Machine Learning Modeling of Aerobic Biodegradation for Azo Dyes and Hexavalent Chromium
Abstract
1. Introduction
2. Methodology
2.1. Culture Medium, Chemicals, and Microorganism
2.2. Evaluation of Klebsiella sp. KOD36 Potential for Biodegradation of Azo Dyes and Chromium (VI) in Single and Combined System
2.3. Estimation of Optimal Growth Fcators for Simultaneous Bioremoval of Azo Dyes and Azo Dyes and Chromium (VI) in Single and Combined System
2.4. Methods of Analysis
Azo Dye and Chromium Concentration Measurement
2.5. Machine Learning Methodolgies
2.5.1. Gene Expression Programming (GEP)
2.5.2. Support Vector Regression
2.5.3. FOA (Fruit Fly Optimization Algorithm)
2.6. Modeling Methodologies
Parameters for Evaluation of Models’ Performance
3. Results and Discussion
3.1. Evaluation of Klebsiella sp. KOD36 for Biodegradation of Azo Dyes and Chromium (VI)
3.1.1. In Single System
3.1.2. In Combined System
3.2. Modeling Outcomes
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Nomenclature
| SBAHC | Simultaneous aerobic biodegradation of azo dyes and hexavalent chromium |
| GEP | Gene expression programming |
| RF | Random forest |
| SVR | Supportive vector regression |
| FOA | Fruit fly optimization algorithm |
| SRM | Structural risk minimization |
| ERM | Empirical risk minimization |
| CC | Correlation coefficient |
| IP | Incubation period |
| DO | Dissolved oxygen |
| DT | Decision tree |
| ML | Machine learning |
| SI | Scattered index |
| WI | Willmott agreement index |
References
- Sandrin, T.R.; Maier, R.M. Impact of metals on the biodegradation of organic pollutants. Environ. Health Perspect. 2003, 111, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, S.K.; Singh, M.P.; Srivastava, A.K.; Pandey, V.K. Azo dye (direct blue 14) decolorization by immobilized extracellular enzymes of Pleurotus species. Cell. Mol. Boil. 2012, 58, 21–25. [Google Scholar]
- Simmons, J.E.; Yang, R.S.H.; Berman, E. Evaluation of the nephrotoxicity of complex mixtures containing organics and metals: Advantages and disadvantages of the use of real-world complex mixtures. Environ. Health Perspect. 1995, 103 (Suppl. 1), 67–71. [Google Scholar] [PubMed]
- Saratale, R.G.; Saratale, G.D.; Chang, J.S.; Govindwar, S. Bacterial decolorization and degradation of azo dyes: A review. J. Taiwan Inst. Chem. Eng. 2011, 42, 138–157. [Google Scholar] [CrossRef]
- Ali, N.; Hameed, A.; Siddiqui, M.; Ghumro, P.; Ahmed, S. Application of Aspergillus niger SA1 for the enhanced bioremoval of azo dyes in simulated textile effluent. Afr. J. Biotechnol. 2009, 8, 3839–3845. [Google Scholar]
- Moawad, W.M.; El-Rahim, W.M.A.; Khalafallah, M. Evaluation of biotoxicity of textile dyes using two bioassays. J. Basic Microbiol. 2003, 43, 218–229. [Google Scholar] [CrossRef]
- Baath, E. Thymidine incorporation into macromolecules of bacteria extracted from soil by homogenization centrifugation. Soil Boil. Biochem. 1992, 24, 1157–1165. [Google Scholar] [CrossRef]
