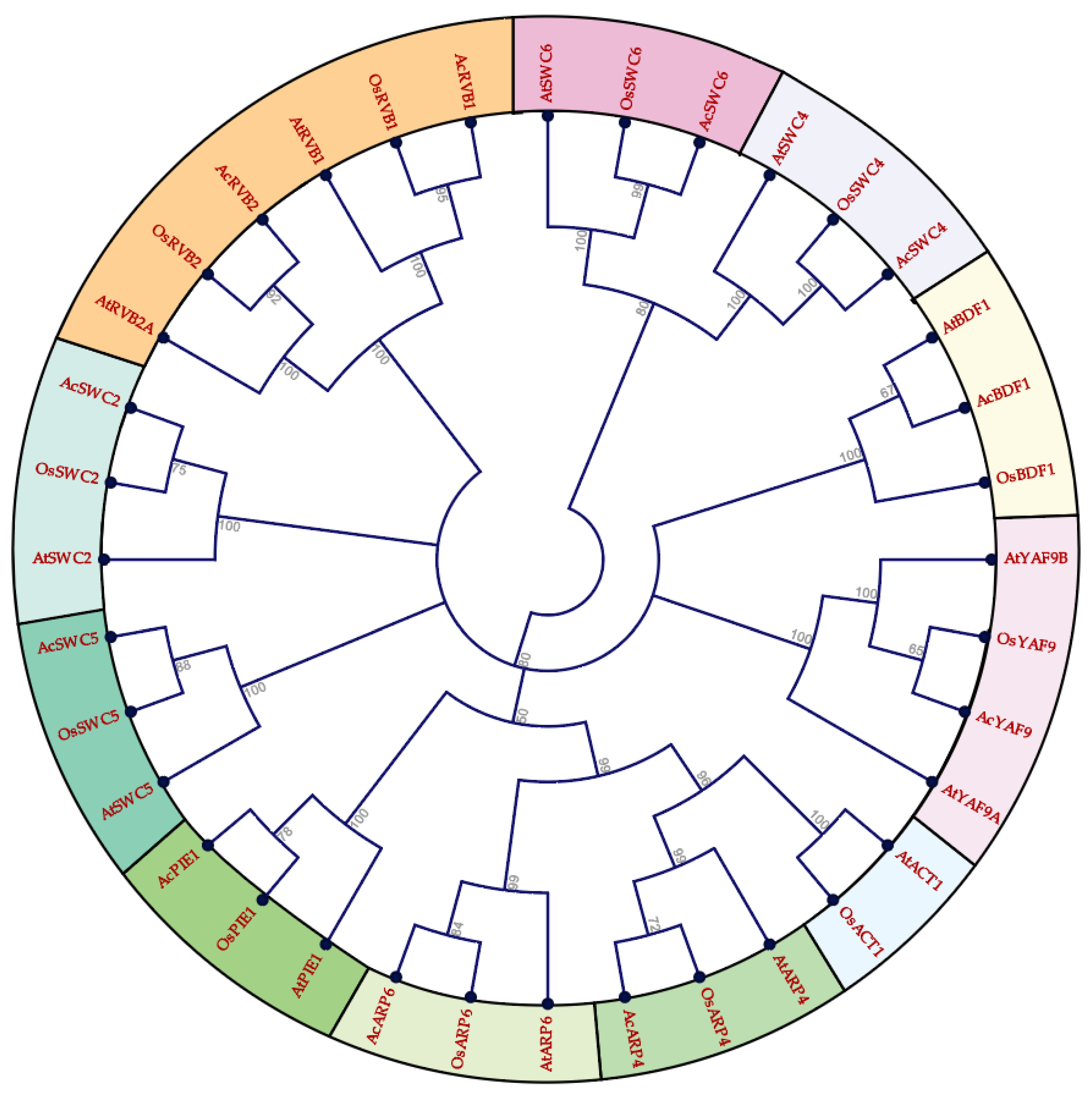

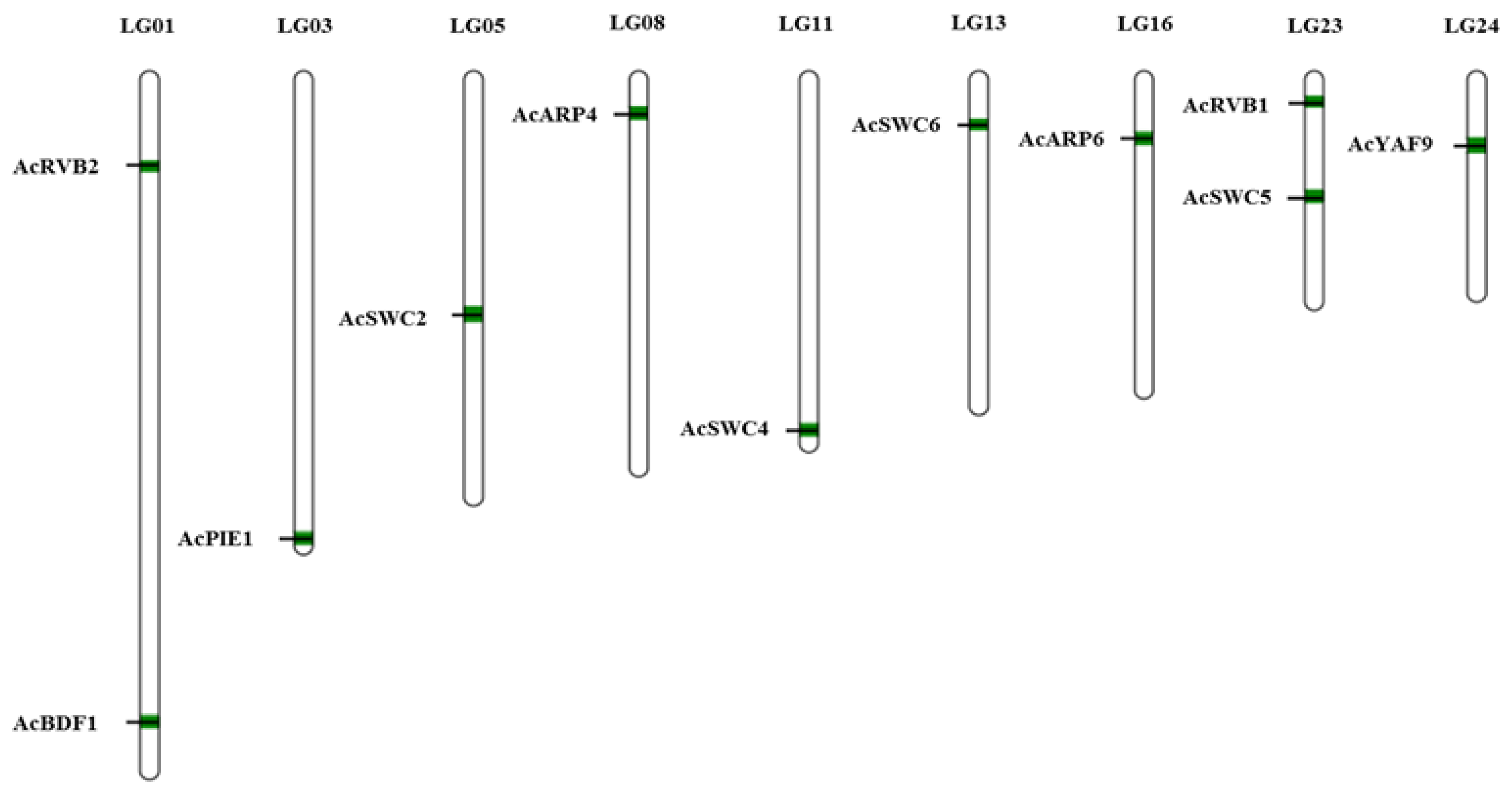
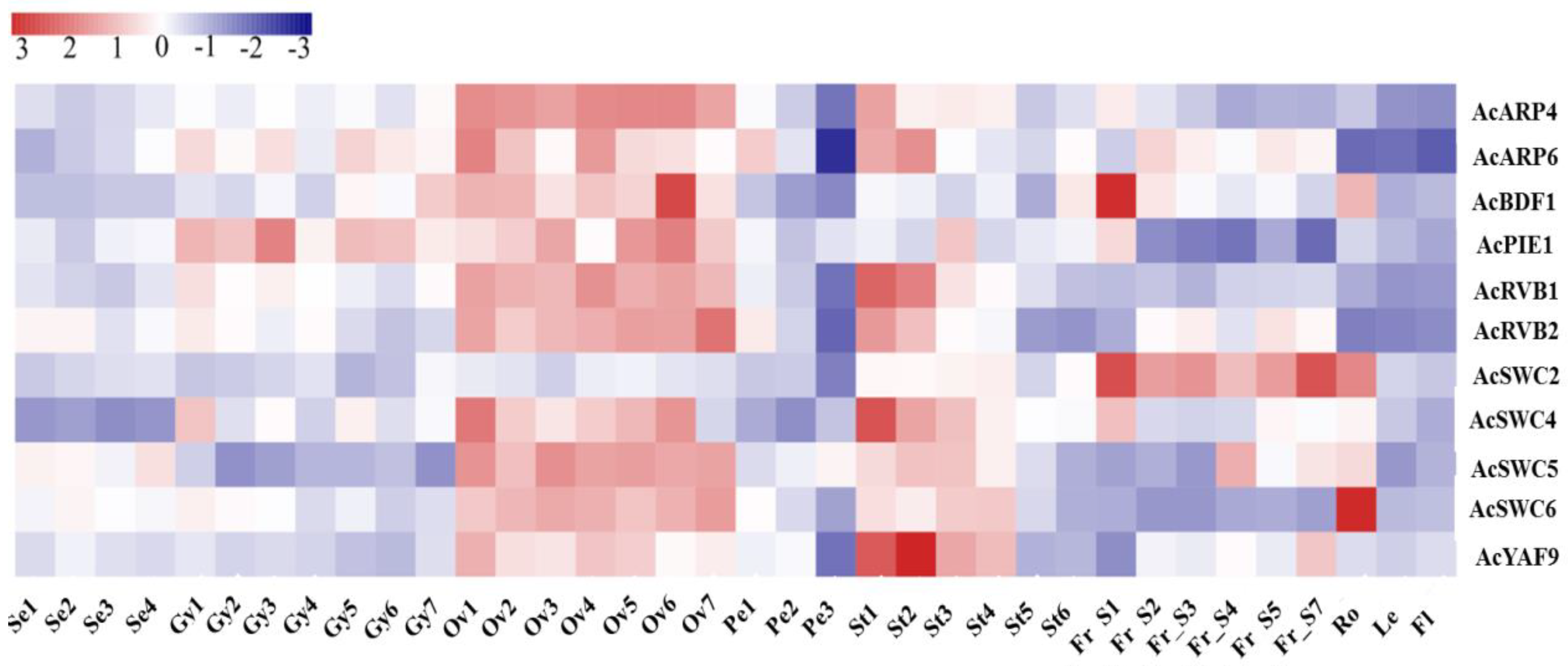
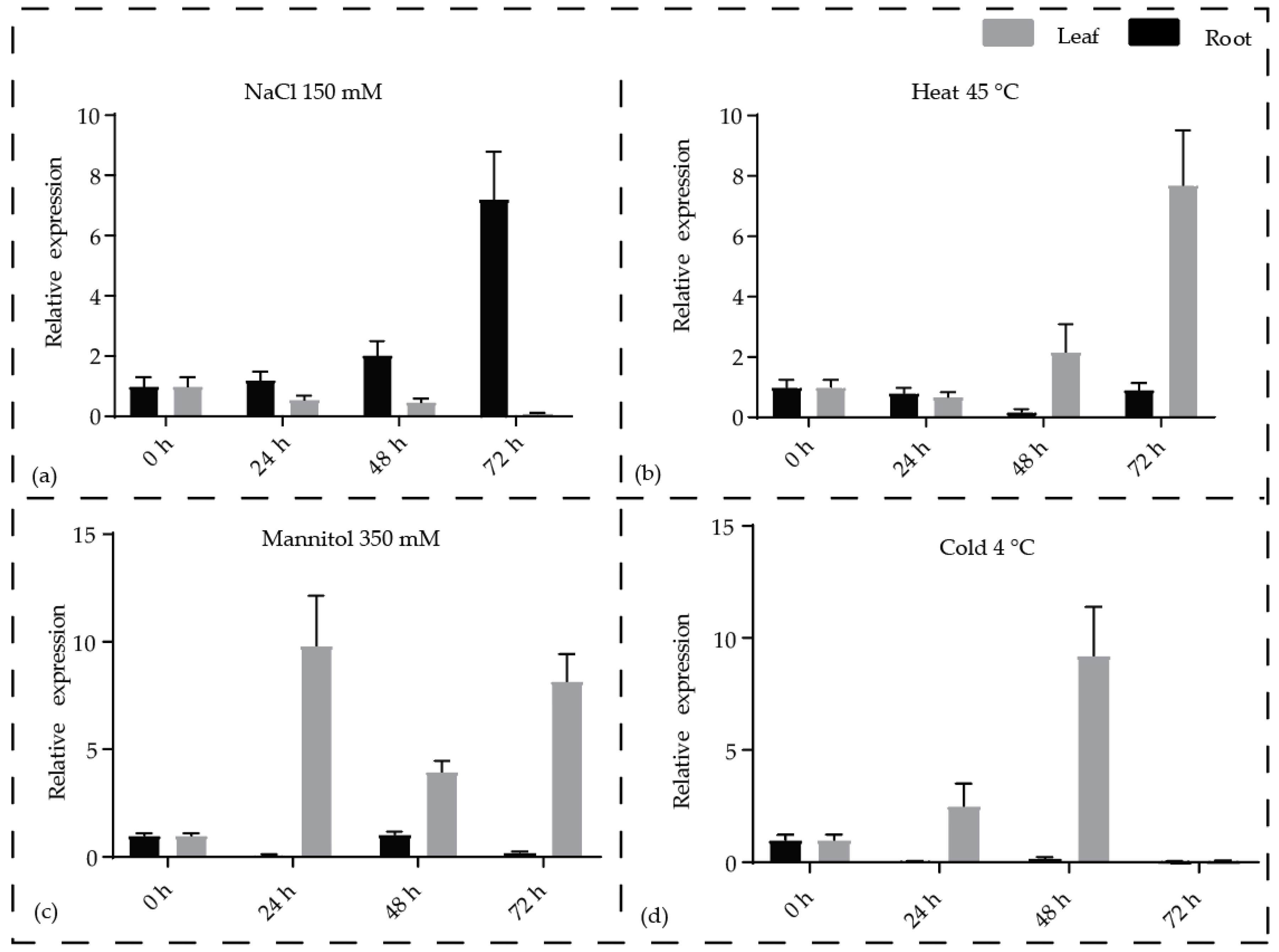




| Gene | Gene ID | MW (kDa) | pI | Amino Acid | Location | ORF | Chr |
|---|---|---|---|---|---|---|---|
| AcPIE1 | Aco017256 | 230.32 | 5.44 | 2021 | 16521947..16538538 | 6066 | 03 |
| AcARP4 | Aco007686 | 48.75 | 5.09 | 444 | 1249997..1258550 | 1335 | 08 |
| AcARP6 | Aco018334 | 42.98 | 5.97 | 382 | 2117532..2122193 | 1149 | 16 |
| AcYAF9 | Aco027612 | 32.38 | 6.72 | 295 | 2382105..2388792 | 888 | 24 |
| AcRVB1 | Aco015484 | 56.28 | 6.94 | 511 | 847770..852493 | 1536 | 23 |
