Integrative Analysis of the Core Fruit Lignification Toolbox in Pear Reveals Targets for Fruit Quality Bioengineering
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Silico Identification of P. bretschneideri Phenylpropanoid/Monolignol-Pathway Genes
2.2. Phylogenetic and Synteny Analyses
2.3. Expression Analysis of P. bretschneideri Phenylpropanoid/Monolignol-Pathway Genes
2.4. BiFC Assays
2.5. Availability of Data and Materials
3. Results and Discussion
3.1. PAL
3.2. The Hydroxylation Steps
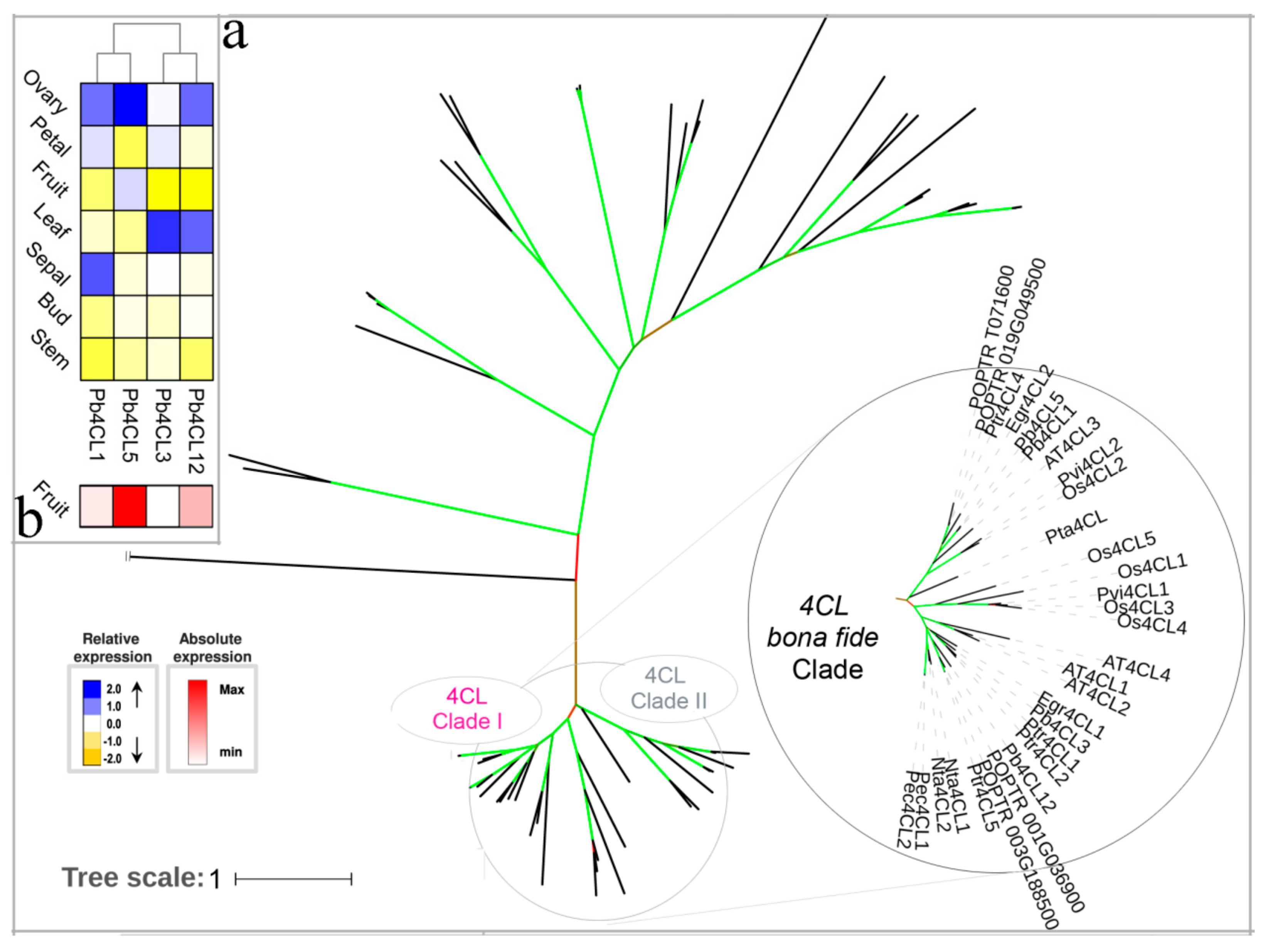
3.3. 4CL
3.4. HCT
3.5. CSE
3.6. The methylation Steps
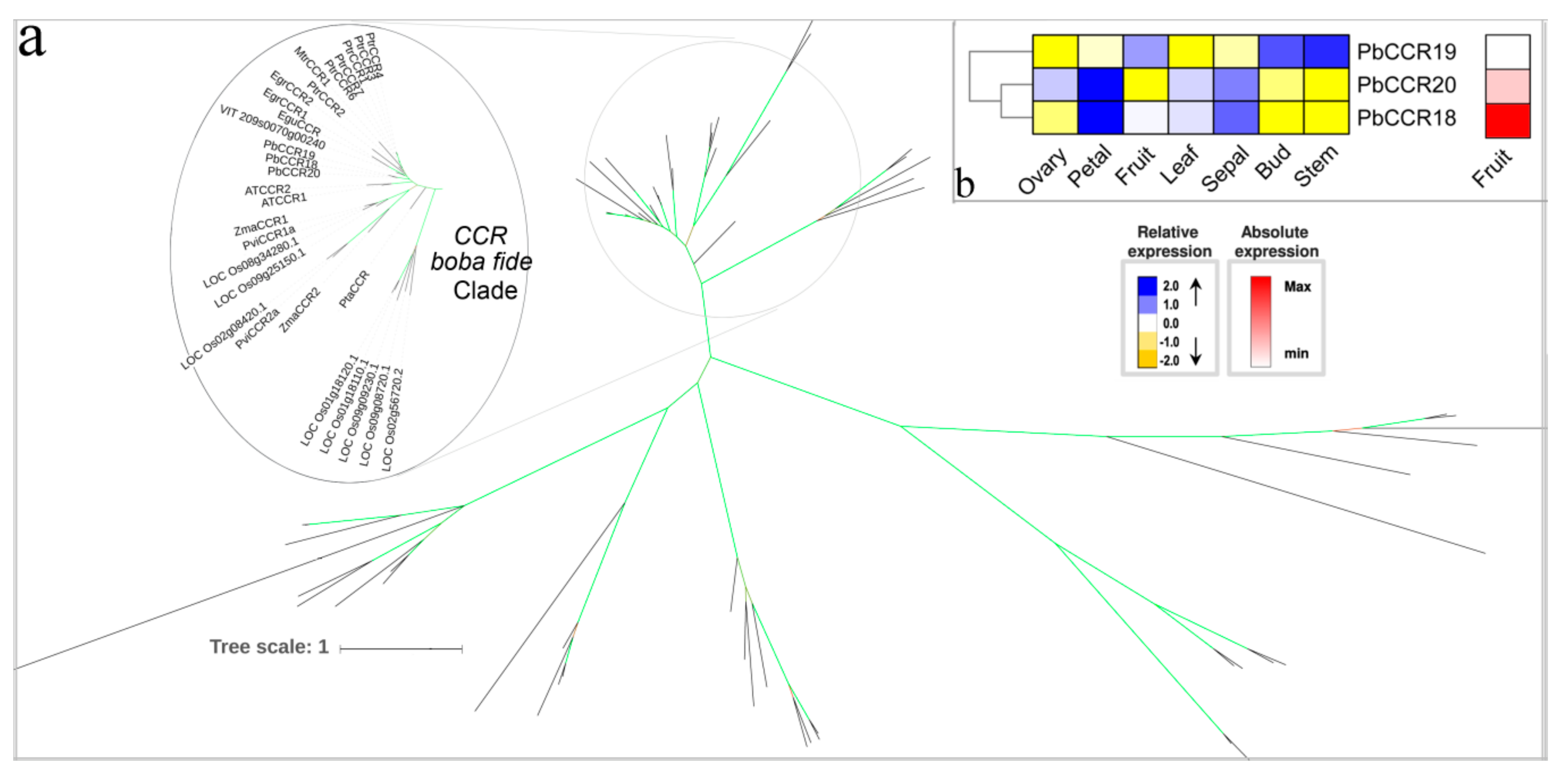
3.7. The Two Last Reductive Steps
3.8. Hypothetical Pathways Involved in the Biosynthesis of Lignin in Pear Fruit
3.9. BiFC Assays
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin biosynthesis and structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Raes, J.; Rohde, A.; Christensen, J.H.; De Peer, Y.V.; Boerjan, W. Genome-wide characterization of the lignification toolbox in arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef] [PubMed]
- Carocha, V.; Soler, M.; Hefer, C.; Cassan-Wang, H.; Fevereiro, P.; Myburg, A.A.; Paiva, J.A.P.; Grima-Pettenati, J. Genome-wide analysis of the lignin toolbox of eucalyptus grandis. New Phytol. 2015, 206, 1297–1313. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [PubMed]
- Ralph, J.; Lapierre, C.; Marita, J.M.; Kim, H.; Lu, F.; Hatfield, R.D.; Ralph, S.A.; Chapple, C.; Franke, R.; Hemm, M.R. Elucidation of new structures in lignins of cad- and comt-deficient plants by nmr. Phytochemistry 2001, 57, 993–1003. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Khan, Y.; Jaishankar, J.; Shweta, S.; Lata, C.; Prasad, M. Integrative analysis and expression profiling of secondary cell wall genes in c4 biofuel model setaria italica reveals targets for lignocellulose bioengineering. Front. Plant Sci. 2015, 6, 965. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, N.H.; Selvaraj, G.; Wei, Y.; King, J.L. Role of lignification in plant defense. Plant Signal. Behav. 2009, 4, 158–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plomion, C.; Leprovost, G.; Stokes, A. Wood formation in trees. Plant Physiol. 2001, 127, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Boudet, A.; Kajita, S.; Grimapettenati, J.; Goffner, D. Lignins and lignocellulosics: A better control of synthesis for new and improved uses. Trend. Plant Sci. 2003, 8, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Grimapettenati, J.; Goffner, D. Lignin genetic engineering revisited. Plant Sci. 1999, 145, 51–65. [Google Scholar] [CrossRef]
- Jin, Q.; Yan, C.; Qiu, J.; Zhang, N.; Lin, Y.; Cai, Y. Structural characterization and deposition of stone cell lignin in dangshan su pear. Sci. Hortic. 2013, 155, 123–130. [Google Scholar] [CrossRef]
