Hyperbolic Representation of the Richards Growth Model
Abstract
1. Introduction
2. Materials and Methods
- (i)
- The normalized standard deviation (NSD):in which and denote the numbers of data and parameters fitted, is the SD of i-th experimental mass , whereas is the theoretically calculated mass by the model function. If reproduction of the data is performed within , then , and the theory reproduces the data with experimental accuracy.
- (ii)
- The coefficient of determination :where the best fits are represented by .
- (iii)
- The standard deviation (SD):applied together with when the experimental SDs of the data are unavailable.
- (iv)
- Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) defined in [16,17], respectively: They have been used to select the best model with the optimal number of parameters fitted and to avoid overfitting. The smaller values of AIC and BIC indicate the model more preferred and statistically reliable. The explicit form of the AIC and BIC formulae used in the calculations is presented in [14].
3. Results
3.1. Sigmoid Richards Growth
3.2. Involuted Richards Growth
3.3. Generalized Universal Growth Curve
3.4. PU-SIR Epidemic Model
4. Discussion
5. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AIC | Akaike Information Criterion |
| BIC | Bayesian Information Criterion |
| CDG | Castorina Delsanto Guiot |
| NSD | Normalized Standard Deviation |
| PU | Phenomenological Universalities |
| PU-SIR | Phenomenological Universalities Susceptible–Infective–Removed |
| SD | Standard Deviation |
| SDE | Stochastic Differential Equation |
| SIR | Susceptible–Infective–Removed |
References
- Gilbert, S.F. Developmental Biology; Sinauer Associates: Sunderland, MA, USA; Swarthmore College: Swarthmore, PA, USA, 2000; ISBN 10:0-87893-243-7. [Google Scholar]
- Bertalanffy, V.L. Problems of organic growth. Nature 1949, 163, 156–158. [Google Scholar] [CrossRef] [PubMed]
