Dynamic Candidate Solution Boosted Beluga Whale Optimization Algorithm for Biomedical Classification
Abstract
1. Introduction
- BWO has a lack of diversity, which could lead to being trapped in local optima.
- OBL can enhance the exploration of BWO by exploring more potential regions that produce BWO.
- DCS and OBWO boost variety and improve the consistency of the selected solution by giving potential candidates a chance to solve the given problem with a high fitness value, which produces the OBWOD approach.
- In comparison to the other seven MH algorithms, OBWOD is used to resolve global optimization issues based on the CEC’22 test suite.
- The OBWOD approach based on the kNN classifier was built for biomedical classification tasks and estimated on ten disease datasets with different dimension sizes extracted from the UCI repository.
- The experimental results demonstrated the superiority of OBWOD over seven other competitors according to the performance evaluation matrix.
2. Literature Reviews
3. Preliminaries
3.1. K-Nearest Neighbor
3.2. Opposition-Based Learning
3.3. Dynamic Candidate Solution
3.4. Beluga Whale Optimization Algorithm
- Exploration phase: Beluga whales’ swimming behavior is considered when establishing the BWO exploring phase. Beluga whales can engage in social–sexual behaviors in various postures, as evidenced by the behaviors observed in beluga whales kept in human care, such as a pair of closely spaced beluga whales swimming in a coordinated manner. As a result, beluga whales’ positions are updated as shown in Equation (7).where T is the current iteration, denotes the next iteration, and denote random numbers between (0, 1), and j indicates the new position of the beluga whale in the dimension.
- Exploitation phase: The Levy flight strategy is employed in BWO’s exploitative phase to improve convergence. With the Levy flight technique as our supposition, the mathematical model is shown in Equation (8).where is calculated by Equation (9), Levy flight function is calculated by Equation (10), and is calculated by Equation (11).where the default constant was set to 1.5 and u and are random values with a normal distribution calculated by Equations (12) and (13) [57].
- Whale fall phase: We used the whale fall probability from the population’s individuals as our arbitrary premise to simulate slight changes in the groups to represent the behavior of whale fall in each iteration. We presumed that these beluga whales have relocated or have been fired at and dropped into the deep ocean. The locations of beluga whales and the magnitude of a whale fall are used to determine the updated position to maintain a steady population size, as calculated by Equation (14).where is calculated by Equation (15).where is calculated by Equation (16).where is calculated by Equation (17).
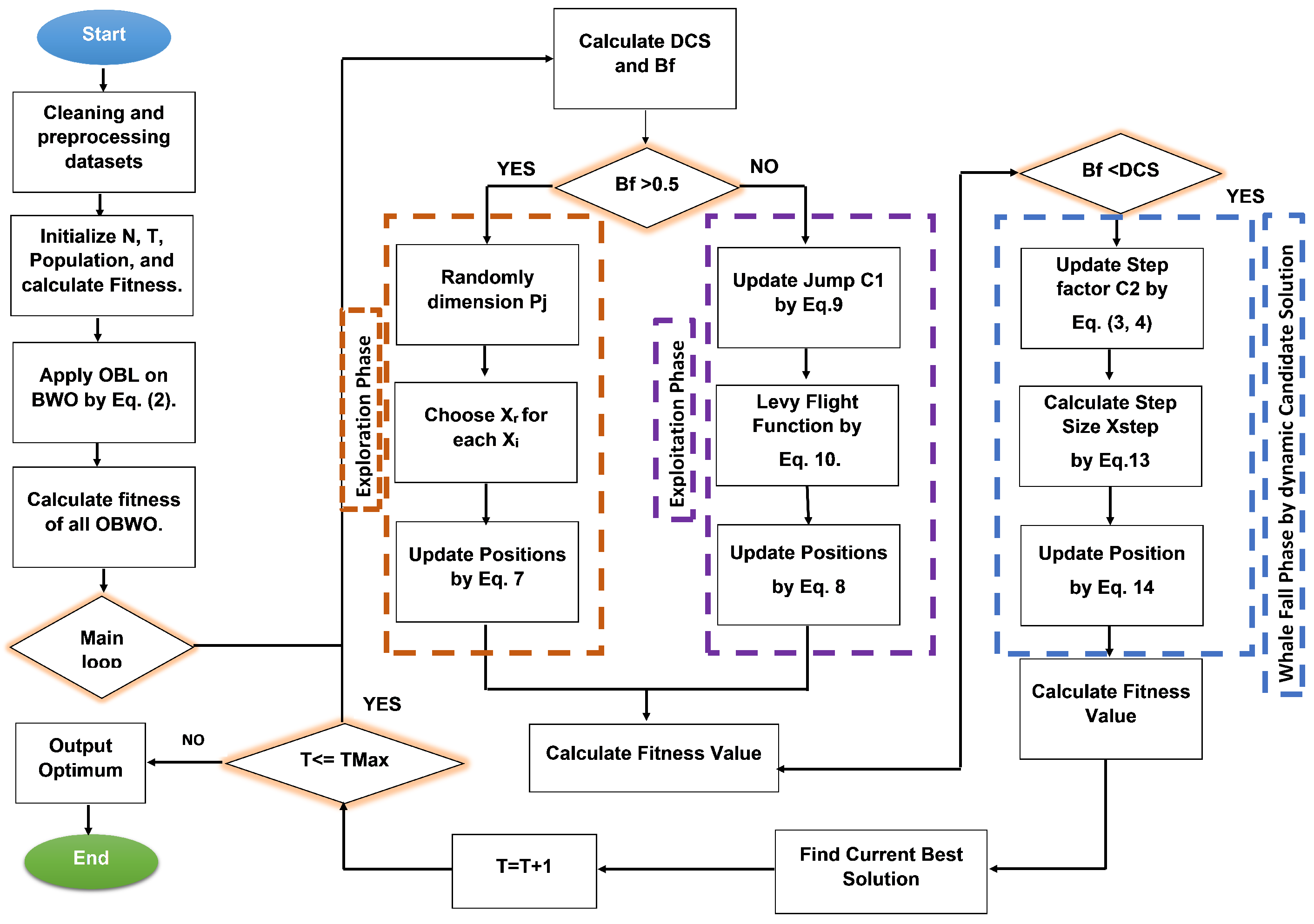
4. The Proposed OBWOD Approach
4.1. Drawbacks of the Original BWO
4.2. Fitness Function
4.3. The Major Stages of the OBWOD
4.3.1. Initialization Phase
4.3.2. Solution Update Phase
- The first step: To select a better generation of candidate solutions for the basic BWO, OBL helps to speed up the search process and improve the learning approach by using Equation (2). We allowed each population to evolve following an OBWOD position update for all individuals to increase the solution accuracy, accelerate convergence, and avoid becoming stuck in the local optimum. The exploratory sub-population increases its location to enlarge the search area and increase its global exploration potential. The exploitative sub-population performs a deep local search close to the current best solution to speed up convergence and improve the quality of the solution. The tiny population revises its position in response to OBL, which produces new individuals to replace those who are less fit and further broadens the population’s genetic variation.
- The second step: We used DCS to increase the variety and improve the consistency of the chosen solution by giving potential candidates a chance to solve the given problem with a high fitness value by using Equations (3) and (4) in the whale fall phase instead of using Equation (14). The fitness values of the new population are then calculated to find the optimum solution. This process is repeated until the termination condition (i.e., the maximum number of iterations).
4.3.3. Classification Phase
4.4. The Computational Complexity of OBWOD
| Algorithm 1 The pseudo-code of the proposed OBWOD algorithm. |
|
5. Experimental Evaluation and Discussion
5.1. Algorithm Configurations and Datasets
5.1.1. Parameter Settings
5.1.2. Dataset Description
5.1.3. Performance Matrix
- Mean accuracy (): For classification algorithms, the accuracy metric is the most-typical performance metric. One way to describe it is the proportion of correct predictions to all predictions made, as shown in Equation (19).is calculated by Equation (20).
- Mean feature selection size (): This metric (), which is denoted by Equation (22), indicates the average size of FS.According to Equation (23), we determined the overall FS ratio by dividing the FS size by the total size of the features F in the original dataset.
- Mean sensitivity (): The proposed model’s sensitivity gauges how well it can identify positive events. Another name for it is the recall or True Positive Rate (). Sensitivity () is used to assess the performance of models since it enables us to count the number of occurrences that the model was able to classify as accurately as positive, as shown in Equation (24).The metric is calculated as shown in Equation (25).