- Cai, Y.; Li, G.; Nie, J.; Lin, Y.; Nie, F.; Zhang, J.; Xu, Y. Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Hortic. 2010, 125, 374–379. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. Myb transcription factors in chinese pear (pyrus bretschneideri rehd.): Genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 2016, 7, 577. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, J.M.; Chapple, C. Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 2002, 5, 224–229. [Google Scholar] [CrossRef]
- Baucher, M.; Halpin, C.; Petit-Conil, M.; Boerjan, W. Lignin: Genetic engineering and impact on pulping. Crit. Rev. Biochem. Mol. Bio. 2003, 38, 305–350. [Google Scholar] [CrossRef] [PubMed]
- Ralph, J.; Lundquist, K.; Brunow, G.; Lu, F.; Kim, H.; Schatz, P.F.; Marita, J.M.; Hatfield, R.D.; Ralph, S.A.; Christensen, J.H. Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenyl- propanoids. Phytochem. Rev. 2004, 3, 29–60. [Google Scholar] [CrossRef]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P. Caffeoyl shikimate esterase (cse) is an enzyme in the lignin biosynthetic pathway in arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef]
- Adams, Z.; Ehlting, J.; Edwards, R. The regulatory role of shikimate in plant phenylalanine metabolism. J. Theor. Bio. 2019, 462, 158–170. [Google Scholar] [CrossRef]
- Ha, C.M.; Escamillatrevino, L.; Yarce, J.C.S.; Kim, H.; Ralph, J.; Chen, F.; Dixon, R.A. An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in medicago truncatula. Plant J. 2016, 86, 363–375. [Google Scholar] [CrossRef]
- Saleme, M.D.L.S.; Cesarino, I.; Vargas, L.; Kim, H.; Vanholme, R.; Goeminne, G.; Van Acker, R.; Fonseca, F.; Pallidis, A.; Voorend, W. Silencing caffeoyl shikimate esterase affects lignification and improves saccharification in poplar. Plant Physiol. 2017, 175, 1040–1057. [Google Scholar] [CrossRef]
- Hamberger, B.; Ellis, M.; Friedmann, M.; de Azevedo Souza, C.; Barbazuk, B.; Douglas, C.J. Genome-wide analyses of phenylpropanoid-related genes in populus trichocarpa, arabidopsis thaliana, and oryza sativa: The populus lignin toolbox and conservation and diversification of angiosperm gene families this article is one of a selection of papers published in the special issue on poplar research in canada. Botany 2007, 85, 1182–1201. [Google Scholar]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H. The genome of the pear (pyrus bretschneideri rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jin, Q.; Lin, Y.; Cai, Y. Structural, evolutionary, and functional analysis of the class iii peroxidase gene family in chinese pear (pyrus bretschneideri). Front. Plant Sci. 2016, 7, 1874. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jiao, C.; Jin, Q.; Lin, Y.; Cai, Y. B-box genes: Genome-wide identification, evolution and their contribution to pollen growth in pear (pyrus bretschneideri rehd.). BMC Plant Biol. 2017, 17, 156. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.P.; Han, Y.; Jin, Q.; Lin, Y.; Cai, Y. Comparative genomic analysis of the grf genes in chinese pear (pyrus bretschneideri rehd), poplar (populous), grape (vitis vinifera), arabidopsis and rice (oryza sativa). Front. Plant Sci. 2016, 7, 1750. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J. The pfam protein families database. Nucleic Acids Res. 2011, 40, D290–D301. [Google Scholar] [CrossRef] [PubMed]
