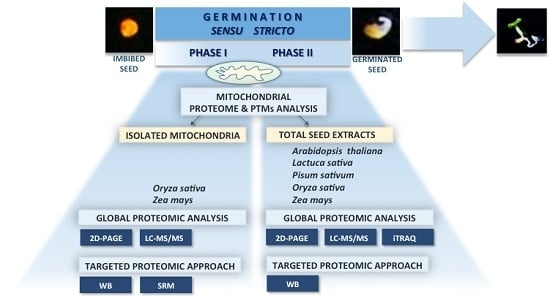
Mitochondrial Proteome Studies in Seeds during Germination
Abstract
:1. Introduction
2. Bioenergetics and Heterogeneity of Seed Mitochondria Structure
3. Experimental Approaches in Seed Mitochondrial Proteome Studies
4. Limitations in Seed Mitochondrial Proteome Studies
5. The Changes in Abundance of Mitochondrial Proteins during the Germination Course
6. Relationship between Proteomic and Transcriptomic Changes in Seeds during Germination
7. Post-Translational Modifications of Mitochondrial Proteins in Seeds
7.1. Carbonylation of Seed Mitochondrial Proteins
7.2. Phosphorylation of Seed Mitochondrial Proteins
7.3. S-Nitrosylation of Seed Mitochondrial Proteins
8. General Outlook and Challenges in Seed Proteomic Studies
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MS | mass spectrometry |
| 2D-PAGE | two-dimensional polyacrylamide gel electrophoresis |
| SRM | selected reaction monitoring |
| PTMs | post-translational modifications |
| SSPs | seed storage proteins |
| ROS | reactive oxygen species |
| RNS | reactive nitrogen species |
| NO | nitric oxide |
References
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [PubMed]
- Nonogaki, H.; Bassel, G.W.; Bewley, J.D. Germination-still a mystery. Plant Sci. 2010, 179, 574–581. [Google Scholar] [CrossRef]
- Logan, D.C. Mitochondrial morphology, dynamics and inheritance. In Plant Mitochondria: From Genome to Function, 1st ed.; Day, D.A., Millar, A.H., Whelan, J., Eds.; Springer: Dordrecht, The Netherlands, 2004; Volume 17, pp. 13–29. [Google Scholar]
- Law, S.; Narsai, R.; Whelan, J. Mitochondrial biogenesis in plants during seed germination. Mitochondrion 2014, 19, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Tzagoloff, A.; Myers, A.M. Genetics of mitochondrial biogenesis. Annu. Rev. Biochem. 1986, 55, 249–285. [Google Scholar] [CrossRef] [PubMed]
- Grivell, L.A. Nucleo-mitochondrial interactions in yeast mitochondrial biogenesis. Eur. J. Biochem. 1989, 182, 477–493. [Google Scholar] [CrossRef] [PubMed]
- Sanger, N.; Strohmeier, R.; Kaufmann, M.; Kuhl, H. Cell cycle-related expression and ligand binding of peripheral benzodiazepine receptor in human breast cancer cell lines. Eur. J. Cancer 2000, 36, 2157–2163. [Google Scholar] [CrossRef]
- Jahnke, V.E.; Sabido, O.; Freyssenet, D. Control of mitochondrial biogenesis, ROS level, and cytosolic Ca2+ concentration during the cell cycle and the onset of differentiation in L6E9 myoblasts. Am. J. Physiol. Cell Physiol. 2009, 296, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Plattner, H.; Salpeter, M.M.; Saltzgaber, J.; Schatz, G. Promitochondria of anaerobically grown yeast. IV. Conversion into respiring mitochondria. Proc. Natl. Acad. Sci. USA 1970, 66, 1252–1259. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Lo, Y.-S.; Jane, W.-N.; Lee, L.-W.; Chiang, K.-S. Population heterogeneity of higher-plant mitochondria in structure and function. Eur. J. Cell Biol. 1998, 75, 198–209. [Google Scholar] [CrossRef]
- Logan, D.C.; Millar, A.H.; Sweetlove, L.J.; Hill, S.A.; Leaver, C.J. Mitochondrial biogenesis during germination in maize embryos. Plant Physiol. 2001, 125, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Howell, K.A.; Millar, A.H.; Whelan, J. Ordered assembly of mitochondria during rice germination begins with promitochondrial structures rich in component of the protein import apparatus. Plant Mol. Biol. 2006, 60, 201–223. [Google Scholar] [CrossRef] [PubMed]
- Law, S.R.; Narsai, R.; Taylor, N.L.; Delannoy, E.; Carrie, C.; Giraud, E.; Millar, A.H.; Small, I.; Whelan, J. Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol. 2012, 158, 1610–1627. [Google Scholar] [CrossRef] [PubMed]
- Carrie, C.; Murcha, M.W.; Giraud, E.; Ng, S.; Zhang, M.F.; Narsai, R.; Whelan, J. How do plants make mitochondria? Planta 2013, 237, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Rajjou, L.; Gallardo, K.; Debeaujon, I.; Vandekerckhove, J.; Job, C.; Job, D. The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 2004, 134, 1598–1613. [Google Scholar] [CrossRef] [PubMed]