- Mean specificity (): As indicated in Equation (26), specificity indicates the percentage of s that the model accurately detects. This implies that an additional percentage of s was predicted to be positive and may be referred to as s. The True Negative Rate () is another name for this percentage. Specificity (actual negative rate)would always equal one. Low specificity indicates that the model mislabels as many negative findings as positive, whereas high specificity means that the model accurately detects the most unfavorable results.The metric is calculated as shown in Equation (27).
- Mean precision (): The precision is measured as the proportion of categorized s to all positive samples (either correctly or incorrectly). The precision gauges how accurately the model classifies a sample as positive, as shown in Equation (28).Equation (29) is used to calculate the metric.
- Standard Deviation (STD): The STD over the many executions determines the result variances for each optimization algorithm. It is calculated using Equation (30).With the aid of , all measurements in the performance matrix were calculated.
- Mean time consumption : Using , each optimization algorithm’s average consumption time (in seconds) was determined, as indicated in Equation (31).
5.2. Experimental Series 1: Global Optimization Using CEC’22 Benchmark Test Functions
5.2.1. Description of the CEC’22 Test Suite
5.2.2. Statistical Results’ Analysis
5.2.3. Convergence Behavior Analysis
5.3. Experimental Series 2: Biomedical Classification Tasks
5.3.1. Best Fitness and Convergence Evaluation
5.3.2. Accuracy and Boxplots’ Evaluation
5.3.3. Analyzing Qualitative Information
- Feature Selection (FS) evaluation:FS, which involves reducing duplicate or unnecessary features from disease datasets, can make classifiers more efficient, quick, and accurate. Regarding medical decisions, FS has an obvious advantage in increasing understandability. The advantages of choosing a subset of all attributes are several; among them, it makes data understanding and visualization more accessible. It decreases the time needed for training and usage and the measurement and storage requirements. The OBWOD algorithm can be fed data with lower dimensionality and produce a more accurate result by applying FS.Table 9 shows that the OBWOD approach outperformed the other optimizers on seven datasets. Meanwhile, the WOA achieved the best results on the Arrhythmia dataset. The second-best optimization method (HHO) had a rank of 3.40, while the highest rank of the FS ratio in the OBWOD was 2.50.
- Sensitivity (SE) evaluation:Sensitivity is a measure of the percentage of positive cases that were misclassified as positive . This suggests that a different proportion of positive cases will occur, but will be misdiagnosed as negative s. An rate can also be used to illustrate this. The sensitivity and rates added together would equal one.Table 10 shows that OBWOD achieved the best results on seven datasets. OBWOD achieved 100% on the Arrhythmia, Leukemia2, and Prostate Tumors datasets, ranging from 99.7% to 99.9% on the CKD, Parkinson’s, and Lymphography datasets.Notably, the Leukemia2 and Prostate Tumors datasets were classified with 100% sensitivity by OBWOD, INFO, HHO, HGS, and MFO (Table 10). Regarding Friedman’s rank sensitivity, OBWOD came in first with a score of 7.30, followed by HHO (6.50).
- Specificity (SP) evaluation:The percentage of s projected as negatives is known as specificity. This proposes that a different percentage of will occur. The specificity plus the rate would always add up to one.Table 11 shows that the OBWOD approach outperformed the other seven optimization algorithms on all datasets. The OBWOD achieved 100% on the Leukemia2 and Prostate Tumors datasets, ranging from 99.7% to 99.9% on the Primary Tumor, Parkinson’s, and Immunotherapy datasets.Notably, the Leukemia2 and Prostate Tumors datasets were classified with 100% specificity by OBWOD, HHO, HGS, and MFO (Table 11). Regarding Friedman’s rank specificity, the OBWOD came in first with a score of 7.30, followed by HHO (6.50).
- Precision (PPV) evaluation:To evaluate the effectiveness of ML models or overall AI solutions, precision is a frequently used statistic. It aids in understanding how accurate the predictions made by models are. The percentage of accurate positive predictions is how precision is calculated. Table 12 shows that the OBWOD approach outperformed the other seven algorithms on all datasets. The OBWOD achieved 100% on the Prostate Tumors dataset and achieved a range from 91.9% to 99.9% on the Primary Tumor, Parkinson’s, and Immunotherapy datasets.Notably, the Prostate Tumors dataset was classified with 100% precision by OBWOD, HHO, HGS, and MFO (Table 12). Regarding Friedman’s rank precision, OBWOD came in first with a score of 7.65, followed by HHO (7.15).
- Time consumption evaluation:Making a precise estimate of the amount of time and money needed to train a proposed model is crucial. This is particularly true when you use a cloud environment to train your model on a sizable amount of data. Knowing the length of the training period will help you make crucial decisions if you are working on a proposed approach.As shown in Table 13, the OBWOD approach outperformed all experiments, including the other seven algorithms, except for the time consumption test.The statistics also showed that OBWOD did an excellent job balancing exploration and extraction. Due to its premature convergence issue, BWO outperformed all other methods in terms of time consumption. This prevented us from discovering the best candidate solution since the algorithms became stuck in a local optimum, as seen in Figure 3.BWO had the highest Friedman’s rank of consumption time and was the fastest optimizer (1). The results also demonstrate that OBWOD ranked eighth.
- Friedman test: To objectively compare the performance of the OBWOD algorithm with that of the other seven algorithms, Friedman’s non-parametric test [62] was used for the mean and STD of the best solutions in Table 6, Table 7, Table 8, Table 9, Table 10, Table 11 and Table 12.In Table 6, for the Friedman test, the worst-performing algorithm is rated highest, and the one with the best performance is listed lowest. Conversely, in Table 8 and Table 10, Table 11 and Table 12, the algorithm that performs the best is rated first, and the algorithm that performs the worst is ranked last.
5.4. Discussion
- OBWOD is well able to resolve global optimization issues based on the CEC’22 test suite. OBWOD generates optimization solutions with better fitness values than the other seven MH algorithms (see Table 6). The proposed hybridization also proves to enhance the convergence ability of the algorithm; see Figure 3.
- The datasets used for this study’s analysis range in feature size from 8 to 10510 features, offering a sufficient testing environment for an optimization technique. Here, OBWOD has the highest rank of the FS ratio of 2.50, on average, on all datasets, which is better than the other seven MH algorithms; see Table 9.
- Any chance to improve OBWOD can be easily implemented because of its straightforward architecture.
- OBWOD proved its superiority compared to the other optimization algorithms on biomedical classification, as illustrated in Table 14.
- The features used by OBWOD may change each time it is run because it is an optimization strategy based on randomization. As a result, there is no assurance that the features subset chosen in one run will be present in another.
- Because OBWOD was developed from BWO, it is computationally more expensive than the other seven MH algorithms (see Table 13).
- Due to kNN’s simplicity, it was used as a learning algorithm. However, kNN has several drawbacks, such as the fact that it is a slow learner and noisy data can make it vulnerable.
6. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Houssein, E.H.; Mohamed, R.E.; Ali, A.A. Machine learning techniques for biomedical natural language processing: A comprehensive review. IEEE Access 2021, 9, 140628–140653. [Google Scholar] [CrossRef]
- Houssein, E.H.; Saber, E.; Ali, A.A.; Wazery, Y.M. Centroid mutation-based Search and Rescue optimization algorithm for feature selection and classification. Expert Syst. Appl. 2022, 191, 116235. [Google Scholar] [CrossRef]
- Chang, P.C.; Lin, J.J.; Liu, C.H. An attribute weight assignment and particle swarm optimization algorithm for medical database classifications. Comput. Methods Programs Biomed. 2012, 107, 382–392. [Google Scholar] [CrossRef]
- Houssein, E.H.; Abdelminaam, D.S.; Hassan, H.N.; Al-Sayed, M.M.; Nabil, E. A hybrid barnacles mating optimizer algorithm with support vector machines for gene selection of microarray cancer classification. IEEE Access 2021, 9, 64895–64905. [Google Scholar] [CrossRef]
- Abubakar, I.R.; Olatunji, S.O. Computational intelligence-based model for diarrhea prediction using Demographic and Health Survey data. Soft Comput. 2020, 24, 5357–5366. [Google Scholar] [CrossRef]
- Ioniţă, I.; Ioniţă, L. Prediction of thyroid disease using data mining techniques. BRAIN. Broad Res. Artif. Intell. Neurosci. 2016, 7, 115–124. [Google Scholar]
- Ganggayah, M.D.; Taib, N.A.; Har, Y.C.; Lio, P.; Dhillon, S.K. Predicting factors for survival of breast cancer patients using machine learning techniques. BMC Med. Inform. Decis. Mak. 2019, 19, 48. [Google Scholar] [CrossRef]
- Marquez, E.; Barrón, V. Artificial intelligence system to support the clinical decision for influenza. In Proceedings of the 2019 IEEE International Autumn Meeting on Power, Electronics and Computing (ROPEC), Ixtapa, Mexico, 13–15 November 2019; pp. 1–5. [Google Scholar]
- Abdelrahim, M.; Merlos, C. Hybrid machine learning approaches: A method to improve expected output of semi-structured sequential data. In Proceedings of the 2016 IEEE Tenth International Conference on Semantic Computing (ICSC), Laguna Hills, CA, USA, 4–6 February 2016; pp. 342–345. [Google Scholar]
- Blei, D.M.; Ng, A.Y.; Jordan, M.I. Latent dirichlet allocation. J. Mach. Learn. Res. 2003, 3, 993–1022. [Google Scholar]
- Mandic, D.; Chambers, J. Recurrent Neural Networks for Prediction: Learning Algorithms, Architectures and Stability; Wiley: Hoboken, NJ, USA, 2001. [Google Scholar]
- Zhang, H. The optimality of naive Bayes. Aa 2004, 1, 3. [Google Scholar]
- Peng, C.Y.J.; Lee, K.L.; Ingersoll, G.M. An introduction to logistic regression analysis and reporting. J. Educ. Res. 2002, 96, 3–14. [Google Scholar] [CrossRef]
- Houssein, E.H.; Emam, M.M.; Ali, A.A.; Suganthan, P.N. Deep and machine learning techniques for medical imaging-based breast cancer: A comprehensive review. Expert Syst. Appl. 2021, 167, 114161. [Google Scholar] [CrossRef]
- Houssein, E.H.; Emam, M.M.; Ali, A.A. Improved manta ray foraging optimization for multi-level thresholding using COVID-19 CT images. Neural Comput. Appl. 2021, 33, 16899–16919. [Google Scholar] [CrossRef]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Calhoun, V.D.; Sui, J. Machine learning in major depression: From classification to treatment outcome prediction. CNS Neurosci. Ther. 2018, 24, 1037–1052. [Google Scholar] [CrossRef]
- Bey, R.; Goussault, R.; Grolleau, F.; Benchoufi, M.; Porcher, R. Fold-stratified cross-validation for unbiased and privacy-preserving federated learning. J. Am. Med. Inform. Assoc. 2020, 27, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Mirjalili, S.; Lewis, A. The whale optimization algorithm. Adv. Eng. Softw. 2016, 95, 51–67. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, H.; Heidari, A.A.; Gandomi, A.H. Hunger games search: Visions, conception, implementation, deep analysis, perspectives, and towards performance shifts. Expert Syst. Appl. 2021, 177, 114864. [Google Scholar] [CrossRef]
- Mirjalili, S. SCA: A sine cosine algorithm for solving optimization problems. Knowl.-Based Syst. 2016, 96, 120–133. [Google Scholar] [CrossRef]
- Heidari, A.A.; Mirjalili, S.; Faris, H.; Aljarah, I.; Mafarja, M.; Chen, H. Harris hawks optimization: Algorithm and applications. Future Gener. Comput. Syst. 2019, 97, 849–872. [Google Scholar] [CrossRef]
- Ahmadianfar, I.; Heidari, A.A.; Noshadian, S.; Chen, H.; Gandomi, A.H. INFO: An Efficient Optimization Algorithm based on Weighted Mean of Vectors. Expert Syst. Appl. 2022, 195, 116516. [Google Scholar] [CrossRef]
- Mirjalili, S. Moth-flame optimization algorithm: A novel nature-inspired heuristic paradigm. Knowl.-Based Syst. 2015, 89, 228–249. [Google Scholar] [CrossRef]
- Zhong, C.; Li, G.; Meng, Z. Beluga whale optimization: A novel nature-inspired metaheuristic algorithm. Knowl.-Based Syst. 2022, 251, 109215. [Google Scholar] [CrossRef]
- Hameed, N.; Shabut, A.; Hossain, M.A. A Computer-aided diagnosis system for classifying prominent skin lesions using machine learning. In Proceedings of the 2018 10th Computer Science and Electronic Engineering (CEEC), Colchester, UK, 19–21 September 2018; pp. 186–191. [Google Scholar]
- Bhattacharya, M.; Lu, D.Y.; Kudchadkar, S.M.; Greenland, G.V.; Lingamaneni, P.; Corona-Villalobos, C.P.; Guan, Y.; Marine, J.E.; Olgin, J.E.; Zimmerman, S.; et al. Identifying ventricular arrhythmias and their predictors by applying machine learning methods to electronic health records in patients with hypertrophic cardiomyopathy (HCM-VAr-risk model). Am. J. Cardiol. 2019, 123, 1681–1689. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, X.; Huo, J.; Xia, L.; Shan, F.; Liu, J.; Mo, Z.; Yan, F.; Ding, Z.; Yang, Q.; Song, B.; et al. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging 2020, 39, 2595–2605. [Google Scholar] [CrossRef]
- Mohan, S.; Thirumalai, C.; Srivastava, G. Effective heart disease prediction using hybrid machine learning techniques. IEEE Access 2019, 7, 81542–81554. [Google Scholar] [CrossRef]
- Ghazal, T.M.; Taleb, N. Feature optimization and identification of ovarian cancer using internet of medical things. Expert Syst. 2022, 39, e12987. [Google Scholar] [CrossRef]