- Zdobnov, E.M.; Apweiler, R. Interproscan—An integration platform for the signature-recognition methods in interpro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: Hmmer3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Soler, M.; Camargo, E.L.O.; Carocha, V.; Cassan-Wang, H.; San Clemente, H.; Savelli, B.; Hefer, C.A.; Paiva, J.A.P.; Myburg, A.A.; Grima-Pettenati, J. The eucalyptus grandis r2r3-myb transcription factor family: Evidence for woody growth-related evolution and function. New Phytol. 2015, 206, 1364–1377. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, D.; Hu, J.; Zhou, X.; Ye, X.; Reichel, K.L.; Stewart, N.R.; Syrenne, R.D.; Yang, X.; Gao, P.; et al. Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinformatics 2009, 10, S3. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Bio. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. Iq-tree: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Bio. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (itol) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. Mcscanx: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Cai, B.; Yang, X.; Tuskan, G.A.; Cheng, Z.-M. Microsyn: A user friendly tool for detection of microsynteny in a gene family. BMC Bioinformatics 2011, 12, 79. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Qiao, X.; Yin, H.; Zhou, Y.; Dong, H.; Qi, K.; Li, L.; Zhang, S. Unbiased subgenome evolution following a recent whole-genome duplication in pear (pyrus bretschneideri rehd.). Hortic. Res. 2019, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of rna-seq experiments with hisat, stringtie and ballgown. Nat. Protoc. 2016, 11, 1650. [Google Scholar] [CrossRef]
- Walter, M.; Chaban, C.; Schütze, K.; Batistic, O.; Weckermann, K.; Näke, C.; Blazevic, D.; Grefen, C.; Schumacher, K.; Oecking, C. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2010, 40, 428–438. [Google Scholar] [CrossRef]
- Liang, X.; Dron, M.; Cramer, C.L.; Dixon, R.A.; Lamb, C.J. Differential regulation of phenylalanine ammonia-lyase genes during plant development and by environmental cues. J. Bio. Chem. 1989, 264, 14486–14492. [Google Scholar]
- Subramaniam, R.; Reinold, S.; Douglas, C.J. Structure, inheritance, and expression of hybrid poplar (populus trichocarpa × populus deltoides) phenylalanine ammonia-lyase genes. Plant Physiol. 1993, 102, 71–83. [Google Scholar] [CrossRef]
- Kao, Y.; Harding, S.A.; Tsai, C. Differential expression of two distinct phenylalanine ammonia-lyase genes in condensed tannin-accumulating and lignifying cells of quaking aspen. Plant Physiol. 2002, 130, 796–807. [Google Scholar] [CrossRef] [PubMed]