- Bader, J.L.; González, G.; Goodell, P.C.; Pillai, S.D.; Ali, A.S.; Pillai, S.D. Chromium-resistant bacterial populations from a site heavily contaminated with hexavalent chromium. Water Air Soil Pollut. 1999, 109, 263–276. [Google Scholar] [CrossRef]
- Huq, S.I. Critical environmental issues relating to tanning industries in Bangladesh. In Proceedings of the ACIAR, Coimbatore, India, 31 January–4 February 1998; pp. 22–28. [Google Scholar]
- Mahmood, S.; Khalid, A.; Mahmood, T.; Arshad, M.; Ahmad, R. Potential of newly isolated bacterial strains for simultaneous removal of hexavalent chromium and reactive black-5 azo dye from tannery effluent. J. Chem. Technol. Biotechnol. 2012, 88, 1506–1513. [Google Scholar] [CrossRef]
- Ho, W.S.W.; Poddar, T.K. New membrane technology for removal and recovery of chromium from waste waters. Environ. Prog. 2001, 20, 44–52. [Google Scholar] [CrossRef]
- Onwosi, O.; Odibo, F.J.C. Use of response surface design in the optimization of starter cultures for enhanced rhamnolipid production by Pseudomonas nitroreducens. Afr. J. Biotechnol. 2013, 12, 2611–2617. [Google Scholar]
- Abbasi, H.; Hamedi, M.M.; Lotfabad, T.B.; Zahiri, H.S.; Sharafi, H.; Masoomi, F.; Moosavi-Movahedi, A.A.; Ortiz, A.; Amanlou, M.; Noghabi, K.A. Biosurfactant-producing bacterium, Pseudomonas aeruginosa MA01 isolated from spoiled apples: Physicochemical and structural characteristics of isolated biosurfactant. J. Biosci. Bioeng. 2012, 113, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Lotfy, W.A.; Ghanem, K.M.; El-Helow, E.R. Citric acid production by a novel Aspergillus niger Klebseilla pneumoniae. II. Optimization of process parameters through statistical experimental designs. Bioresour. Technol. 2007, 98, 3470–3477. [Google Scholar] [CrossRef] [PubMed]
- Tanyildizi, S.; Ozer, D.; Elibol, M. Optimization of a-amylase production by Bacillus sp. using response surface methodology. Process Biochem. 2005, 40, 2291–2296. [Google Scholar] [CrossRef]
- Locner, H.; Matar, J.E. Designing for Quality; Productivity Press: New York, NY, USA, 1990. [Google Scholar]
- Yang, W.; Tarng, Y. Design optimization of cutting parameters for turning operations based on the Taguchi method. J. Mater. Process. Technol. 1998, 84, 122–129. [Google Scholar] [CrossRef]
- Kalyani, L.T.; Sireesha, G.N.; Aditya, A.K.G.; Sankar, G.G.; Prabhakar, T. Production optimization of rhamnolipid biosurfactant by Streptomyces coelicoflavus (NBRC 15399T) using Plackett–Burman design. Eur. J. Biotechnol. Biosci. 2014, 1, 7–13. [Google Scholar]
- Zhao, F.; Mandlaa, M.; Hao, J.; Liang, X.; Shi, R.; Han, S.; Zhang, Y. Optimization of culture medium for anaerobic production of rhamnolipid by recombinant Pseudomonas stutzeri Rhl for microbial enhanced oil recovery. Lett. Appl. Microbiol. 2014, 59, 231–237. [Google Scholar] [CrossRef]