- Calp, M.H. Medical diagnosis with a novel SVM-CoDOA based hybrid approach. arXiv 2019, arXiv:1902.00685. [Google Scholar]
- Rahman, F.; Mahmood, M. A Dynamic Approach to Identify the Most Significant Biomarkers for Heart Disease Risk Prediction Utilizing Machine Learning Techniques. In Proceedings of the International Conference on Bangabandhu and Digital Bangladesh, Dhaka, Bangladesh, 30 December 2021; Springer: Berlin/Heidelberg, Germany, 2022; pp. 12–22. [Google Scholar]
- Hashim, F.A.; Hussain, K.; Houssein, E.H.; Mabrouk, M.S.; Al-Atabany, W. Archimedes optimization algorithm: A new metaheuristic algorithm for solving optimization problems. Appl. Intell. 2021, 51, 1531–1551. [Google Scholar] [CrossRef]
- Hashim, F.A.; Houssein, E.H.; Hussain, K.; Mabrouk, M.S.; Al-Atabany, W. Honey Badger Algorithm: New metaheuristic algorithm for solving optimization problems. Math. Comput. Simul. 2022, 192, 84–110. [Google Scholar] [CrossRef]
- Houssein, E.H.; Hosney, M.E.; Mohamed, W.M.; Ali, A.A.; Younis, E.M. Fuzzy-based hunger games search algorithm for global optimization and feature selection using medical data. Neural Comput. Appl. 2022, 1–25. [Google Scholar] [CrossRef]
- Fajri, D.M.N.; Mahmudy, W.F.; Anggodo, Y.P. Optimization of FIS Tsukamoto using particle swarm optimization for dental disease identification. In Proceedings of the 2017 International Conference on Advanced Computer Science and Information Systems (ICACSIS), Jakarta, Indonesia, 28–29 October 2017; pp. 261–268. [Google Scholar]
- Ahmad, G.N.; Fatima, H.; Ullah, S.; Saidi, A.S.; Imdadullah. Efficient medical diagnosis of human heart diseases using machine learning techniques with and without GridSearchCV. IEEE Access 2022, 10, 80151–80173. [Google Scholar] [CrossRef]
- Jalali, S.M.J.; Ahmadian, M.; Ahmadian, S.; Khosravi, A.; Alazab, M.; Nahavandi, S. An oppositional-Cauchy based GSK evolutionary algorithm with a novel deep ensemble reinforcement learning strategy for COVID-19 diagnosis. Appl. Soft Comput. 2021, 111, 107675. [Google Scholar] [CrossRef]
- Hsu, C.H.; Chen, X.; Lin, W.; Jiang, C.; Zhang, Y.; Hao, Z.; Chung, Y.C. Effective multiple cancer disease diagnosis frameworks for improved healthcare using machine learning. Measurement 2021, 175, 109145. [Google Scholar] [CrossRef]
- Cai, X.; Li, X.; Razmjooy, N.; Ghadimi, N. Breast cancer diagnosis by convolutional neural network and advanced thermal exchange optimization algorithm. Comput. Math. Methods Med. 2021, 2021, 5595180. [Google Scholar] [CrossRef]
- Bharti, R.; Khamparia, A.; Shabaz, M.; Dhiman, G.; Pande, S.; Singh, P. Prediction of heart disease using a combination of machine learning and deep learning. Comput. Intell. Neurosci. 2021, 2021, 8387680. [Google Scholar] [CrossRef]
- Ghosh, D.; Ghosh, E. Breast Mammography-based Tumor Detection and Classification Using Fine-Tuned Convolutional Neural Networks. Int. J. Adv. Res. Radiol. Imaging Sci. 2022, 1. [Google Scholar]
- Nadakinamani, R.G.; Reyana, A.; Kautish, S.; Vibith, A.; Gupta, Y.; Abdelwahab, S.F.; Mohamed, A.W. Clinical Data Analysis for Prediction of Cardiovascular Disease Using Machine Learning Techniques. Comput. Intell. Neurosci. 2022, 2022, 2973324. [Google Scholar] [CrossRef]
- Houssein, E.H.; Emam, M.M.; Ali, A.A. An optimized deep learning architecture for breast cancer diagnosis based on improved marine predators algorithm. Neural Comput. Appl. 2022, 34, 18015–18033. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Jimenez, G. Optimal diagnosis of the skin cancer using a hybrid deep neural network and grasshopper optimization algorithm. Open Med. 2022, 17, 508–517. [Google Scholar] [CrossRef]
- Reddy, K.V.V.; Elamvazuthi, I.; Abd Aziz, A.; Paramasivam, S.; Chua, H.N.; Pranavanand, S. Prediction of Heart Disease Risk Using Machine Learning with Correlation-based Feature Selection and Optimization Techniques. In Proceedings of the 2021 7th International Conference on Signal Processing and Communication (ICSC), Noida, India, 25–27 November 2021; pp. 228–233. [Google Scholar]
- Mehta, P.; Petersen, C.A.; Wen, J.C.; Banitt, M.R.; Chen, P.P.; Bojikian, K.D.; Egan, C.; Lee, S.I.; Balazinska, M.; Lee, A.Y.; et al. Automated detection of glaucoma with interpretable machine learning using clinical data and multimodal retinal images. Am. J. Ophthalmol. 2021, 231, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Polat, K.; Güneş, S. Automatic determination of diseases related to lymph system from lymphography data using principles component analysis (PCA), fuzzy weighting pre-processing and ANFIS. Expert Syst. Appl. 2007, 33, 636–641. [Google Scholar] [CrossRef]
- Zhang, S.; Li, X.; Zong, M.; Zhu, X.; Wang, R. Efficient knn classification with different numbers of nearest neighbors. IEEE Trans. Neural Netw. Learn. Syst. 2017, 29, 1774–1785. [Google Scholar] [CrossRef] [PubMed]
- El-Kenawy, E.S.; Eid, M. Hybrid gray wolf and particle swarm optimization for feature selection. Int. J. Innov. Comput. Inf. Control 2020, 16, 831–844. [Google Scholar]
- Mafarja, M.; Aljarah, I.; Heidari, A.A.; Faris, H.; Fournier-Viger, P.; Li, X.; Mirjalili, S. Binary dragonfly optimization for feature selection using time-varying transfer functions. Knowl.-Based Syst. 2018, 161, 185–204. [Google Scholar] [CrossRef]
- Subasi, A. Use of artificial intelligence in Alzheimer’s disease detection. In Artificial Intelligence in Precision Health; Elsevier: Amsterdam, The Netherlands, 2020; pp. 257–278. [Google Scholar]
- Tizhoosh, H.R. Opposition-based learning: A new scheme for machine intelligence. In Proceedings of the International Conference on Computational Intelligence for Modelling, Control and Automation and International Conference on Intelligent Agents, Web Technologies and Internet Commerce (CIMCA-IAWTIC’06), Vienna, Austria, 28–30 November 2005; Volume 1, pp. 695–701. [Google Scholar]
- Houssein, E.H.; Ibrahim, I.E.; Neggaz, N.; Hassaballah, M.; Wazery, Y.M. An Efficient ECG Arrhythmia Classification Method Based on Manta Ray Foraging Optimization. Expert Syst. Appl. 2021, 181, 115131. [Google Scholar] [CrossRef]