| AcRVB2 | Aco012319 | 55.51 | 5.69 | 507 | 3080033..3084927 | 1524 | 01 |
| AcSWC2 | Aco016372 | 43.23 | 5.69 | 376 | 8471728..8501804 | 1131 | 05 |
| AcSWC4 | Aco005752 | 50.48 | 9.49 | 449 | 12625273..12633418 | 1350 | 11 |
| AcSWC5 | Aco007381 | 28.71 | 8.65 | 256 | 4261067..4266596 | 771 | 23 |
| AcSWC6 | Aco012501 | 28.51 | 9.03 | 252 | 1632792..1637849 | 759 | 13 |
| AcBDF1 | Aco006628 | 126.34 | 6.93 | 1127 | 23130832..23140236 | 3384 | 01 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jakada, B.H.; Aslam, M.; Fakher, B.; Greaves, J.G.; Li, Z.; Li, W.; Lai, L.; Ayoade, O.A.; Cheng, Y.; Cao, S.; et al. Identification of SWI2/SNF2-Related 1 Chromatin Remodeling Complex (SWR1-C) Subunits in Pineapple and the Role of Pineapple SWR1 COMPLEX 6 (AcSWC6) in Biotic and Abiotic Stress Response. Biomolecules 2019, 9, 364. https://doi.org/10.3390/biom9080364
Jakada BH, Aslam M, Fakher B, Greaves JG, Li Z, Li W, Lai L, Ayoade OA, Cheng Y, Cao S, et al. Identification of SWI2/SNF2-Related 1 Chromatin Remodeling Complex (SWR1-C) Subunits in Pineapple and the Role of Pineapple SWR1 COMPLEX 6 (AcSWC6) in Biotic and Abiotic Stress Response. Biomolecules. 2019; 9(8):364. https://doi.org/10.3390/biom9080364
Chicago/Turabian StyleJakada, Bello Hassan, Mohammad Aslam, Beenish Fakher, Joseph G. Greaves, Zeyun Li, Weimin Li, Linyi Lai, Oyekunle Adenike Ayoade, Yan Cheng, Shijiang Cao, and et al. 2019. "Identification of SWI2/SNF2-Related 1 Chromatin Remodeling Complex (SWR1-C) Subunits in Pineapple and the Role of Pineapple SWR1 COMPLEX 6 (AcSWC6) in Biotic and Abiotic Stress Response" Biomolecules 9, no. 8: 364. https://doi.org/10.3390/biom9080364
APA StyleJakada, B. H., Aslam, M., Fakher, B., Greaves, J. G., Li, Z., Li, W., Lai, L., Ayoade, O. A., Cheng, Y., Cao, S., Li, G., Hu, J.-M., & Qin, Y. (2019). Identification of SWI2/SNF2-Related 1 Chromatin Remodeling Complex (SWR1-C) Subunits in Pineapple and the Role of Pineapple SWR1 COMPLEX 6 (AcSWC6) in Biotic and Abiotic Stress Response. Biomolecules, 9(8), 364. https://doi.org/10.3390/biom9080364