- Mabrouk, M.E.; Youssif, E.M.; Sabry, S.A. Biosurfactant production by a newly Klebseilla pneumoniaed soft coral-associated marine Bacillus sp. E34: Statistical optimization and characterization. Life Sci. J. 2014, 11, 756–768. [Google Scholar]
- Amodu, O.S.; Ntwampe, S.K.; Ojumu, T.V. Optimization of biosurfactant production by Bacillus licheniformis STK 01 grown exclusively on Beta vulgaris waste using response surface methodology. BioResources 2014, 9, 5045–5065. [Google Scholar] [CrossRef]
- Chandankere, R.; Yao, J.; Masakorala, K.; Jain, A.K.; Kumar, R. Enhanced production and characterization of biosurfactant produced by a newly isolated Bacillus amyloliquefaciens USTBb using response surface methodology. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 66–80. [Google Scholar]
- Sen, R.; Swaminathan, T. Application of response-surface methodology to evaluate the optimum environmental conditions for the enhanced production of surfactin. Appl. Microbiol. Biotechnol. 1997, 47, 358–363. [Google Scholar] [CrossRef]
- Mnif, I.; Sahnoun, R.; Ellouze-Chaabouni, S.; Ghribi, D. Evaluation of B. subtilis SPB1 biosurfactants’ potency for diesel-contaminated soil washing: Optimization of oil desorption using Taguchi design. Environ. Sci. Pollut. Res. 2014, 21, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Mutalik, S.R.; Vaidya, B.K.; Joshi, R.M.; Desai, K.M.; Nene, S.N. Use of response surface optimization for the production of biosurfactant from Rhodococcus spp. MTCC 2574. Bioresour. Technol. 2008, 99, 7875–7880. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.C. Optimizing the concentrations of carbon, nitrogen and phosphorous in a citric acid fermentation with response surface method. Food Biotechnol. 1996, 10, 13–27. [Google Scholar] [CrossRef]
- Zounemat-Kermani, M.; Rajaee, T.; Ramezani-Charmahineh, A.; Adamowski, J.F. Estimating the aeration coefficient and air demand in bottom outlet conduits of dams using GEP and decision tree methods. Flow Meas. Instrum. 2017, 54, 9–19. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cheng, K.; Lu, Z. Adaptive sparse polynomial chaos expansions for global sensitivity analysis based on support vector regression. Comput. Struct. 2018, 194, 86–96. [Google Scholar] [CrossRef]
- Pan, W.T. A new fruit fly optimization algorithm: Taking the financial distress model as an example. Knowl.-Based Syst. 2012, 26, 69–74. [Google Scholar] [CrossRef]
- Abidin, Z.; Hamzah, M.; Arshad, M.; Ngah, U. A calibration framework for swarming ASVs’ system design. Indian J. Mar. Sci. 2012, 41, 581–588. [Google Scholar]
- Liu, Y.; Wang, X.; Li, Y. A modified fruit-fly optimization algorithm aided PID controller designing. In Proceedings of the Intelligent Control and Automation (WCICA), 10th World Congress on IEEE, Beijing, China, 6–8 July 2012; pp. 233–238. [Google Scholar]
- Ahmad, Z.; Arshad, M.; Asghar, H.N.; Sheikh, M.A.; Crowley, D.E. Isolation, screening and functional characterization of biosurfactant producing bacteria isolated from crude oil contaminated site. Int. J. Agric. Boil. 2016, 18, 542–548. [Google Scholar]