- Khodadadi, N.; Snasel, V.; Mirjalili, S. Dynamic arithmetic optimization algorithm for truss optimization under natural frequency constraints. IEEE Access 2022, 10, 16188–16208. [Google Scholar] [CrossRef]
- Mantegna, R.N. Fast, accurate algorithm for numerical simulation of Levy stable stochastic processes. Phys. Rev. E 1994, 49, 4677. [Google Scholar] [CrossRef]
- Frank. Machine Learning Repository. 2010. Available online: https://archive.ics.uci.edu/ml/index.php (accessed on 25 December 2022).
- Yan, F.; Xu, X.; Xu, J. Grey wolf optimizer with a novel weighted distance for global optimization. IEEE Access 2020, 8, 120173–120197. [Google Scholar] [CrossRef]
- Gajendra, E.; Kumar, J. A novel approach of ECG classification for diagnosis of heart diseases: Review. Int. J. Adv. Res. Comput. Eng. Technol. (IJARCET) 2015, 4, 4096–4100. [Google Scholar]
- Ahrari, A.; Elsayed, S.; Sarker, R.; Essam, D.; Coello, C.A.C. Problem Definition and Evaluation Criteria for the CEC’2022 Competition on Dynamic Multimodal Optimization. In Proceedings of the IEEE World Congress on Computational Intelligence (IEEE WCCI 2022), Padua, Italy, 18–23 July 2022; pp. 1–10. [Google Scholar]
- Friedman, M. A comparison of alternative tests of significance for the problem of m rankings. Ann. Math. Stat. 1940, 11, 86–92. [Google Scholar] [CrossRef]
- Vijayashree, J.; Sultana, H.P. A machine learning framework for feature selection in heart disease classification using improved particle swarm optimization with support vector machine classifier. Program. Comput. Softw. 2018, 44, 388–397. [Google Scholar] [CrossRef]
- Ismaeel, S.; Miri, A.; Chourishi, D. Using the Extreme Learning Machine (ELM) technique for heart disease diagnosis. In Proceedings of the 2015 IEEE Canada International Humanitarian Technology Conference (IHTC2015), Ottawa, ON, Canada, 31 May–4 June 2015; pp. 1–3. [Google Scholar]
- Tuncer, T.; Dogan, S.; Acharya, U.R. Automated detection of Parkinson’s disease using minimum average maximum tree and singular value decomposition method with vowels. Biocybern. Biomed. Eng. 2020, 40, 211–220. [Google Scholar] [CrossRef]
- Sharma, S.R.; Singh, B.; Kaur, M. Classification of Parkinson disease using binary Rao optimization algorithms. Expert Syst. 2021, 38, e12674. [Google Scholar] [CrossRef]
- Polat, K. A hybrid approach to Parkinson disease classification using speech signal: The combination of smote and random forests. In Proceedings of the 2019 Scientific Meeting on Electrical-Electronics & Biomedical Engineering and Computer Science (EBBT), Istanbul, Turkey, 24–26 April 2019; pp. 1–3. [Google Scholar]
- Wazery, Y.M.; Saber, E.; Houssein, E.H.; Ali, A.A.; Amer, E. An efficient slime mold algorithm combined with k-nearest neighbor for medical classification tasks. IEEE Access 2021, 9, 113666–113682. [Google Scholar] [CrossRef]
- Houssein, E.H.; Saber, E.; Ali, A.A.; Wazery, Y.M. Opposition-based learning tunicate swarm algorithm for biomedical classification. In Proceedings of the 2021 17th International Computer Engineering Conference (ICENCO), Cairo, Egypt, 29–30 December 2021; pp. 1–6. [Google Scholar]




| Ref. | Datasets | Used Techniques | Results |
|---|---|---|---|
| [37] | Dental disease | Using OBL, Cauchy mutation operators, and a modified version of Gaining– Sharing Knowledge (GSK). | Average accuracy = 88%. |
| [38] | Human heart diseases | GridSearchCV based on extreme gradient boosting classifier | Accuracy = 99.03%. |
| [39] | COVID-19 | Tsukamoto Fuzzy inference system with PSO | Accuracy = 0.9914, precision = 0.9935, recall = 0.9814, F-measure = 0.9896, and AUC = 0.9903. |
| [40] | Breast, cervical, and lung cancer | A machine-learning-based feature modeling | Breast accuracy = 99.62%, cervical accuracy = 96.88%, and lung accuracy = 98.21%. |
| [41] | Breast cancer | Convolutional neural networks and advanced thermal exchange optimization algorithm | Accuracy = 93.79%, specificity = 67.7%, and recall = 96.89%. |
| [42] | Heart disease | kNN with ANN | Accuracy = 94.2%. |
| [43] | Breast cancer | Visual Geometry Group (VGG)-16 and VGG-19 pre-trained CNNs | Accuracy = 97.1%, specificity = 97.9%, and recall = 96.3%. |
| [44] | Cardiovascular disease | REP tree, M5P tree, random tree, linear regression, NB, J48, and JRIP | Random tree’s accuracy = 100%. |
| [45] | Breast cancer | Optimized deep residual learning model, Improved Marine Predators Algorithm (IMPA), IMPA-ResNet50 | Accuracy = 98.32%. |
| [46] | Skin cancer | Grasshopper Optimization Algorithm (GOA) with AlexNet and extreme learning machine network | Accuracy = 98% and sensitivity = 93%. |
| [47] | Heart disease | SVM algorithm with statistical FS | Accuracy = 89.7%. |
| [48] | Glaucoma clinical data | DL model built on CFP classified | Accuracy = 97%. |
| [49] | Lymphography | Principle Component Analysis (PCA), fuzzy weighting pre-processing, and Adaptive Neuro-Fuzzy Inference System (ANFIS) | Accuracy = 88.83%. |
| Algorithm | Parameter | Value |
|---|---|---|
| INFO [24] | c | 2 |
| d | 4 | |
| WOA [20] | [2, 0] | |
| [−1, 1] | ||
| MFO [25] | [−1, −2] | |
| SCA [22] | [2, 0] | |
| HHO [23] | escaping energy | [0.5, 0.5] |
| BWO [26] | whale fall | [0.1, 0.05] |
| 0.99 | ||
| 0.01 | ||
| HGS [21] | R | |
| , | [0, 1] |
| Item | Setting |
|---|---|
| 0 | |
| 1 | |
| 100 | |
| N | 20 |
| M | 20 |
| k | 5 |
| Dataset | Total Features | Total Patients | Category | Feature Types | Size |
|---|---|---|---|---|---|
| Arrhythmia | 279 | 452 | Classification | Categorical, Integer, Real | High |
| Leukemia2 | 11,226 | 72 | Classification | Integer, Real | High |
| Prostate Tumors | 10,510 | 102 | Classification | Integer, Real | High |
| Statlog (Heart) | 13 | 270 | Classification | Categorical, Real | Medium |
| Chronic Kidney Disease (CKD) | 25 | 400 | Classification | Real | Medium |
| Parkinson’s | 23 | 197 | Classification | Real | Medium |
| Pima Indians Diabetes (Pima) | 8 | 768 | Classification | Integer, Real | Medium |
| Primary Tumor | 17 | 339 | Classification | Categorical | low |
| Lymphography | 18 | 148 | Classification | Categorical | low |