- Galland, M.; Huguet, R.; Aec, E.; Cueff, G.; Job, D.; Rajjou, L. Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol. Cell Proteom. 2014, 13, 252–268. [Google Scholar] [CrossRef] [PubMed]
- Finnie, C.; Svensson, B. Barley seed proteomics from spots to structures. J. Proteom. 2009, 72, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Miernyk, J.A.; Hajduk, M. Seed proteomics. J. Proteom. 2011, 74, 389–400. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Yang, P. Proteomics of rice seed germination. Front. Plant Sci. 2013, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Chen, S.; Wang, T.; Dai, S. Proteomic insights into seed germination in response to environmental factors. Proteomics 2013, 13, 1850–1870. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-Q.; Liu, S.-J.; Song, S.-Q.; Møller, I.M. Proteomics of seed development, desiccation tolerance, germination and vigor. Plant Physiol. Biochem. 2015, 86, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Attucci, S.; Carde, J.P.; Raymond, P.; Saint-Ges, V.; Spiteri, A.; Pradet, A. Oxidative phosphorylation by mitochondria extracted from dry sunflower seeds. Plant Physiol. 1991, 95, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Howell, K.A.; Cheng, K.; Murcha, M.W.; Jenkin, L.E.; Millar, A.H.; Whelan, J. Oxygen initiation of respiration and mitochondrial biogenesis in rice. J. Biol. Chem. 2007, 282, 15619–15619. [Google Scholar] [CrossRef] [PubMed]
- Ehrenshaft, M.; Brambl, R. Respiration and mitochondrial biogenesis in germinating embryos of maize. Plant Physiol. 1990, 93, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Benamar, A.; Tallon, C.; Macherel, D. Membrane integrity and oxidative properties of mitochondria isolated from imbibing pea seeds after priming or accelerated ageing. Seed Sci. Res. 2003, 13, 35–45. [Google Scholar] [CrossRef]
- Taylor, N.L.; Howell, K.A.; Heazlewood, J.L.; Tan, T.Y.W.; Narsai, R.; Huang, S.; Whelan, J.; Millar, A.H. Analysis of the rice mitochondrial carrier family reveals anaerobic accumulation of a basic amino acid carrier involved in arginine metabolism during seed germination. Plant Physiol. 2010, 154, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Wang, B.-C.; Jin, X.; Li, H.-B.; Han, P.; Wei, K.-H.; Zhang, X.-M.; Zhu, Y.-X. Proteomic analysis and extensive protein identification from dry, germinating Arabidopsis seed and young seedlings. J. Biochem. Mol. Biol. 2005, 38, 650–660. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-Q.; Møller, I.M.; Song, S.-Q. Proteomic analysis of embryonic axis of Pisum sativum seeds during germination and identification of proteins associated with loss of desiccation tolerance. J. Proteom. 2012, 77, 68–86. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; He, D.; Li, M.; Yang, P. In-depth proteomic analysis of rice embryo reveals its important roles in seed germination. Plant Cell Physiol. 2014, 55, 1826–1847. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-Q.; Song, B.-Y.; Deng, Z.-Y.; Wang, Y.; Liu, S.-J.; Møller, I.M.; Song, S.-Q. Proteomic analysis of lettuce seed germination and thermoinhibition by sampling of individual seeds at germination and removal of storage proteins by polyethylene glycol fractionation. Plant Physiol. 2015, 167, 1332–1350. [Google Scholar] [CrossRef] [PubMed]
- Rogowska-Wrzesinska, A.; Le Bihan, M.-C.; Thaysen-Andersen, M.; Roepstorff, P. 2D gels still have a niche in proteomics. J. Proteom. 2013, 88, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Bensimon, A.; Heck, A.J.R.; Aebersold, R. Mass spectrometry-based proteomics and network biology. Annu. Rev. Biochem. 2012, 81, 379–405. [Google Scholar] [CrossRef] [PubMed]