- Desai, C.; Jain, K.; Patel, B.; Madamwar, D. Efficacy of bacterial consortium-AIE2 for contemporaneous Cr (VI) and azo dye bioremediation in batch and continuous bioreactor systems, monitoring steady-state bacterial dynamics using qPCR assays. Biodegradation 2009, 20, 813. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C. Gene Expression Programming in Problem Solving, in Soft Computing and Industry; Springer: New York, NY, USA, 2002; pp. 635–653. [Google Scholar]
- Baghban, A.; Mosavi, A. Insight into the antiviral activity of synthesized schizonepetin derivatives: A theoretical investigation. Sci. Rep. 2020, 25, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Suykens, J.; Vandewalle, J. Least squares support vector machine classifiers. Neural Process. Lett. 1999, 9, 293–300. [Google Scholar] [CrossRef]
- Karballaeezadeh, N.; Mohammadzadeh, S.D.; Shamshirband, S.; Hajikhodaverdikhan, P.; Mosavi, A.; Chau, K.W. Prediction of remaining service life of pavement using an optimized support vector machine (case study of Semnan–Firuzkuh road). Eng. Appl. Comput. Fluid Mech. 2019, 13, 188–198. [Google Scholar] [CrossRef]
- Ebtehaj, I.; Bonakdari, H.; Zaji, A.H.; Azimi, H.; Sharifi, A. Gene expression programming to predict the discharge coefficient in rectangular side weirs. Appl. Soft Comput. 2015, 35, 618–628. [Google Scholar] [CrossRef]
- Lopes, H.S.; Weinert, W.R. EGIPSYS: An enhanced gene expression programming approach for symbolic regression problems. Int. J. Appl. Math. Comput. Sci. 2004, 14, 375–384. [Google Scholar]
- Ferreira, C. Gene Expression Programming: Mathematical Modeling by an Artificial Intelligence; Springer: New York, NY, USA, 2006; Volume 21. [Google Scholar]
- Yadav, B.; Ch, S.; Mathur, S.; Adamowski, J. Estimation of in-situ bioremediation system cost using a hybrid Extreme Learning Machine (ELM)-particle swarm optimization approach. J. Hydrol. 2016, 543, 373–385. [Google Scholar] [CrossRef]
- Cao, Q.; Leung, K.M. Prediction of chemical biodegradability using support vector classifier optimized with differential evolution. J. Chem. Inf. Model. 2014, 54, 2515–2523. [Google Scholar] [CrossRef]
- Tang, L.; Zhang, M.; Wen, L. Support vector machine classification of seismic events in the Tianshan orogenic belt. J. Geophys. Res. Solid Earth 2020, 125, e2019JB018132. [Google Scholar] [CrossRef]
- Xiao, C.; Hao, K.; Ding, Y. An improved fruit fly optimization algorithm inspired from cell communication mechanism. Math. Probl. Eng. 2015, 2015, 492195. [Google Scholar] [CrossRef]
- Nabipour, N.; Daneshfar, R.; Rezvanjou, O.; Mohammadi-Khanaposhtani, M.; Baghban, A.; Xiong, Q.; Li, L.K.; Habibzadeh, S.; Doranehgard, M.H. Estimating biofuel density via a soft computing approach based on intermolecular interactions. Renew. Energy 2020, 152, 1086–1098. [Google Scholar] [CrossRef]