- Bertalanffy, V.L. Quantitative laws in metabolism and growth. Q. Rev. Biol. 1957, 32, 217–231. [Google Scholar] [CrossRef]
- Richards, F.J. A flexible growth function for empirical use. J. Exp. Bot. 1959, 10, 290–300. [Google Scholar] [CrossRef]
- West, G.B.; Brown, J.H.; Enquist, B.J. A general model for ontogenetic growth. Nature 2001, 413, 628–631. [Google Scholar] [CrossRef]
- Castorina, P.; Delsanto, P.P.; Guiot, C. Classification Scheme for Phenomenological Universalities in Growth Problems in Physics and Other Sciences. Phys. Rev. Lett. 2006, 96, 188701. [Google Scholar] [CrossRef]
- Gliozzi, A.S.; Guiot, C.; Delsanto, P.P. A new computational tool for the phenomenological analysis of multipassage tumor growth curves. PLoS ONE 2009, 4, e5358. [Google Scholar] [CrossRef]
- Barberis, L.; Condat, C.A.; Gliozzi, A.S.; Delsanto, P.P. Concurrent growth of phenotypic features: A phenomenological universalities approach. J. Theor. Biol. 2010, 264, 123–129. [Google Scholar] [CrossRef]
- Barberis, L.; Condat, C.A. Describing interactive growth using vector universalities. Ecol. Model. 2012, 227, 56–63. [Google Scholar] [CrossRef]
- Molski, M. Extended class of phenomenological universalities. Phys. Lett. A 2017, 381, 2629–2635. [Google Scholar] [CrossRef]
- Molski, M. Quantum and classical phenomenological universalities. Phys. Lett. A 2018, 382, 79–84. [Google Scholar] [CrossRef]
- Golmankhaneh, A.K.; Tunç, S.; Schlichtinger, A.M.; Asanza, D.M.; Golmankhaneh, A.K. Modeling tumor growth using fractal calculus: Insights into tumor dynamics. Biosystems 2024, 235, 105071. [Google Scholar] [CrossRef] [PubMed]
- Tjørve, E.; Tjørve, K.M.C. A unified approach to the Richards model family for use in growth analysis: Why we need only two model forms. J. Theor. Biol. 2010, 267, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Molski, M. Theoretical modeling of pre and postnatal growth. J. Theor. Biol. 2018, 458, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Laird, A.K. Dynamics of tumor growth. Br. J. Cancer 1964, 18, 490–502. [Google Scholar] [CrossRef] [PubMed Central]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Myhrvold, N.P. Revisiting the estimation of dinosaur growth rates. PLoS ONE 2013, 8, e81917. [Google Scholar] [CrossRef]
- Rhouma, K.B.; Sakly, M. Involution of rat thymus: Characterization of cytoplasmic glucocorticoid receptors, evidence of glucocorticoid resistant dexamethasone receptor-positive cells. Arch. Int. Physiol. Biochim. Biophys. 1994, 102, 97–102. [Google Scholar] [CrossRef]
- He, M.; Liang, X.; Wang, K.; Pu, H.; Hu, Y.; Ye, G.; Li, X.; Liu, L. Age-related development and histomorphological observation of bursa of Fabricius in yellow quails. Can. J. Anim. Sci. 2015, 95, 487–491. [Google Scholar] [CrossRef]
- Dampin, N.; Tarnchalanukit, W.; Chunkao, K.; Maleewong, M. Fish growth model for Nile Tilapia (Oreochromis niloticus). Procedia Environ. Sci. 2012, 13, 513–524. [Google Scholar] [CrossRef]
- Aggrey, S.E. Comparison of three nonlinear and spline regression models for describing chicken growth curves. Poult. Sci. 2002, 81, 1782–1788. [Google Scholar] [CrossRef] [PubMed]
- Smuts, G.L. Pre- and post-natal growth phenoena of Burchell’s zebra Equus burchelli antiquorum. Koedoe 1975, 18, 69–102. [Google Scholar] [CrossRef]
- Kempes, C.P.; Dutkiewicz, S.; Follows, M.J. Growth, metabolic partitioning, and the size of microorganisms. Proc. Natl. Acad. Sci. USA 2012, 109, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, K.; Benedict, S.R. The influence of certain diets upon tumor susceptibility and growth in albino rats. J. Cancer Res. 1920, 5, 373–401. [Google Scholar]
- Lee, S.A. Incubation times of dinosaur eggs via embryonic metabolism. Phys. Rev. E 2016, 94, 022402. [Google Scholar] [CrossRef]
- Hethcote, H.W. The Mathematics of Infectious Diseases. SIAM Rev. 2000, 42, 599–653. [Google Scholar] [CrossRef]