- Piechota, J.; Bereza, M.; Sokolowska, A.; Suszynski, K.; Lech, K.; Janska, H. Unraveling the functions of type II-prohibitins in Arabidopsis mitochondria. Plant Mol. Biol. 2015, 88, 249–267. [Google Scholar] [CrossRef] [PubMed]
- Thelen, J.J.; Peck, S.C. Quantitative proteomics in plants: Choices in abundance. Plant Cell 2007, 19, 3339–3346. [Google Scholar] [CrossRef] [PubMed]
- Picotti, P.; Aebersold, R. Selected reaction monitoring-based proteomics: Workflows, potential, pitfalls and future directions. Nat. Methods 2012, 9, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.L.; Heazlewood, J.L.; Millar, A.H. The Arabidopsis thaliana 2-D gel mitochondrial proteome: Refining the value of reference maps for assessing protein abundance, contaminants and post-translational modifications. Proteomics 2011, 11, 1720–1733. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.L.; Fenske, R.; Castleden, I.; Tomaz, T.; Nelson, C.J.; Millar, A.H. Selected reaction monitoring to determine protein abundance in Arabidopsis using the Arabidopsis proteotypic predictor. Plant Physiol. 2014, 164, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Taylor, N.L.; Ströher, E.; Fenske, R.; Millar, A.H. Succinate dehydrogenase assembly factor 2 is needed for assembly and activity of mitochondrial complex II and for normal root elongation in Arabidopsis. Plant J. 2013, 73, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Czarna, M.; Domanski, D.; Kwasniak-Owczarek, M.; Skibior, R.; Dadlez, M.; Janska, H. Role of AtFtsH4 protease in biogenesis of mitochondria during Arabidopsis seed germination. In Proceedings of the 9th International Conference for Plant Mitochondrial Biology, Wroclaw, Poland, 17–22 May 2015; University of Wroclaw: Wroclaw, Poland, 2015; p. 164. [Google Scholar]
- Zhang, H.; He, D.; Yu, J.; Li, M.; Damaris, R.N.; Gupta, R.; Kim, S.T.; Yang, P. Analysis of dynamic protein carbonylation in rice embryo during germination through AP-SWATH. Proteomics 2016, 16, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-T.; Meng, L.-B.; Yang, C.-P.; Liu, G.-F.; Liu, G.-J.; Ma, W.; Wang, B.-C. A shotgun phosphoproteomics analysis of embryos in germinated maize seeds. Planta 2008, 228, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Yang, P.; Sakata, K.; Komatsu, S. Quantitative proteomics reveals the role of protein phosphorylation in rice embryos during early stages of germination. J. Proteome Res. 2014, 13, 1766–1782. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yin, X.; Sakata, K.; Yang, P.; Komatsu, S. Proteomic analysis of phosphoproteins in the rice nucleus during the early stage of seed germination. J. Proteome Res. 2015, 14, 2884–2896. [Google Scholar] [CrossRef] [PubMed]
- Macherel, D.; Benamar, A.; Avelange-Macherel, M.-H.; Tolleter, D. Function and stress tolerance of seed mitochondria. Physiol. Plant. 2007, 129, 233–241. [Google Scholar] [CrossRef]
- Nawa, Y.; Asahi, T. Rapid development of mitochondria in pea cotyledons during the early stage of germination. Plant Physiol. 1971, 48, 671–674. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.B.; Bonner, W.D. Studies of electron transport in dry and imbibed peanut embryo. Plant Physiol. 1971, 48, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Asahi, T. Biochemical properties of mitochondrial membrane from dry pea seeds and changes in the properties during imbibition. Plant Physiol. 1975, 56, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.; Fu, Y.-B. An improved method with a wider applicability to isolate plant mitochondria. Plant Methods 2015, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, K.; Job, C.; Groot, S.P.C.; Puype, M.; Demol, H.; Vandekerckhove, J.; Job, D. Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol. 2001, 126, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Bewley, D.J. Seed germination and dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, H.; Meguro, N.; Suzuki, Y.; Tsutsumi, N.; Hirai, A.; Nakazono, M. Induction of mitochondrial aldehyde dehydrogenase by submergence facilitates oxidation of acetaldehyde during re-aeration in rice. FEBS Lett. 2003, 546, 369–373. [Google Scholar] [CrossRef]