- Qasem, S.N.; Samadianfard, S.; Kheshtgar, S.; Jarhan, S.; Kisi, O.; Shamshirband, S.; Chau, K.W. Modeling monthly pan evaporation using wavelet support vector regression and wavelet artificial neural networks in arid and humid climates. Eng. Appl. Comput. Fluid 2019, 13, 177–187. [Google Scholar] [CrossRef]
- Samadianfard, S.; Majnooni-Heris, A.; Qasem, S.N.; Kisi, O.; Shamshirband, S.; Chau, K.W. Daily global solar radiation modeling using data-driven techniques and empirical equations in a semi-arid climate. Eng. Appl. Comput. Fluid Mech. 2019, 13, 142–157. [Google Scholar] [CrossRef]
- Liu, W.; Liu, C.; Liu, L.; You, Y.; Jiang, J.; Zhou, Z.; Dong, Z. Simultaneous decolorization of sulfonated azo dyes and reduction of hexavalent chromium under high salt condition by a newly isolated salt-tolerant strain Bacillus circulans BWL1061. Ecotoxicol. Environ. Saf. 2017, 141, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Thacker, U.; Madamwar, D. Reduction of toxic chromium and partial localization of chromium reductase activity in bacterial isolate DM1. World J. Microbiol. Biotechnol. 2005, 21, 891–899. [Google Scholar] [CrossRef]
- Halmi, M.I.E.; Abdullah, S.R.S.; Shukor, M.Y. Characterization of Chromate Reducing Pseudomonas Aeruginosa Strain Mie3 Isolated from Juru River Sludge and its Potential on Azo Dye Decolorization. J. Chem. Pharmac. Sci. 2017, 10, 522–526. [Google Scholar]
- Chang, B.V.; Chao, W.L.; Yeh, S.L.; Kuo, D.L.; Yang, C.W. Biodegradation of Sulfamethoxazole in Milkfish (Chanos chanos) Pond Sediments. Appl. Sci. 2019, 9, 4000. [Google Scholar] [CrossRef]
- Shamshirband, S.; Hadipoor, M.; Baghban, A.; Mosavi, A.; Bukor, J.; Várkonyi-Kóczy, A.R. Developing an ANFIS-PSO model to predict mercury emissions in combustion flue gases. Mathematics 2019, 7, 965. [Google Scholar] [CrossRef]
- Nabipour, N.; Mosavi, A.; Baghban, A.; Shamshirband, S.; Felde, I. Extreme learning machine-based model for Solubility estimation of hydrocarbon gases in electrolyte solutions. Processes 2020, 8, 92. [Google Scholar] [CrossRef]
- Amato, F.; Moscato, V.; Picariello, A.; Sperli’ì, G. Extreme events management using multimedia social networks. Future Gener. Comput. Syst. 2019, 94, 444–452. [Google Scholar] [CrossRef]








| Levels | ||||
|---|---|---|---|---|
| Independent Variables | Unit | −1 | 0 | 1 |
| Temperature | °C | 30 | 35 | 40 |
| pH | Unit less | 3 | 6 | 9 |
| IP | h | 8 | 16 | 24 |
| Carbon source | 20 mg/L | Glucose | Sucrose | Starch |
| Nitrogen source | 2 mg/L | Yeast | Ammonium sulfate | Urea |
| Shaking | rpm | 150 | 200 | 250 |
| Temperature °C | pH | IP (Hours) | Source of Carbon (20 mg/L) | Source of Nitrogen (2 mg/L) | Shaking (rpm) | Simultaneous Reduction of Azo Dye and Chromium (%) |
|---|---|---|---|---|---|---|
| 30 | 3 | 8 | 1 | 1 | 1 | 43 |
| 30 | 6 | 8 | 1 | 2 | 2 | 93 |
| 30 | 9 | 8 | 1 | 3 | 3 | 84 |
| 30 | 3 | 16 | 2 | 1 | 1 | 63 |
| 30 | 6 | 16 | 2 | 2 | 2 | 10 |
| 30 | 9 | 16 | 2 | 3 | 3 | 45 |
| 30 | 3 | 24 | 3 | 1 | 1 | 11 |