- Rohde, A.; Morreel, K.; Ralph, J.; Goeminne, G.; Hostyn, V.; De Rycke, R.; Kushnir, S.; Van Doorsselaere, J.; Joseleau, J.; Vuylsteke, M. Molecular phenotyping of the pal1 and pal2 mutants of arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 2004, 16, 2749–2771. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.; Yu, J.; Chen, Z. Functional analysis of the arabidopsis pal gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Ehlting, J.; Hamberger, B.; Millionrousseau, R.; Werckreichhart, D. Cytochromes p450 in phenolic metabolism. Phytochem. Rev. 2006, 5, 239–270. [Google Scholar] [CrossRef]
- Ro, D.K.; Douglas, C.J. Reconstitution of the entry point of plant phenylpropanoid metabolism in yeast (saccharomyces cerevisiae): Implications for control of metabolic flux into the phenylpropanoid pathway. J. Bio. Chem. 2004, 279, 2600–2607. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.A.; Collins, R.E.; Anterola, A.M.; Cochrane, F.C.; Davin, L.B.; Lewis, N.G. An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in arabidopsis thaliana and limitations thereof. Phytochemistry 2003, 64, 1097–1112. [Google Scholar] [CrossRef]
- Schoch, G.A.; Goepfert, S.; Morant, M.; Hehn, A.; Meyer, D.; Ullmann, P.; Werckreichhart, D. Cyp98a3 from arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Bio. Chem. 2001, 276, 36566–36574. [Google Scholar] [CrossRef] [PubMed]
- Franke, R.; Humphreys, J.M.; Hemm, M.R.; Denault, J.W.; Ruegger, M.O.; Cusumano, J.C.; Chapple, C. The arabidopsis ref8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 2002, 30, 33–45. [Google Scholar] [CrossRef]
- Abdulrazzak, N.; Pollet, B.; Ehlting, J.; Larsen, K.; Asnaghi, C.; Ronseau, S.; Proux, C.; Erhardt, M.; Seltzer, V.; Renou, J. A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta -hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol. 2005, 140, 30–48. [Google Scholar] [CrossRef]
- Chapple, C.C.S.; Vogt, T.; Ellis, B.E.; Somerville, C. An arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 1992, 4, 1413–1424. [Google Scholar]
- Osakabe, K.; Tsao, C.C.; Li, L.; Popko, J.L.; Umezawa, T.; Carraway, D.T.; Smeltzer, R.H.; Joshi, C.P.; Chiang, V.L. Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. USA 1999, 96, 8955–8960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, K.; Shirley, A.M.; Cusumano, J.C.; Belllelong, D.A.; Chapple, C. Lignin monomer composition is determined by the expression of a cytochrome p450-dependent monooxygenase in arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 6619–6623. [Google Scholar] [CrossRef] [PubMed]
- Marita, J.M.; Ralph, J.; Hatfield, R.D.; Chapple, C. Nmr characterization of lignins in arabidopsis altered in the activity of ferulate 5-hydroxylase. Proc. Natl. Acad. Sci. USA 1999, 96, 12328–12332. [Google Scholar] [CrossRef]