- Grelet, J.; Benamar, A.; Teyssier, E.; Avelange-Macherel, M.H.; Grunwald, D.; Macherel, D. Identification pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiol. 2005, 137, 157–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailly, C.; El-Maarouf-Bouteau, H.; Corbineau, F. From intracellular signaling networks to cell death: The dual role of reactive oxygen species in seed physiology. C. R. Biol. 2008, 331, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, K.; Okamoto, M.; Koshiba, T.; Kamiya, Y.; Nambara, E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 2005, 41, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasulu, N.; Usadel, B.; Winter, A.; Radchuk, V.; Scholz, U.; Stein, N.; Weschke, W.; Strickert, M.; Close, T.J.; Stitt, M.; et al. Barley grain maturation and germination: Metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol. 2008, 146, 1738–1758. [Google Scholar] [CrossRef] [PubMed]
- Howell, K.A.; Narsai, R.; Carroll, A.; Ivanova, A.; Lohse, M.; Usadel, B.; Millar, A.H.; Whelan, J. Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol. 2009, 149, 961–980. [Google Scholar] [PubMed]
- Kimura, M.; Nambara, E. Stored and neosynthesized mRNA in Arabidopsis seeds: Effects of cycloheximide and controlled deterioration treatment on the resumption of transcription during imbibition. Plant Mol. Biol. 2010, 73, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Narsai, R.; Law, S.R.; Carrie, C.; Xu, L.; Whelan, J. In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol. 2011, 157, 1342–1362. [Google Scholar] [CrossRef] [PubMed]
- Weitbrecht, K.; Müller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.M.; Jensen, P.E.; Hansson, A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef] [PubMed]
- Job, C.; Rajjou, L.; Lovigny, Y.; Belghazi, M.; Job, D. Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol. 2005, 138, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Johansson, E.; Olsson, O.; Nyström, T. Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 22204–22208. [Google Scholar] [CrossRef] [PubMed]
- Oracz, K.; Bouteau, H.E.M.; Farrant, J.M.; Cooper, K.; Belghazi, M.; Job, C.; Job, D.; Corbineau, F.; Bailly, C. ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J. 2007, 50, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Rajjou, L.; Lovigny, Y.; Groot, S.P.; Belghazi, M.; Job, C.; Job, D. Proteome-wide characterization of seed aging in Arabidopsis: A comparison between artificial and natural aging protocols. Plant Physiol. 2008, 148, 620–641. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Meng, X.; Wang, Q.; Tian, S. Oxidative damage of mitochondrial proteins contributes to fruit senescence: A redox proteomics analysis. J. Prot. Res. 2009, 8, 2449–2462. [Google Scholar] [CrossRef] [PubMed]
- Bartoli, C.G.; Gómez, F.; Martinez, D.E.; Guiamet, J.J. Mitochondria are the main target for oxidative damage in leaves of wheat. J. Exp. Bot. 2004, 55, 1663–1669. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.T.; Donaldson, R.P. Metal-catalyzed oxidation induces carbonylation of peroxisomal proteins and loss of enzymatic activities. Arch. Biochem. Biophys. 2005, 439, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Matamoros, M.A.; Fernández-García, N.; Wienkoop, S.; Loscos, J.; Saiz, A.; Becana, M. Mitochondria are an early target of oxidative modifications in senescing legume nodules. New Phytol. 2013, 197, 873–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smakowska, E.; Czarna, M.; Janska, H. Mitochondrial-ATP-dependent proteases in protection against accumulation of carbonylated proteins. Mitochondrion 2014, 19, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Bowler, C.; Montagu, M.V.; Inze, D. Superoxide dismutase and stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 83–116. [Google Scholar] [CrossRef]