| 30 | 6 | 24 | 3 | 2 | 2 | 90 |
| 30 | 9 | 24 | 3 | 3 | 3 | 35 |
| 35 | 3 | 8 | 1 | 1 | 1 | 66 |
| 35 | 6 | 8 | 1 | 2 | 2 | 80 |
| 35 | 9 | 8 | 1 | 3 | 3 | 38 |
| 35 | 3 | 16 | 2 | 1 | 1 | 25 |
| 35 | 6 | 16 | 2 | 2 | 2 | 74 |
| 35 | 9 | 16 | 2 | 3 | 3 | 30 |
| 35 | 3 | 24 | 3 | 1 | 1 | 69 |
| 35 | 6 | 24 | 3 | 2 | 2 | 44 |
| 35 | 9 | 24 | 3 | 3 | 3 | 86 |
| 40 | 3 | 8 | 1 | 1 | 1 | 16 |
| 40 | 6 | 8 | 1 | 2 | 2 | 64 |
| 40 | 9 | 8 | 1 | 3 | 3 | 95 |
| 40 | 3 | 16 | 2 | 1 | 1 | 99 |
| 40 | 6 | 16 | 2 | 2 | 2 | 98 |
| 40 | 9 | 16 | 2 | 3 | 3 | 60 |
| 40 | 3 | 24 | 3 | 1 | 1 | 20 |
| 40 | 6 | 24 | 3 | 2 | 2 | 50 |
| 40 | 9 | 24 | 3 | 3 | 3 | 46 |
| Variable | Mean | Minimum | Maximum | Standard Deviation | Coefficient of Variation | Skewness | Correlation with Simultaneous Degradation of Azo Dyes And chromium (%) |
|---|---|---|---|---|---|---|---|
| Shaking (rpm) | 200.0 | 150.0 | 250.0 | 41.6 | 0.21 | 0.0 | 0.176 |
| pH | 6.0 | 3.0 | 9.0 | 2.50 | 0.42 | 0.0 | 0.176 |
| IP (h) | 16.0 | 8.0 | 24.0 | 6.66 | 0.42 | 0.0 | −0.210 |
| Temperature (C) | 35.0 | 30.0 | 40.0 | 4.16 | 0.12 | 0.0 | 0.121 |
| Simultaneous degradation of azo dyes and chromium (%) | 56.8 | 10.0 | 99.0 | 28.2 | 0.50 | −0.08 | 1 |
| Parameter | SVR | SVR-FOA |
|---|---|---|
| C | 1.0000 | 1.7242 |
| γ | 0.0100 | 0.0517 |
| ε | 0.0010 | 0.0468 |
| Parameter | Value |
|---|---|
| Head size | 8 |
| Linking Function | Addition (+) |
| Number of Genes | 3 |
| Chromosomes | 30 |
| Mutation Rate | 0.044 |
| Inversion Rate | 0.1 |
| One-Point RR | 0.3 |
| Two-Point RR | 0.3 |
| Gene RR | 0.1 |
| Gene Transposition Rate | 0.1 |
| Used functions | +, −, ×, ÷, power |
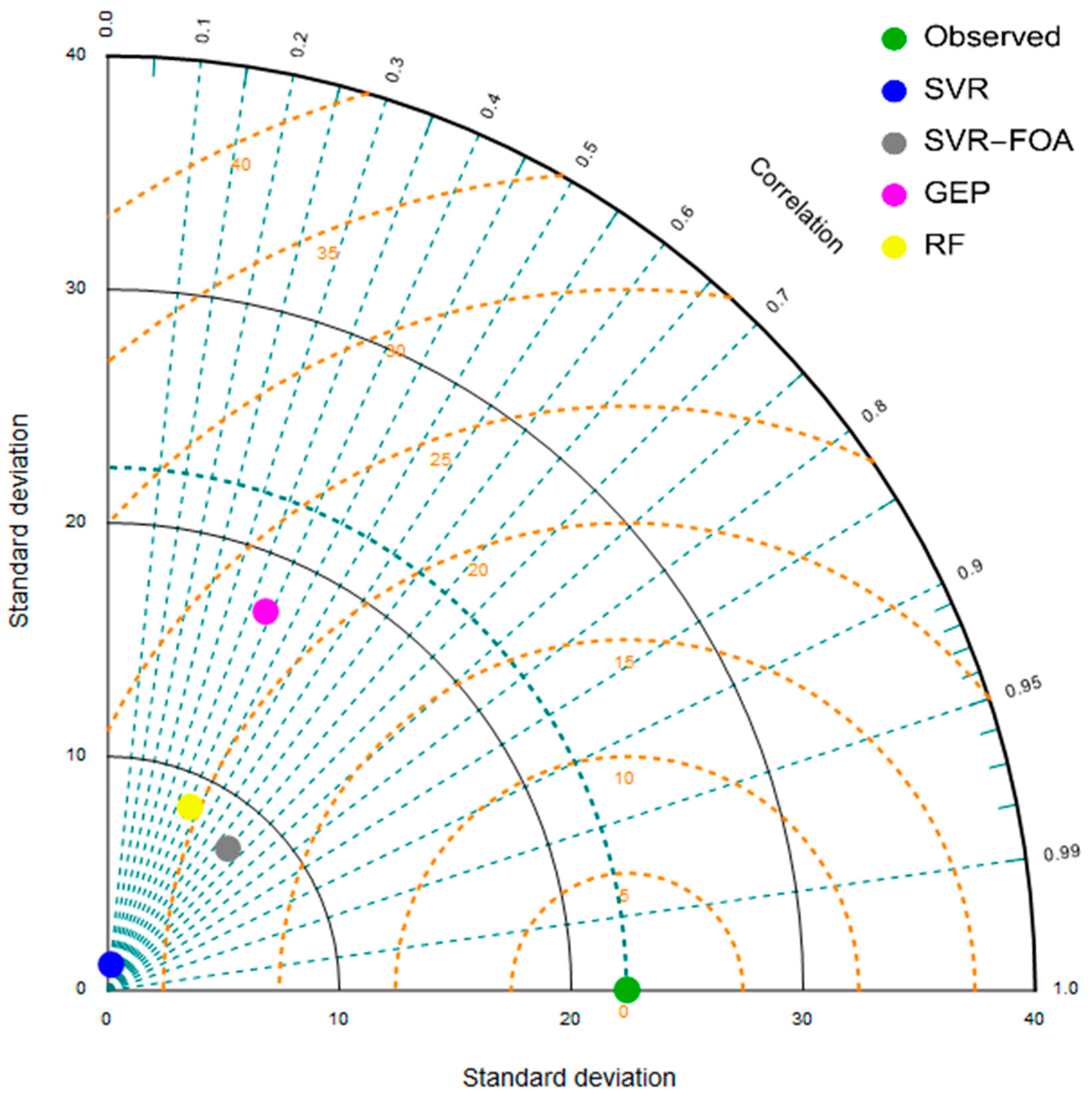
| Parameter | SVR | SVR-FOA | GEP | RF |
|---|---|---|---|---|
| CC | 0.146 | 0.644 | 0.387 | 0.41 |
| SI | 0.473 | 0.374 | 0.647 | 0.519 |
| WI | 0.408 | 0.607 | 0.456 | 0.507 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, Z.; Zhong, H.; Mosavi, A.; Sadiq, M.; Saleem, H.; Khalid, A.; Mahmood, S.; Nabipour, N. Machine Learning Modeling of Aerobic Biodegradation for Azo Dyes and Hexavalent Chromium. Mathematics 2020, 8, 913. https://doi.org/10.3390/math8060913
Ahmad Z, Zhong H, Mosavi A, Sadiq M, Saleem H, Khalid A, Mahmood S, Nabipour N. Machine Learning Modeling of Aerobic Biodegradation for Azo Dyes and Hexavalent Chromium. Mathematics. 2020; 8(6):913. https://doi.org/10.3390/math8060913
Chicago/Turabian StyleAhmad, Zulfiqar, Hua Zhong, Amir Mosavi, Mehreen Sadiq, Hira Saleem, Azeem Khalid, Shahid Mahmood, and Narjes Nabipour. 2020. "Machine Learning Modeling of Aerobic Biodegradation for Azo Dyes and Hexavalent Chromium" Mathematics 8, no. 6: 913. https://doi.org/10.3390/math8060913
APA StyleAhmad, Z., Zhong, H., Mosavi, A., Sadiq, M., Saleem, H., Khalid, A., Mahmood, S., & Nabipour, N. (2020). Machine Learning Modeling of Aerobic Biodegradation for Azo Dyes and Hexavalent Chromium. Mathematics, 8(6), 913. https://doi.org/10.3390/math8060913