- Kröger, M.; Schlickeiser, R. Analytical solution of the SIR-model for the temporal evolution of epidemics. Part A: Time-independent reproduction factor. J. Phys. Math. Theor. 2020, 53, 505601. [Google Scholar] [CrossRef]
- Schlickeiser, R.; Kröger, M. Analytical solution of the SIR-model for the temporal evolution of epidemics. Part B: Semi-time case. J. Phys. Math. Theor. 2021, 54, 175601. [Google Scholar] [CrossRef]
- Sivaraman, N.K.; Gaur, M.; Baijal, S.; Muthiah, S.B.; Sheth, A. Exo-SIR: An epidemiological model to analyze the impact of exogenous spread of infection. Int. J. Data Sci. Anal. 2025, 19, 303–318. [Google Scholar] [CrossRef]
- Wanga, X.-S.; Wu, J.; Yang, Y. Richards model revisited: Validation by and application to infection dynamics. J. Theor. Biol. 2012, 313, 12–19. [Google Scholar] [CrossRef]
- van den Driessche, P. Reproduction numbers of infectious disease models. Infect. Dis. Model. 2017, 2, 288–303. [Google Scholar] [CrossRef]
- Biazae, J. Solution of the epidemic model by Adomian decomposition method. Appl. Math. Comput. 2006, 173, 1101–1106. [Google Scholar] [CrossRef]
- Rafei, H.; Daniali, H.; Ganji, D.D. Variational iteration method for solving the epidemic model and the prey and predator problem. Appl. Math. Comput. 2007, 186, 1701–1709. [Google Scholar] [CrossRef]
- Rafei, H.; Ganji, D.D.; Daniali, H. Solution of the epidemic model by homotopy perturbation method. Appl. Math. Comput. 2007, 187, 1056–1062. [Google Scholar] [CrossRef]
- Engida, H.A.; Theuri, D.M.; Gathungu, D.; Gachohi, J.; Alemneh, H.T. A mathematical model analysis for the transmission dynamics of leptospirosis disease in human and rodent populations. Comput. Math. Methods Med. 2022, 1, 1806585. [Google Scholar] [CrossRef] [PubMed]
- Kapur, P.K.; Anand, S.; Yamada, S.; Yadavalli, V.S.S. Stochastic Differential Equation-Based Flexible Software Reliability Growth Model. Math. Probl. Eng. 2009, 2009, 581383. [Google Scholar] [CrossRef]