- Finkemeier, I.; Goodman, M.; Lamkemer, P.; Kandlbinder, A.; Sweetlove, L.J.; Dietz, K.J. The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 2005, 280, 12168–12180. [Google Scholar] [CrossRef] [PubMed]
- Bender, T.; Lewrenz, I.; Franken, S.; Baitzel, C.; Voos, W. Mitochondrial enzymes are protected from stress-induced aggregation by mitochondrial chaperones and the Pim/LON protease. Mol. Biol. Cell 2011, 22, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Sweetlove, L.J.; Heazlewood, J.L.; Herald, V.; Holtzapffel, R.; Day, D.A.; Leaver, C.J.; Millar, A.H. The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 2002, 32, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Arc, E.; Galland, M.; Cueff, G.; Godin, B.; Lounifi, I.; Job, D.; Rajjou, L. Reboot the system thanks to protein post-translational modifications and proteome diversity: How quiescent seeds restart their metabolism to prepare seedling establishment. Proteomics 2011, 11, 1606–1618. [Google Scholar] [CrossRef] [PubMed]
- Irar, S.; Oliveira, E.; Pages, M.; Goday, A. Towards the identification of late-embryogenic-abundant phosphoproteome in Arabidopsis by 2-DE and MS. Proteomics 2006, 6, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Ross, A.R.; Yang, J.; Hegedus, D.D.; Kermode, A.R. Phosphorylation of the 12S globulin cruciferin in wild-type and abi1-1 mutant Arabidopsis thaliana (thale cress) seeds. Biochem. J. 2007, 404, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Le, H.; Browning, K.S.; Gallie, D.R. The phosphorylation state of the wheat translation initiation factors eIF4B, eIF4A, and eIF2 is differentially regulated during seed development and germination. J. Biol. Chem. 1998, 273, 20084–20089. [Google Scholar] [CrossRef] [PubMed]
- Montoya-Garcia, L.; Munoz-Ocotero, V.; Aguilar, R.; de Jimenez, E.S. Regulation of acidic ribosomal protein expression and phosphorylation in maize. Biochemistry 2002, 41, 10166–10172. [Google Scholar] [CrossRef] [PubMed]
- Juszczuk, I.M.; Bykova, N.; Møller, I.M. Protein phosphorylation in plant mitochondria. Physiol. Plant. 2007, 129, 90–103. [Google Scholar] [CrossRef]
- Havelund, J.F.; Thelen, J.J.; Møller, I.M. Biochemistry, proteomics, and phosphoproteomics of plant mitochondria from non-photosynthetic cells. Front. Plant Sci. 2013, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bardel, J.; Louwagie, M.; Jaquinod, M.; Jourdain, A.; Luche, S.; Rabilloud, T.; Macherel, D.; Garin, J.; Bourguignon, J. A survey of plant mitochondria proteome in relation with development. Proteomics 2002, 2, 880–898. [Google Scholar] [CrossRef]
- Koc, E.C.; Koc, H. Regulation of mammalian mitochondrial translation by post-translational modifications. Biochim. Biophys. Acta 2012, 1819, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Lindermayr, C.; Saalbach, G.; Durner, J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2005, 137, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, M.C.; Lindermayr, C.; Bauwe, H.; Steinhauser, C.; Durner, J. Regulation of plant glycine decarboxylase by S-nitrosylation and glutathionylation. Plant Physiol. 2010, 152, 1514–1528. [Google Scholar] [CrossRef] [PubMed]
- Fares, A.; Rossignol, M.; Peltier, J.B. Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem. Biophys. Res. Commun. 2011, 416, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Alconada, A.M.; Echevarria-Zomeno, S.; Lindermayr, C.; Redondo-Lopez, I.; Durner, J.; Jorrin-Novo, J.V. Proteomic analysis of Arabidopsis protein S-nitrosylation in response to inoculation with Pseudomonas syringae. Acta Physiol. Plant. 2011, 33, 1493–1514. [Google Scholar] [CrossRef]
- Romero-Puertas, M.C.; Campostrini, N.; Matte, A.; Righetti, P.G.; Perazzolli, M.; Zolla, L.; Roepstorff, P.; Delledonne, M. Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 2008, 8, 1459–1469. [Google Scholar] [CrossRef] [PubMed]