| Immunotherapy | 8 | 90 | Classification | Integer, Real | low |
| Type | Function | Description | Search Range | |
|---|---|---|---|---|
| Unimodal Function | CEC-01 | Shifted and fully rotated Zakharov function. | 300 | |
| Basic Functions | CEC-02 | Shifted and fully rotated Rosenbrock function. | 400 | |
| Basic Functions | CEC-03 | Shifted and fully rotated expanded Schaffer f6 Function. | 600 | |
| basic Functions | CEC-04 | Shifted and fully rotated non-continuous Rastrigin function. | 800 | |
| Basic Functions | CEC-05 | Shifted and fully rotated Levy function. | 900 | |
| Hybrid Functions | CEC-06 | Hybrid Function 1 (N = 3) | 1800 | |
| Hybrid Functions | CEC-07 | Hybrid Function 2 (N = 6). | 2000 | |
| Hybrid Functions | CEC-08 | Hybrid Function 3 (N = 5). | 2200 | |
| Composition Functions | CEC-09 | Composition Function 1 (N = 5) | 2300 | |
| Composition Functions | CEC-10 | Composition Function 2 (N = 4) | 2400 |
| CEC Functions | Measure | BWO | OBWOD | INFO | HHO | HGS | SCA | WOA | MFO |
|---|---|---|---|---|---|---|---|---|---|
| CEC-01 | 2.494610 × 10 | 2.152710 × 10 | 5.979706 × 10 | 7.571684 × 10 | 9.463143 × 10 | 9.558730 × 10 | 8.52330 × 10 | 3.087340 × 10 | |
| 1.953938 × 10 | 1.064252 × 10 | 2.311781 × 10 | 2.539657 × 10 | 9.558730 × 10 | 7.5746858 × 10 | 5.851858 × 10 | 1.947247 × 10 | ||
| CEC-02 | 6.564376 × 10 | 5.086089 × 10 | 1.508763 × 10 | 7.756663 × 10 | 7.433591 × 10 | 7.508678 × 10 | 7.69828 × 10 | 6.151887 × 10 | |
| 9.464901 × 10 | 1.439867 × 10 | 8.297016 × 10 | 1.040701 × 10 | 7.508678 × 10 | 9.14076 × 10 | 9.5896 × 10 | 3.057235 × 10 | ||
| CEC-03 | 6.303864 × 10 | 6.287108 × 10 | 6.668264 × 10 | 6.408925 × 10 | 7.527101 × 10 | 7.603132 × 10 | 7.569872 × 10 | 6.421989 × 10 | |
| 1.900605 × 10 | 1.244823 × 10 | 1.042105 × 10 | 2.797057 × 10 | 7.603132 × 10 | 4.570383 × 10 | 3.72589 × 10 | 2.024130 × 10 | ||
| CEC-04 | 8.965367 × 10 | 8.449059 × 10 | 9.676970 × 10 | 9.052959 × 10 | 1.066585 × 10 | 1.077359 × 10 | 1.188301 × 10 | 9.001926 × 10 | |
| 4.459035 × 10 | 2.369911 × 10 | 3.657403 × 10 | 4.566980 × 10 | 1.077359 × 10 | 2.371115 × 10 | 4.371115 × 10 | 4.345011 × 10 | ||
| CEC-05 | 2.363152 × 10 | 2.020499 × 10 | 3.100224 × 10 | 3.369690 × 10 | 1.038756 × 10 | 1.049249 × 10 | 3.887064 × 10 | 3.666241 × 10 | |
| 1.041466 × 10 | 5.239663 × 10 | 1.281234 × 10 | 1.946050 × 10 | 1.049249 × 10 | 7.312613 × 10 | 3.329509 × 10 | 8.850809 × 10 | ||
| CEC-06 | 6.790918 × 10 | 1.521993 × 10 | 1.513433 × 10 | 1.377625 × 10 | 8.770613 × 10 | 8.859205 × 10 | 1.263948 × 10 | 2.595407 × 10 | |
| 4.237511 × 10 | 1.003401 × 10 | 8.375683 × 10 | 4.769067 × 10 | 8.859205 × 10 | 1.341870 × 10 | 1.541873 × 10 | 1.113193 × 10 | ||
| CEC-07 | 2.123416 × 10 | 2.008917 × 10 | 2.205811 × 10 | 2.094071 × 10 | 2.664960 × 10 | 2.691879 × 10 | 3.265476 × 10 | 3.452825 × 10 | |
| 5.428055 × 10 | 4.403649 × 10 | 4.602230 × 10 | 7.801352 × 10 | 2.691879 × 10 | 2.691879 × 10 | 4.627033 × 10 | 4.452825 × 10 | ||
| CEC-08 | 2.251655 × 10 | 2.208762 × 10 | 2.280427 × 10 | 2.362581 × 10 | 2.230307 × 10 | 2.69876 × 10 | 2.252836 × 10 | 2.254398 × 10 | |
| 6.435948 × 10 | 4.460133 × 10 | 1.178931 × 10 | 6.281607 × 10 | 2.252836 × 10 | 3.78027 × 10 | 1.755027 × 10 | 4.393503 × 10 | ||
| CEC-09 | 2.544037 × 10 | 2.305232 × 10 | 2.782409 × 10 | 2.533574 × 10 | 6.551957 × 10 | 6.618138 × 10 | 6.618138 × 10 | 2.509326 × 10 | |
| 1.294362 × 10 | 5.583183 × 10 | 2.330702 × 10 | 1.580436 × 10 | 6.618138 × 10 | 6.618138 × 10 | 6.618138 × 10 | 7.913255 × 10 | ||
| CEC-10 | 4.133289 × 10 | 2.803257 × 10 | 6.313881 × 10 | 4.372988 × 10 | 1.081208 × 10 | 3.45629 × 10 | 1.092129 × 10 | 2.526238 × 10 | |
| 1.211022 × 10 | 1.037671 × 10 | 9.866894 × 10 | 1.439390 × 10 | 1.092129 × 10 | 1.828153 × 10 | 1.828153 × 10 | 1.725860 × 10 | ||
| Friedman Rank | 2.65 | 1.10 | 4.70 | 4.00 | 6.30 | 7.20 | 6.80 | 3.25 | |
| 4.75 | 2.40 | 4.70 | 6.00 | 6.40 | 4.15 | 4.35 | 3.25 |
| Datasets | BWO | OBWOD | INFO | HHO | HGS | SCA | WOA | MFO | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arrhythmia | 0.2465 | 0.0000 | 0.2038 | 0.0000 | 0.0670 | 0.2455 | 0.0001 | 0.0013 | 0.2201 | 0.0021 | 0.4480 | 0.4982 | 0.4910 | 0.5008 | 0.4480 | 0.4982 |
| Leukemia2 | 0.0601 | 0.0307 | 0.0560 | 0.0005 | 0.1399 | 0.0606 | 0.0977 | 0.0333 | 0.1099 | 8.0834 | 0.2440 | 0.0246 | 0.2448 | 0.0006 | 0.2454 | 0.0000 |
| Prostate Tumors | 0.0524 | 0.0538 | 0.0011 | 0.0250 | 0.2133 | 0.0828 | 0.1558 | 0.0402 | 0.1781 | 0.0918 | 0.2700 | 0.0272 | 0.2992 | 0.0039 | 0.3138 | 0.0061 |
| Statlog (Heart) | 0.2149 | 0.0095 | 0.1854 | 0.0005 | 0.2439 | 0.0422 | 0.2357 | 0.0238 | 0.2243 | 0.0496 | 0.2851 | 0.0288 | 0.2709 | 0.0061 | 0.3124 | 0.0007 |
| CKD | 0.2137 | 0.0266 | 0.2037 | 0.0014 | 0.2891 | 0.0563 | 0.3120 | 0.0268 | 0.2481 | 0.0685 | 0.3627 | 0.0366 | 0.3562 | 0.0067 | 0.3994 | 0.0025 |
| Parkinson’s | 0.0926 | 0.0141 | 0.0810 | 0.0016 | 0.1375 | 0.0355 | 0.1312 | 0.0313 | 0.1327 | 0.0497 | 0.1517 | 0.0153 | 0.2025 | 0.0046 | 0.1876 | 0.0446 |
| Pima | 0.2567 | 0.0004 | 0.2006 | 0000 | 0.2988 | 0.1715 | 0.2657 | 0.0269 | 0.2649 | 0.0354 | 0.2972 | 0.0300 | 0.3182 | 0.0078 | 0.2911 | 0.0072 |
| Primary Tumor | 0.1891 | 0.0420 | 0.1801 | 0.0008 | 0.2251 | 0.0040 | 0.2219 | 0.0025 | 0.2275 | 0.5291 | 0.3112 | 0.0324 | 0.2790 | 0.0009 | 0.5793 | 0.0005 |
| Lymphography | 0.1227 | 0.0128 | 0.1167 | 0.0198 | 0.1290 | 0.0070 | 0.1802 | 0.0199 | 0.1318 | 0.0206 | 0.1451 | 0.0315 | 0.1849 | 0.0143 | 0.4516 | 0.0188 |
| Immunotherapy | 0.1533 | 0.0171 | 0.1219 | 0.0002 | 0.1404 | 0.0474 | 0.0129 | 0.0384 | 0.1602 | 0.0538 | 0.2234 | 0.0225 | 0.1907 | 0.0007 | 0.1198 | 0.0020 |
| Friedman Rank | 2.60 | 3.85 | 1.40 | 1.85 | 4.40 | 6.00 | 3.30 | 5.00 | 4.00 | 7.40 | 6.45 | 5.55 | 7.00 | 3.20 | 6.85 | 3.15 |
| Rank | 2 | 1 | 5 | 3 | 4 | 6 | 8 | 7 | ||||||||
| Datasets | BWO | OBWOD | INFO | HHO | HGS | SCA | WOA | MFO | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arrhythmia | 0.4816 | 0.0775 | 0.6590 | 0.0014 | 0.5731 | 0.0126 | 0.5865 | 0.0212 | 0.5222 | 0.0008 | 0.4551 | 0.0487 | 0.2263 | 0.1940 | 0.4778 | 0.01949 |