- Samoletov, A.; Vasiev, B. A mathematical framework for the statistical interpretation of biological growth models. BioSystems 2024, 246, 105342. [Google Scholar] [CrossRef]
- Baltazar-Larios, F.; Delgado-Vences, F.; Ornelas, A. Parameter estimation and model selection for stochastic differential equations for biological growth. Environ. Ecol. Stat. 2025, 32, 195–227. [Google Scholar] [CrossRef]
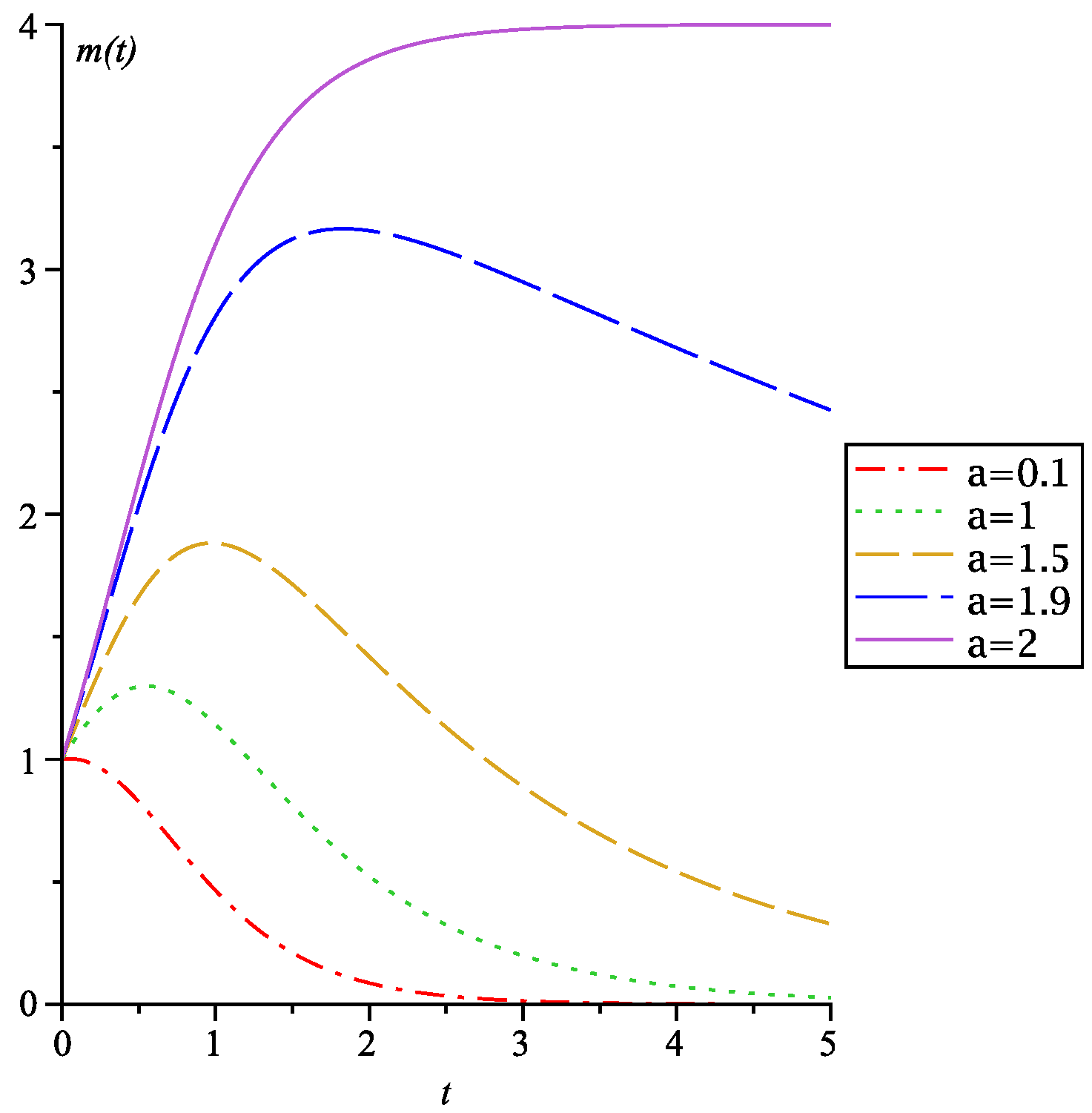
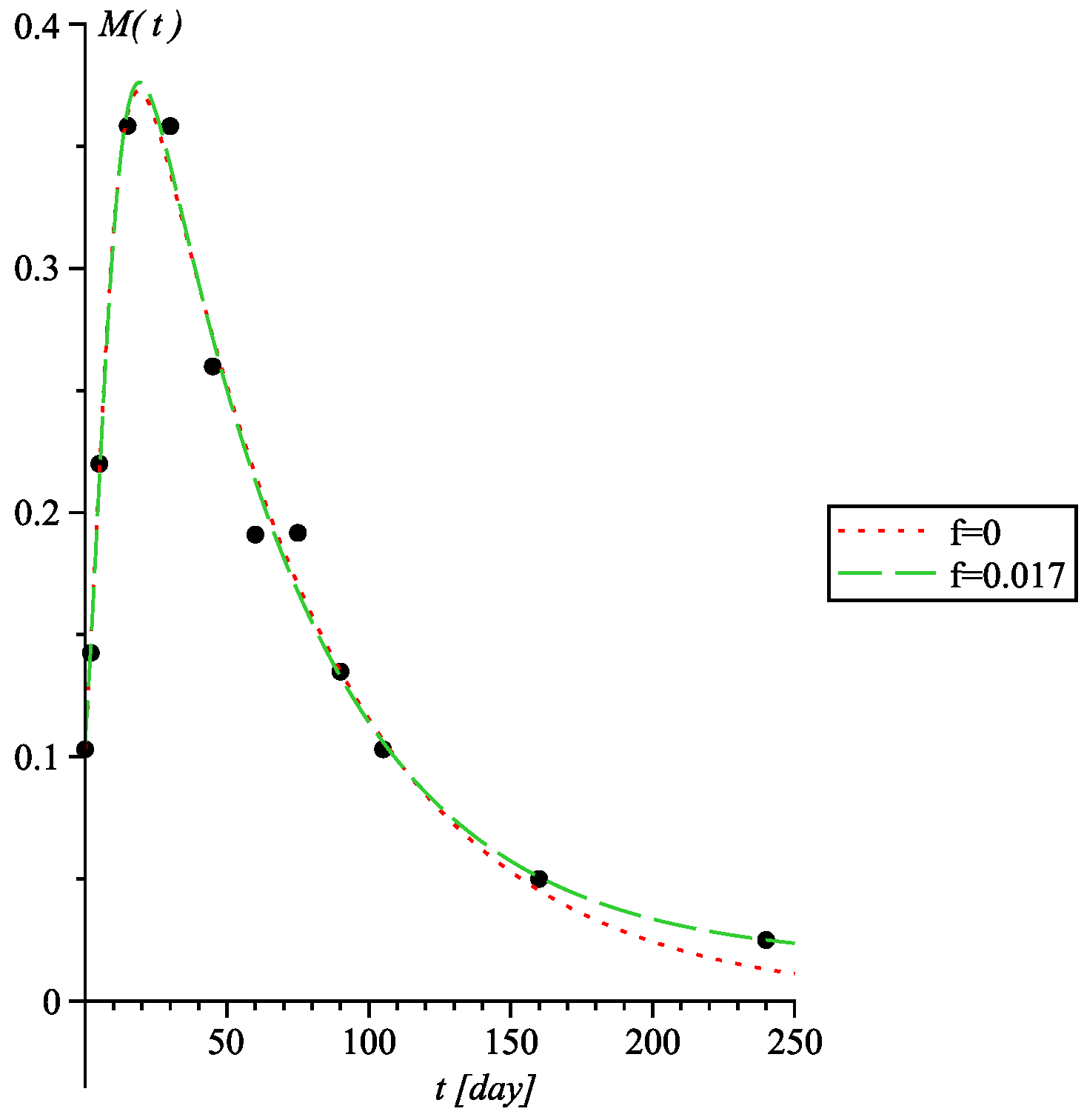

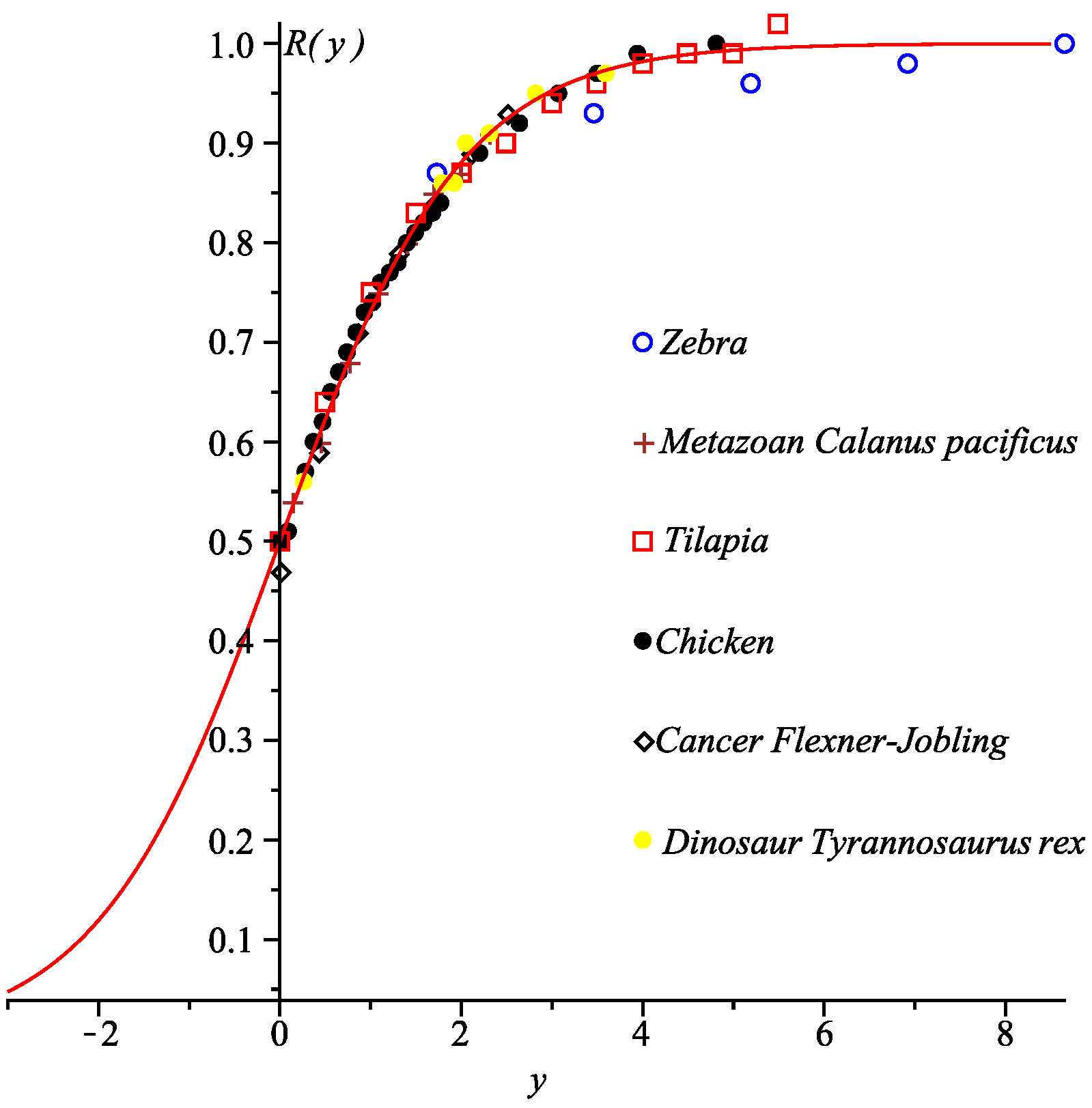

| Parameter | C. pacificus | FJ Cancer | T. Rex |
|---|---|---|---|
| m0 | 4.83(80) [g × 10−6] | 0.0199(116) [g] | 2.04(87) [Kg] |
| c [d−1] | 0.0873(65) | 0.0303(35) | 0.0747(48) [y−1] |
| b | 6.20(18) | 10.99(64) | 11.77(61) |
| SD | 4.568 [g × 10−6] | 0.438 [g] | 283 [Kg] |
| R2 | 0.996 | 0.995 | 0.979 |
| ND | 8 | 7 | 8 |
| Chicken | Zebra | Tilapia | |
| m0 [g] | 35.5(1.1) | 33,864(1858) | 5.83(44) |
| c [d−1] | 0.0155(4) | 0.0024(2) | 0.0089(5) |
| b | 5.953(61) | 3.22(81) | 5.93(12) |
| NSD | 0.600 | 0.669 | 0.611 |
| R2 | 0.993 | 0.950 | 0.951 |
| ND | 28 | 17 | 13 |
| Thymus | Bursa of Fabricius | |||
|---|---|---|---|---|
| Parameter | Equation (35) | Equation (37) | Equation (35) | Equation (37) |
| m0 | 0.103(11) | 0.106(11) | 4.60(3.90) | 9.01(2.01) |
| a | 2.24(16) | 2.44(28) | 4.86(1.25) | 7.16(1.89) |
| b | 2.42(16) | 2.65(30) | 5.12(1.22) | 7.69(1.86) |
| c [d−1] | 0.086(12) | 0.083(11) | 0.0613(193) | 0.0501(99) |
| d = b − a | 0.182(35) | 0.210(50) | 0.26(10) | 0.53(17) |
| f | 0 | 0.017(16) | 0 | 7.58(1.54) |
| NSD | 1.0684 | 1.0707 | 2.1895 | 1.3924 |
| R2 | 0.9860 | 0.9877 | 0.9453 | 0.9815 |
| AIC | −94 | −91 | 58 | 50 |
| BIC | −96 | −95 | 56 | 46 |
| f = 0 | f = 0.017(16) | ||||
|---|---|---|---|---|---|
| Time [d] | |||||
| 0 | 0.1031 | 0.0264 | 0.1027 | −0.1746 | 0.1055 |
| 2 | 0.1426 | −0.2036 | 0.1455 | −0.1913 | 0.1453 |