- Franke, R.; Mcmichael, C.M.; Meyer, K.; Shirley, A.M.; Cusumano, J.C.; Chapple, C. Modified lignin in tobacco and poplar plants over-expressing the arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 2000, 22, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.J.; Akiyama, T.; Chapple, C.; Ralph, J.; Mansfield, S.D. The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar1. Plant Physiol. 2009, 150, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Ehlting, J.; Shin, J.J.K.; Douglas, C.J. Identification of 4-coumarate:Coenzyme a ligase (4cl) substrate recognition domains. Plant J. 2001, 27, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Ehlting, J.; Buttner, D.; Wang, Q.; Douglas, C.J.; Somssich, I.E.; Kombrink, E. Three 4-coumarate:Coenzyme a ligases in arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999, 19, 9–20. [Google Scholar] [CrossRef]
- De Azevedo Souza, C.; Barbazuk, B.; Ralph, S.G.; Bohlmann, J.; Hamberger, B.; Douglas, C.J. Genome-wide analysis of a land plant-specific acyl: Coenzymea synthetase (acs) gene family in arabidopsis, poplar, rice and physcomitrella. New Phytol. 2008, 179, 987–1003. [Google Scholar]
- Hoffmann, L.; Maury, S.; Martz, F.; Geoffroy, P.; Legrand, M. Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J. Bio. Chem. 2003, 278, 95–103. [Google Scholar] [CrossRef]
- Hoffmann, L.; Besseau, S.; Geoffroy, P.; Ritzenthaler, C.; Meyer, D.; Lapierre, C.; Pollet, B.; Legrand, M. Silencing of hydroxycinnamoyl-coenzyme a shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 2004, 16, 1446–1465. [Google Scholar] [CrossRef]
- Ye, Z.H.; Varner, J.E. Differential expression of two o-methyltransferases in lignin biosynthesis in zinnia elegans. Plant Physiol. 1995, 108, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Ann. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Jouanin, L.; Goujon, T.; de Nadaï, V.; Martin, M.-T.; Mila, I.; Vallet, C.; Pollet, B.; Yoshinaga, A.; Chabbert, B.; Petit-Conil, M.; et al. Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol. 2000, 123, 1363–1374. [Google Scholar] [CrossRef] [PubMed]
- Sarni, F.; Grand, C.; Boudet, A. Purification and properties of cinnamoyl-coa reductase and cinnamyl alcohol dehydrogenase from poplar stems (populus x euramericana). FEBS J. 1984, 139, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Lauvergeat, V.; Lacomme, C.; Lacombe, E.; Lasserre, E.; Roby, D.; Grimapettenati, J. Two cinnamoyl-coa reductase (ccr) genes from arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 2001, 57, 1187–1195. [Google Scholar] [CrossRef]