| Leukemia2 | 0.7832 | 0.0132 | 0.9883 | 0.00294 | 0.8902 | 0.0460 | 0.9555 | 0.0442 | 0.8356 | 0.0328 | 0.7779 | 0.0782 | 0.4944 | 0.2184 | 0.7857 | 0.0320 |
| Prostate Tumors | 0.9091 | 0.0981 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 0.9807 | 0.1015 | 0.5126 | 0.2067 | 1.0000 | 0.0000 |
| Statlog (Heart) | 0.5535 | 0.1065 | 0.8813 | 0.0004 | 0.8786 | 0.0238 | 0.8622 | 0.0279 | 0.7593 | 0.0050 | 0.4617 | 0.0706 | 0.4744 | 0.0649 | 0.5912 | 0.0891 |
| CKD | 0.5231 | 0.1914 | 0.7817 | 0.00171 | 0.7022 | 0.0149 | 0.7038 | 0.0175 | 0.6339 | 0.0056 | 0.5595 | 0.0617 | 0.2124 | 0.2207 | 0.6066 | 0.0164 |
| Parkinson’s | 0.7684 | 0.1090 | 0.9983 | 0.0006 | 0.9956 | 0.0110 | 0.9944 | 0.0152 | 0.9113 | 0.0129 | 0.6764 | 0.0987 | 0.6827 | 0.0615 | 0.8284 | 0.0521 |
| Pima | 0.6109 | 0.0537 | 0.8545 | 0.0006 | 0.8079 | 0.0094 | 0.8113 | 0.0098 | 0.7792 | 0.0020 | 0.5816 | 0.0625 | 0.6005 | 0.0330 | 0.6516 | 0.0604 |
| Primary Tumor | 0.6171 | 0.0398 | 0.8876 | 0.0003 | 0.8574 | 0.0348 | 0.8816 | 0.0368 | 0.7692 | 0.0750 | 0.4943 | 0.0874 | 0.5638 | 0.0525 | 0.5936 | 0.0682 |
| Lymphography | 0.5269 | 0.0133 | 0.5702 | 0.0010 | 0.5889 | 0.0229 | 0.5288 | 0.0359 | 0.5325 | 0.0520 | 0.5672 | 0.0572 | 0.5707 | 0.0070 | 0.05918 | 0.0082 |
| Immunotherapy | 0.7037 | 0.0502 | 0.9089 | 0.0001 | 0.8889 | 0.0320 | 0.8889 | 0.0420 | 0.8889 | 0.0320 | 0.6317 | 0.0700 | 0.6336 | 0.0413 | 0.6908 | 0.0879 |
| Friedman mean rank | 3.10 | 5.80 | 7.20 | 1.30 | 6.40 | 3.25 | 6.30 | 4.40 | 5.20 | 3.35 | 2.10 | 7.00 | 2.10 | 5.90 | 3.60 | 5.00 |
| Rank | 6 | 1 | 2 | 3 | 4 | 7 | 8 | 5 | ||||||||
| Datasets | BWO | OBWOD | INFO | HHO | HGS | SCA | WOA | MFO | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arrhythmia | 0.5100 | 0.4999 | 1.0000 | 0.0000 | 0.5035 | 0.5000 | 0.4956 | 0.4572 | 0.4996 | 0.5000 | 0.4973 | 0.5000 | 0.4985 | 0.5000 | 0.5210 | 0.4996 |
| Leukemia2 | 0.0448 | 0.0030 | 0.0280 | 0.0000 | 0.0529 | 0.2161 | 0.0083 | 0.0014 | NaN | NaN | 0.4976 | 0.5000 | 0.4919 | 0.5000 | 0.4942 | 0.5000 |
| Prostate Tumors | 0.0402 | 0.0000 | 0.0215 | 0.0000 | 0.0052 | 0.0278 | 0.0000 | 0.0000 | 0.3214 | 0.1259 | 0.4976 | 0.5000 | 0.4942 | 0.5000 | 0.4970 | 0.5000 |
| Statlog (Heart) | 0.2838 | 0.0890 | 0.2149 | 0.0000 | 0.2949 | 0.2758 | 0.2910 | 0.0000 | 0.2321 | 0.0081 | 0.4615 | 0.5189 | 0.3846 | 0.5064 | 0.3846 | 0.5064 |
| CKD | 0.2949 | 0.0320 | 0.2288 | 0.0000 | 0.2882 | 0.2706 | 0.0000 | 0.0006 | 0.2723 | 0.0251 | 0.4086 | 0.4925 | 0.4875 | 0.5007 | 0.5018 | 0.5009 |
| Parkinson’s | 0.1779 | 0.0090 | 0.0779 | 0.0000 | 0.1817 | 0.3945 | 0.1800 | 0.0000 | 0.1023 | 0.2769 | 0.2727 | 0.4558 | 0.3182 | 0.4767 | 0.3636 | 0.4924 |
| Pima | 0.2562 | 0.0200 | 0.2062 | 0.0000 | 0.2935 | 0.4198 | 0.3210 | 0.0800 | 0.2365 | 0.0021 | 0.3750 | 0.5175 | 0.1250 | 0.3536 | 0.3750 | 0.5175 |
| Primary Tumor | 0.1938 | 0.0200 | 0.1728 | 0.0000 | 0.1943 | 0.2961 | 0.0137 | 0.1966 | 0.1823 | 0.0020 | 0.4118 | 0.5073 | 0.4706 | 0.5145 | 0.2941 | 0.4697 |
| Lymphography | 0.4590 | 0.0300 | 0.3685 | 0.0000 | 0.3594 | 0.4486 | 0.46100 | 0.0230 | 0.4250 | 0.0230 | 0.3833 | 0.4851 | 0.3889 | 0.5016 | 0.3933 | 0.4851 |
| Immunotherapy | 0.1406 | 0.0000 | 0.1206 | 0.0000 | 0.3449 | 0.0000 | 0.3780 | 0.1303 | 0.3449 | 0.2857 | 0.4880 | 0.4286 | 0.5345 | 0.0000 | 0.1429 | 0.0000 |
| Friedman Rank | 4.20 | 3.40 | 2.50 | 1.50 | 4.15 | 4.95 | 3.40 | 2.85 | 3.35 | 3.60 | 6.25 | 6.95 | 5.65 | 6.50 | 6.50 | 6.25 |
| Rank | 5 | 1 | 4 | 3 | 2 | 7 | 6 | 8 | ||||||||
| Datasets | BWO | OBWOD | INFO | HHO | HGS | SCA | WOA | MFO | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arrhythmia | 0.9178 | 0.1368 | 1.0000 | 0.0000 | 0.9178 | 0.1368 | 0.9998 | 0.0017 | 0.9848 | 0.0000 | 0.8974 | 0.0951 | 0.3851 | 0.4423 | 0.9292 | 0.0233 |
| Leukemia2 | 0.9814 | 0.0093 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 0.9900 | 0.0995 | 0.7722 | 0.2715 | 1.0000 | 0.0000 |
| Prostate Tumors | 0.8614 | 0.1959 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 0.9900 | 0.0995 | 0.3840 | 0.2943 | 1.0000 | 0.0000 |
| Statlog (Heart) | 0.5432 | 0.2887 | 0.9565 | 0.0178 | 0.5432 | 0.2887 | 0.9545 | 0.0203 | 0.8519 | 0.0000 | 0.1629 | 0.2547 | 0.2866 | 0.2853 | 0.6453 | 0.1052 |
| CKD | 0.7915 | 0.3283 | 0.9999 | 0.0014 | 0.9915 | 0.3283 | 0.9996 | 0.0029 | 0.9771 | 0.0044 | 0.9060 | 0.0973 | 0.2236 | 0.3740 | 0.9479 | 0.0227 |
| Parkinson’s | 0.6138 | 0.2166 | 0.9992 | 0.0006 | 0.9902 | 0.0092 | 0.9981 | 0.0138 | 0.9000 | 0.0050 | 0.4692 | 0.1913 | 0.3518 | 0.1520 | 0.6899 | 0.1290 |
| Pima | 0.8995 | 0.0023 | 0.8345 | 0.0370 | 0.8965 | 0.0073 | 0.7242 | 0.0119 | 0.7093 | 0.0714 | 0.7179 | 0.0088 | 0.7177 | 0.0066 | 0.8868 | 0.0156 |
| Primary Tumor | 0.5655 | 0.1354 | 0.7995 | 0.0090 | 0.6021 | 0.2031 | 0.7544 | 0.0856 | 0.5000 | 0.0750 | 0.0081 | 0.0418 | 0.0095 | 0.0617 | 0.1830 | 0.1446 |
| Lymphography | 0.6654 | 0.0116 | 0.9977 | 0.0157 | 0.7454 | 0.0326 | 0.9952 | 0.0355 | 0.7804 | 0.0508 | 0.5413 | 0.0862 | 0.4923 | 0.1694 | 0.6154 | 0.0554 |
| Immunotherapy | 0.5446 | 0.1418 | 0.7959 | 0.0283 | 0.5626 | 0.1418 | 0.7885 | 0.0466 | 0.6000 | 0.0000 | 0.5608 | 0.0141 | 0.0086 | 0.0688 | 0.1186 | 0.2228 |
| Friedman Rank | 3.60 | 5.60 | 7.30 | 2.45 | 5.40 | 5.00 | 6.50 | 3.60 | 5.10 | 3.15 | 2.40 | 5.00 | 1.30 | 6.20 | 4.40 | 5.00 |
| Rank | 6 | 1 | 3 | 2 | 4 | 7 | 8 | 5 | ||||||||
| Datasets | BWO | OBWOD | INFO | HHO | HGS | SCA | WOA | MFO | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arrhythmia | 0.1010 | 0.0194 | 0.2734 | 0.0095 | 0.1230 | 0.0175 | 0.6367 | 0.3527 | 0.1441 | 0.0047 | 0.0481 | 0.0210 | 0.0571 | 0.0328 | 0.0638 | 0.0281 |
| Leukemia2 | 0.9873 | 0.0426 | 1.0000 | 0.0000 | 0.9763 | 0.0526 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 0.9822 | 0.1028 | 0.6197 | 0.2744 | 1.0000 | 0.0000 |
| Prostate Tumors | 0.9884 | 0.0464 | 1.0000 | 0.0000 | 0.9754 | 0.0454 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 0.9900 | 0.0995 | 0.6188 | 0.3106 | 1.0000 | 0.0000 |
| Statlog (Heart) | 0.4403 | 0.1805 | 0.8327 | 0.0724 | 0.4563 | 0.1725 | 0.8809 | 0.0806 | 0.6667 | 0.0000 | 0.2354 | 0.1136 | 0.3174 | 0.0939 | 0.4889 | 0.1096 |