| 5 | 0.2200 | 0.2981 | 0.2158 | 0.5025 | 0.2130 |
| 15 | 0.3584 | −0.3778 | 0.3637 | −0.4991 | 0.3654 |
| 30 | 0.3583 | 1.3569 | 0.3393 | 1.1510 | 0.3422 |
| 45 | 0.2599 | −0.8691 | 0.2721 | −0.8170 | 0.2713 |
| 60 | 0.1910 | −1.7526 | 0.2155 | −1.5633 | 0.2129 |
| 75 | 0.1917 | 1.5076 | 0.1706 | 1.7176 | 0.1677 |
| 90 | 0.1349 | −0.0080 | 0.1350 | 0.1476 | 0.1328 |
| 105 | 0.1031 | −0.2679 | 0.1069 | −0.2100 | 0.1060 |
| 160 | 0.0500 | 0.3344 | 0.0453 | −0.0587 | 0.0508 |
| 240 | 0.0250 | 0.8561 | 0.0130 | −0.0047 | 0.0251 |
| f = 0 | f = 7.58(1.54) | ||||
|---|---|---|---|---|---|
| Time [d] | |||||
| 0 | 5.28(2.15) | 0.3143 | 4.6041 | −1.7372 | 9.0148 |
| 7 | 25.97(4.92) | 0.3915 | 24.0437 | 1.5245 | 18.4694 |
| 21 | 70.77(7.71) | −1.1872 | 79.9236 | −0.6558 | 75.8266 |
| 35 | 100.23(6.47) | 2.2507 | 85.6682 | 0.3164 | 98.1829 |
| 63 | 63.21(3.90) | 1.2845 | 58.2003 | 0.6778 | 60.5664 |
| 91 | 24.43(3.88) | −3.2998 | 37.2332 | −2.1928 | 32.9379 |
| 119 | 19.43(1.96) | −2.2175 | 23.7763 | −0.0738 | 19.5746 |
| 147 | 15.23(1.75) | 0.0273 | 15.1822 | 1.1271 | 13.2599 |
| 175 | 12.83(2.54) | 1.2345 | 9.6944 | 1.0092 | 10.2576 |
| 203 | 9.98(3.52) | 1.0766 | 6.1903 | 0.3159 | 8.8678 |
| 224 | 8.15(1.21) | 3.0811 | 4.4218 | −0.1455 | 8.3261 |
| 252 | 7.45(3.28) | 1.4105 | 2.8235 | −0.1517 | 7.9477 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molski, M. Hyperbolic Representation of the Richards Growth Model. Mathematics 2025, 13, 1316. https://doi.org/10.3390/math13081316
Molski M. Hyperbolic Representation of the Richards Growth Model. Mathematics. 2025; 13(8):1316. https://doi.org/10.3390/math13081316
Chicago/Turabian StyleMolski, Marcin. 2025. "Hyperbolic Representation of the Richards Growth Model" Mathematics 13, no. 8: 1316. https://doi.org/10.3390/math13081316
APA StyleMolski, M. (2025). Hyperbolic Representation of the Richards Growth Model. Mathematics, 13(8), 1316. https://doi.org/10.3390/math13081316