- Barakat, A.; Yassin, N.B.M.; Park, J.S.; Choi, A.; Herr, J.; Carlson, J.E. Comparative and phylogenomic analyses of cinnamoyl-coa reductase and cinnamoyl-coa-reductase-like gene family in land plants. Plant Sci. 2011, 181, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, E.; Hawkins, S.; Van Doorsselaere, J.; Piquemal, J.; Goffner, D.; Poeydomenge, O.; Boudet, A.; Grimapettenati, J. Cinnamoyl coa reductase, the first committed enzyme of the lignin branch biosynthetic pathway: Cloning, expression and phylogenetic relationships. Plant J. 1997, 11, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Sibout, R.; Eudes, A.; Mouille, G.; Pollet, B.; Lapierre, C.; Jouanin, L.; Seguin, A. Cinnamyl alcohol dehydrogenase-c and -d are the primary genes involved in lignin biosynthesis in the floral stem of arabidopsis. Plant Cell 2005, 17, 2059–2076. [Google Scholar] [CrossRef]
- Guo, D.; Ran, J.; Wang, X. Evolution of the cinnamyl/sinapyl alcohol dehydrogenase (cad/sad) gene family: The emergence of real lignin is associated with the origin of bona fide cad. J. Mol. Evol. 2010, 71, 202–218. [Google Scholar] [CrossRef]
- Barakat, A.; Bagniewskazadworna, A.; Choi, A.; Plakkat, U.; Diloreto, D.S.; Yellanki, P.; Carlson, J.E. The cinnamyl alcohol dehydrogenase gene family in populus: Phylogeny, organization, and expression. BMC Plant Bio. 2009, 9, 26. [Google Scholar] [CrossRef]
- Sibout, R.; Eudes, A.; Pollet, B.; Goujon, T.; Mila, I.; Granier, F.; Seguin, A.; Lapierre, C.; Jouanin, L. Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiol. 2003, 132, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Li, Q.; Shuford, C.M.; Liu, J.; Muddiman, D.C.; Sederoff, R.R.; Chiang, V.L. Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, 21253–21258. [Google Scholar] [CrossRef] [PubMed] [Green Version]











| Enzymes | Domains | E-Value |
|---|---|---|
| PAL * | PF00221/TIGR01226 | 1.00 × 10−30 |
| C4H | PTHR19383:SF33 | 1.00 × 10−30 |
| 4CL | PTHR11968:SF43 | 1.00 × 10−30 |
| HCT | PF02458 | 1.00 × 10−30 |
| C3H | PTHR19383:SF44 | 1.00 × 10−30 |
| CSE | PF03552 | 1.00 × 10−30 |
| CCOAMT | PTHR10509 | 1.00 × 10−30 |
| CCR ** | PTHR10366:SF9 | 1.00 × 10−24 |
| F5H | PTHR19383:SF46 | 1.00 × 10−30 |
| COMT *** | PIRSF005739 | 1.00 × 10−8 |
| CAD | PTHR11695:SF38 | 1.00 × 10−30 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Li, X.; Jiang, L. Integrative Analysis of the Core Fruit Lignification Toolbox in Pear Reveals Targets for Fruit Quality Bioengineering. Biomolecules 2019, 9, 504. https://doi.org/10.3390/biom9090504
Cao Y, Li X, Jiang L. Integrative Analysis of the Core Fruit Lignification Toolbox in Pear Reveals Targets for Fruit Quality Bioengineering. Biomolecules. 2019; 9(9):504. https://doi.org/10.3390/biom9090504
Chicago/Turabian StyleCao, Yunpeng, Xiaoxu Li, and Lan Jiang. 2019. "Integrative Analysis of the Core Fruit Lignification Toolbox in Pear Reveals Targets for Fruit Quality Bioengineering" Biomolecules 9, no. 9: 504. https://doi.org/10.3390/biom9090504
APA StyleCao, Y., Li, X., & Jiang, L. (2019). Integrative Analysis of the Core Fruit Lignification Toolbox in Pear Reveals Targets for Fruit Quality Bioengineering. Biomolecules, 9(9), 504. https://doi.org/10.3390/biom9090504