- Abat, J.K.; Mattoo, A.K.; Deswal, R. S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata-ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 2008, 275, 2862–2872. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Takemoto, D.; Kawakita, K. Proteomic analysis of S-nitrosylated proteins in potato plant. Physiol. Plant. 2013, 148, 371–386. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Wang, Y.; Tang, J.; Xue, P.; Li, C.; Liu, L.; Hu, B.; Yang, F.; Loake, G.J.; Chu, C. Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 2012, 158, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Galisteo, A.P.; Rodriguez-Serrano, M.; Pazmino, D.M.; Gupta, D.K.; Sandalio, L.M.; Romero-Puertas, M.C. S-nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: Changes under abiotic stress. J. Exp. Bot. 2012, 63, 2089–2103. [Google Scholar] [CrossRef] [PubMed]
- Camejo, D.; Romero-PuertasMdel, C.; Rodriguez-Serrano, M.; Sandalio, L.M.; Lazaro, J.J.; Jimenez, A.; Sevilla, F. Salinity-induced changes in S-nitrosylation of pea mitochondrial proteins. J. Proteom. 2013, 79, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Chakia, M.; Shekariesfahlana, A.; Ageevaa, A.; Mengela, A.; von Toerneb, C.; Durnera, J.; Lindermayr, C. Identification of nuclear target proteins for S-nitrosylation in pathogen-treated Arabidopsis thaliana cell cultures. Plant Sci. 2015, 238, 15–126. [Google Scholar] [CrossRef] [PubMed]
- Bethke, P.C.; Badger, M.R.; Jones, R.L. Apoplastic synthesis of nitric oxide by plant tissues. Plant Cell 2004, 16, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Bethke, P.C.; Gubler, F.; Jacobsen, J.V.; Jones, R.L. Dormancy of Arabidopsis seeds and barley can be broken by nitric oxide. Planta 2004, 219, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Albertos, P.; Romero-Puertas, M.C.; Tatematsu, K.; Mateos, I.; Sanchez-Vicente, I.; Nambara, E.; Lorenzo, O. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 2015, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Borisjuk, L.; Rolletschek, H. The oxygen status of the developing seed. New Phytol. 2009, 182, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Bykova, N.V.; Shah, J.K.; Hill, R.D. Anoxic nitric oxide cycling in plants: Participating reactions and possible mechanisms. Physiol. Plant. 2010, 138, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.-H.; Hood, B.L.; Kim, B.-J.; Hardwick, J.P.; Conrads, T.P.; Veenstra, T.D.; Song, B.J. Inactivation of oxidized and S-nitrosylated mitochondrial proteins in alcoholic fatty liver of rats. Hepatology 2006, 44, 1218–1230. [Google Scholar] [CrossRef] [PubMed]
- Kaspar, S.; Peukert, M.; Svatos, A.; Matros, A.; Mock, H.-P. MALDI-imaging mass spectrometry—An emerging technique in plant biology. Proteomics 2011, 11, 1840–1850. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Li, B.; Malitsky, S.; Rogachev, I.; Aharoni, A.; Kaftan, F.; Svatos, A.; Franceschi, P. Sample preparation for mass spectrometry imaging of plant tissue: A review. Front. Plant Sci. 2016, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czarna, M.; Kolodziejczak, M.; Janska, H. Mitochondrial Proteome Studies in Seeds during Germination. Proteomes 2016, 4, 19. https://doi.org/10.3390/proteomes4020019
Czarna M, Kolodziejczak M, Janska H. Mitochondrial Proteome Studies in Seeds during Germination. Proteomes. 2016; 4(2):19. https://doi.org/10.3390/proteomes4020019
Chicago/Turabian StyleCzarna, Malgorzata, Marta Kolodziejczak, and Hanna Janska. 2016. "Mitochondrial Proteome Studies in Seeds during Germination" Proteomes 4, no. 2: 19. https://doi.org/10.3390/proteomes4020019
APA StyleCzarna, M., Kolodziejczak, M., & Janska, H. (2016). Mitochondrial Proteome Studies in Seeds during Germination. Proteomes, 4(2), 19. https://doi.org/10.3390/proteomes4020019