| CKD | 0.1846 | 0.0175 | 0.4247 | 0.0099 | 0.1846 | 0.0175 | 0.5838 | 0.2678 | 0.2474 | 0.0152 | 0.1333 | 0.0332 | 0.1118 | 0.0535 | 0.1373 | 0.0262 |
| Parkinson’s | 0.8130 | 0.0842 | 0.9972 | 0.0096 | 0.8330 | 0.0952 | 0.9971 | 0.0111 | 0.9392 | 0.0147 | 0.7092 | 0.0868 | 0.7256 | 0.0549 | 0.8776 | 0.0583 |
| Pima | 0.2778 | 0.0883 | 0.7794 | 0.0003 | 0.2638 | 0.0436 | 0.6736 | 0.0338 | 0.5105 | 0.0025 | 0.2130 | 0.0437 | 0.2170 | 0.0560 | 0.3430 | 0.0918 |
| Primary Tumor | 0.8667 | 0.0473 | 0.9994 | 0.0003 | 0.7452 | 0.0321 | 0.9992 | 0.0068 | 0.9458 | 0.0213 | 0.6710 | 0.1143 | 0.7680 | 0.0683 | 0.7619 | 0.0889 |
| Lymphography | 0.1380 | 0.1188 | 0.6093 | 0.0081 | 0.12410 | 0.3028 | 0.6621 | 0.0765 | 0.4286 | 0.0000 | 0.0512 | 0.0486 | 0.0191 | 0.0628 | 0.1142 | 0.1176 |
| Immunotherapy | 0.9129 | 0.0334 | 0.9999 | 0.0024 | 0.9129 | 0.0334 | 0.9992 | 0.0077 | 0.9261 | 0.0150 | 0.7287 | 0.1010 | 0.7640 | 0.0703 | 0.8808 | 0.0683 |
| Friedman Rank | 4.00 | 5.60 | 7.30 | 1.60 | 3.50 | 5.30 | 7.29 | 3.80 | 6.10 | 2.10 | 1.70 | 6.20 | 1.80 | 6.10 | 4.30 | 5.30 |
| Rank | 5 | 1 | 6 | 2 | 3 | 8 | 7 | 4 | ||||||||
| Datasets | BWO | OBWOD | INFO | HHO | HGS | SCA | WOA | MFO | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arrhythmia | 0.4043 | 0.1798 | 0.6347 | 0.0090 | 0.5243 | 0.2172 | 0.5442 | 0.0126 | 0.5039 | 0.0100 | 0.4632 | 0.0479 | 0.1922 | 0.2312 | 0.4806 | 0.0095 |
| Leukemia2 | 0.6214 | 0.0173 | 0.7765 | 0.0020 | 0.6214 | 0.0173 | 0.8605 | 0.0770 | 0.6796 | 0.0435 | 0.6188 | 0.0622 | 0.3786 | 0.1708 | 0.6250 | 0.0034 |
| Prostate Tumors | 0.9830 | 0.0766 | 1.0000 | 0.0000 | 0.9740 | 0.0526 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 0.9900 | 0.0995 | 0.6746 | 0.2313 | 1.0000 | 0.0000 |
| Statlog (Heart) | 0.5508 | 0.0943 | 0.8322 | 0.0015 | 0.6608 | 0.0783 | 0.8514 | 0.0366 | 0.7195 | 0.0029 | 0.4619 | 0.0628 | 0.4965 | 0.0554 | 0.5808 | 0.0718 |
| CKD | 0.4851 | 0.2389 | 0.6706 | 0.0080 | 0.4851 | 0.2389 | 0.6659 | 0.0085 | 0.622 | 0.0032 | 0.5763 | 0.0598 | 0.2284 | 0.2786 | 0.6075 | 0.0078 |
| Parkinson’s | 0.8279 | 0.0757 | 0.9994 | 0.0004 | 0.8529 | 0.0432 | 0.9992 | 0.0052 | 0.9637 | 0.0007 | 0.7934 | 0.0961 | 0.7930 | 0.0375 | 0.8833 | 0.0373 |
| Pima | 0.3637 | 0.1146 | 0.7593 | 0.0004 | 0.3637 | 0.1146 | 0.7561 | 0.0161 | 0.7273 | 0.0008 | 0.2768 | 0.0685 | 0.3172 | 0.0751 | 0.4515 | 0.1420 |
| Primary Tumor | 0.5431 | 0.1414 | 0.9995 | 0.0091 | 0.6241 | 0.2030 | 0.9971 | 0.0216 | 0.8535 | 0.0525 | 0.0585 | 0.1587 | 0.1385 | 0.2114 | 0.3940 | 0.2076 |
| Lymphography | 0.2397 | 0.0017 | 0.3738 | 0.0014 | 0.2397 | 0.0017 | 0.3703 | 0.0233 | 0.2595 | 0.0131 | 0.1940 | 0.0320 | 0.1783 | 0.0449 | 0.2166 | 0.0181 |
| Immunotherapy | 0.7239 | 0.0354 | 0.9198 | 0.0004 | 0.7239 | 0.0354 | 0.9191 | 0.0132 | 0.8667 | 0.0300 | 0.6908 | 0.0703 | 0.7041 | 0.0220 | 0.7290 | 0.0555 |
| Friedman Rank | 3.25 | 5.55 | 7.65 | 1.35 | 3.95 | 5.55 | 7.15 | 3.65 | 5.95 | 2.85 | 2.10 | 6.00 | 1.40 | 6.50 | 4.55 | 4.55 |
| Rank | 6 | 1 | 5 | 2 | 3 | 7 | 8 | 4 | ||||||||
| Datasets | BWO | OBWOD | INFO | HHO | HGS | SCA | WOA | MFO |
|---|---|---|---|---|---|---|---|---|
| Arrhythmia | 10.6230 | 17.3304 | 11.0516 | 12.3240 | 10.1320 | 9.3268 | 10.6587 | 10.6861 |
| Leukemia2 | 22.5840 | 51.7540 | 45.5153 | 32.6532 | 30.4671 | 46.0079 | 29.2826 | 51.6296 |
| Prostate Tumors | 42.1054 | 43.1311 | 62.3347 | 32.3657 | 42.6201 | 58.2549 | 44.8264 | 66.4606 |
| Statlog (Heart) | 3.4428 | 8.4855 | 7.6711 | 6.2339 | 7.0938 | 7.6333 | 7.6686 | 7.6215 |
| CKD | 7.9837 | 17.7139 | 11.1429 | 9.2948 | 8.8317 | 9.3986 | 8.8126 | 10.9263 |
| Parkinson’s | 7.3120 | 8.2712 | 7.8990 | 7.2244 | 8.0921 | 7.5191 | 7.5082 | 7.3047 |
| Pima | 7.1146 | 9.3084 | 8.7200 | 9.2549 | 8.1086 | 8.1127 | 8.1264 | 8.3065 |
| Primary Tumor | 6.0160 | 8.2361 | 7.3495 | 0.2445 | 7.0904 | 7.4478 | 7.3979 | 7.1740 |
| Lymphography | 6.3034 | 8.4789 | 7.5684 | 7.2203 | 0.0926 | 7.5350 | 7.5396 | 7.3110 |
| Immunotherapy | 6.0871 | 8.1276 | 7.5058 | 7.2314 | 7.0917 | 7.2710 | 7.2860 | 7.2984 |
| Friedman Rank | 1.70 | 7.60 | 6.30 | 3.30 | 2.90 | 4.70 | 4.40 | 5.10 |
| Rank | 1 | 8 | 7 | 3 | 2 | 5 | 4 | 6 |
| Ref. | Datasets | Used Techniques | Comparative Algorithm Accuracy | OBWOD Accuracy |
|---|---|---|---|---|
| [63] | Heart disease | SVM with PSO called (PSO-SVM) | 88.13% | 88.13% |
| [2] | Leukemia2 | Centroid-mutation-based search-and-rescue optimization algorithm | 95.60% | 98.83% |
| [64] | Heart disease | Extreme Learning Machine (ELM) algorithm | 80% | 88.13% |
| [65] | Parkinson’s disease | Minimum average maximum tree and SVD with kNN classifier | 92.46% | 99.83% |
| [66] | Parkinson’s disease | Binary Rao1 with a kNN classifier | 96.47% | 99.83% |
| [2] | Primary Tumor | Centroid-mutation-based search-and-rescue optimization algorithm | 44.46% | 88.76% |
| [67] | Parkinson’s disease | SMOTE + random forests | 98.32% | 99.83% |
| [68] | Leukemia2 | Slime Mold Algorithm (SMA) integrated with OBL based on kNN classifier | 90.59% | 98.83% |
| [2] | Statlog (Heart) | Centroid-mutation-based search-and-rescue optimization algorithm | 86.67% | 88.13% |
| [69] | Primary Tumor | Tunicate Swarm Algorithm (TSA) integrated with OBL based on the kNN classifier | 82.87% | 88.76% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Houssein, E.H.; Sayed, A. Dynamic Candidate Solution Boosted Beluga Whale Optimization Algorithm for Biomedical Classification. Mathematics 2023, 11, 707. https://doi.org/10.3390/math11030707
Houssein EH, Sayed A. Dynamic Candidate Solution Boosted Beluga Whale Optimization Algorithm for Biomedical Classification. Mathematics. 2023; 11(3):707. https://doi.org/10.3390/math11030707
Chicago/Turabian StyleHoussein, Essam H., and Awny Sayed. 2023. "Dynamic Candidate Solution Boosted Beluga Whale Optimization Algorithm for Biomedical Classification" Mathematics 11, no. 3: 707. https://doi.org/10.3390/math11030707
APA StyleHoussein, E. H., & Sayed, A. (2023). Dynamic Candidate Solution Boosted Beluga Whale Optimization Algorithm for Biomedical Classification. Mathematics, 11(3), 707. https://doi.org/10.3390/math11030707







