In Search of Complex Disease Risk through Genome Wide Association Studies
Abstract
:1. Introduction
- Complex trait or disease: A multifactorial phenotype resulting from the combination of numerous environmental and genetic factors.
- Genome-Wide Association Study (GWAS): A statistical method to discover the genomic variability that is associated with a complex trait or disease.
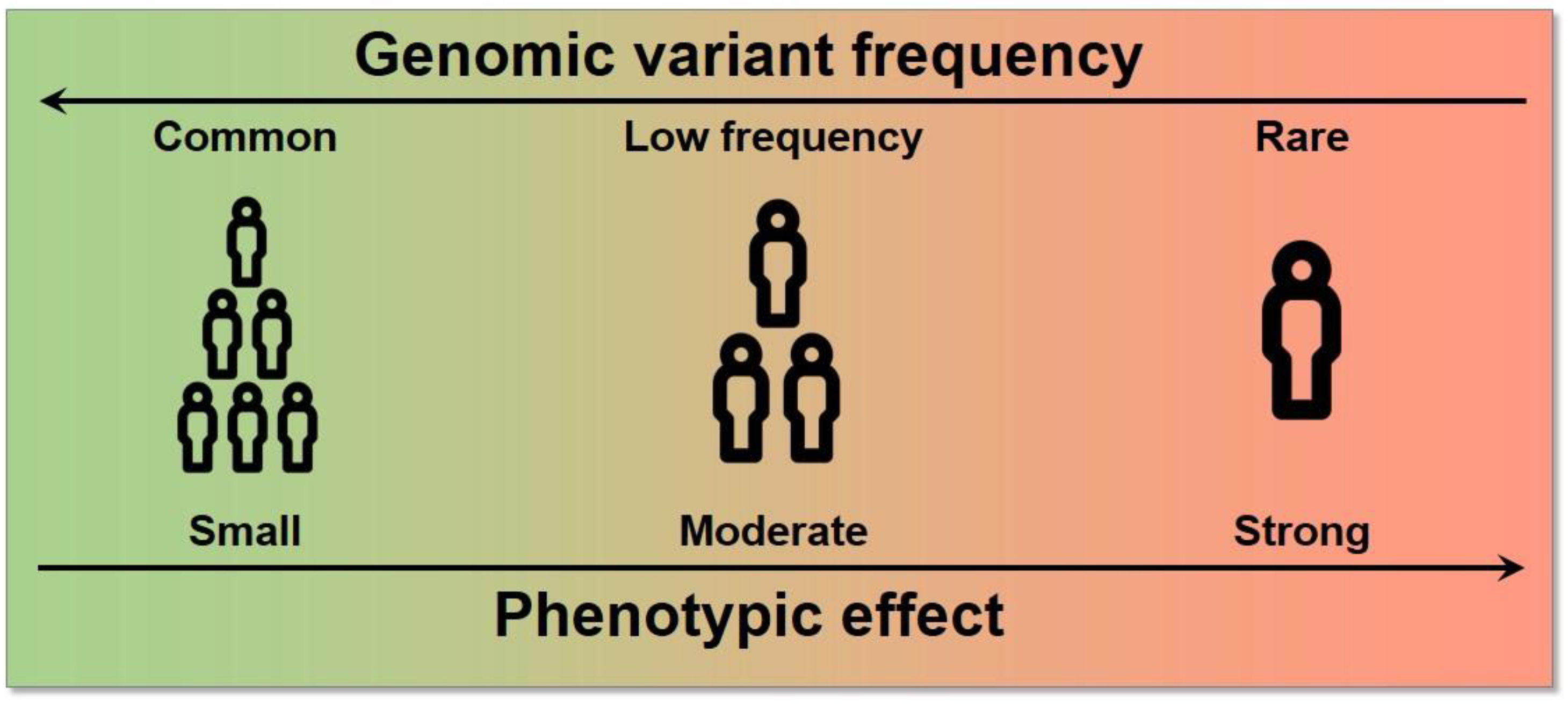
- Genomic or genetic variant: A genomic location known to present variability within a population.
- Personalised medicine: The application of preventive and treatment protocols adjusted to the patient’s genomic profile.
- Phenotype: A measurable characteristic in the individuals of a population, such as height, eye colour, blood pressure, or disease state.
2. Preliminary Genome Biology Concepts
- Allele: One of the possible genomic sequences that exist in a population for a given locus.
- Allelic Frequency: The frequency in which a certain allele is found within a population.
- Genomic locus: A region of the genome.
- Genomic marker: A specific variant that is used as a proxy for nearby variants in high linkage disequilibrium.
- Genotype: The specific combination of alleles of an individual. When compared to a reference genome, the genotype of a variant may be reference homozygous, heterozygous, or alternate homozygous.
- Haplotype: The list of alleles that are present in the same homologous chromosome.
- Inheritance model: A quantitative model for how the genotype of a variant might contribute to the phenotype. The most frequently used is the additive model, but the dominant, recessive, and heterodominant models are also utilized.
- Linkage Disequilibrium (LD): When alleles are inherited together in an individual more often than expected by chance. This is a consequence of the inheritance of these alleles in haplotype blocks instead of them being independent of each other.
- Single nucleotide variant/polymorphism (SNV/SNP): The most frequent type of genomic variant, in which the alleles differ in a single nucleotide position. SNPs are SNVs with a frequency of >1%.
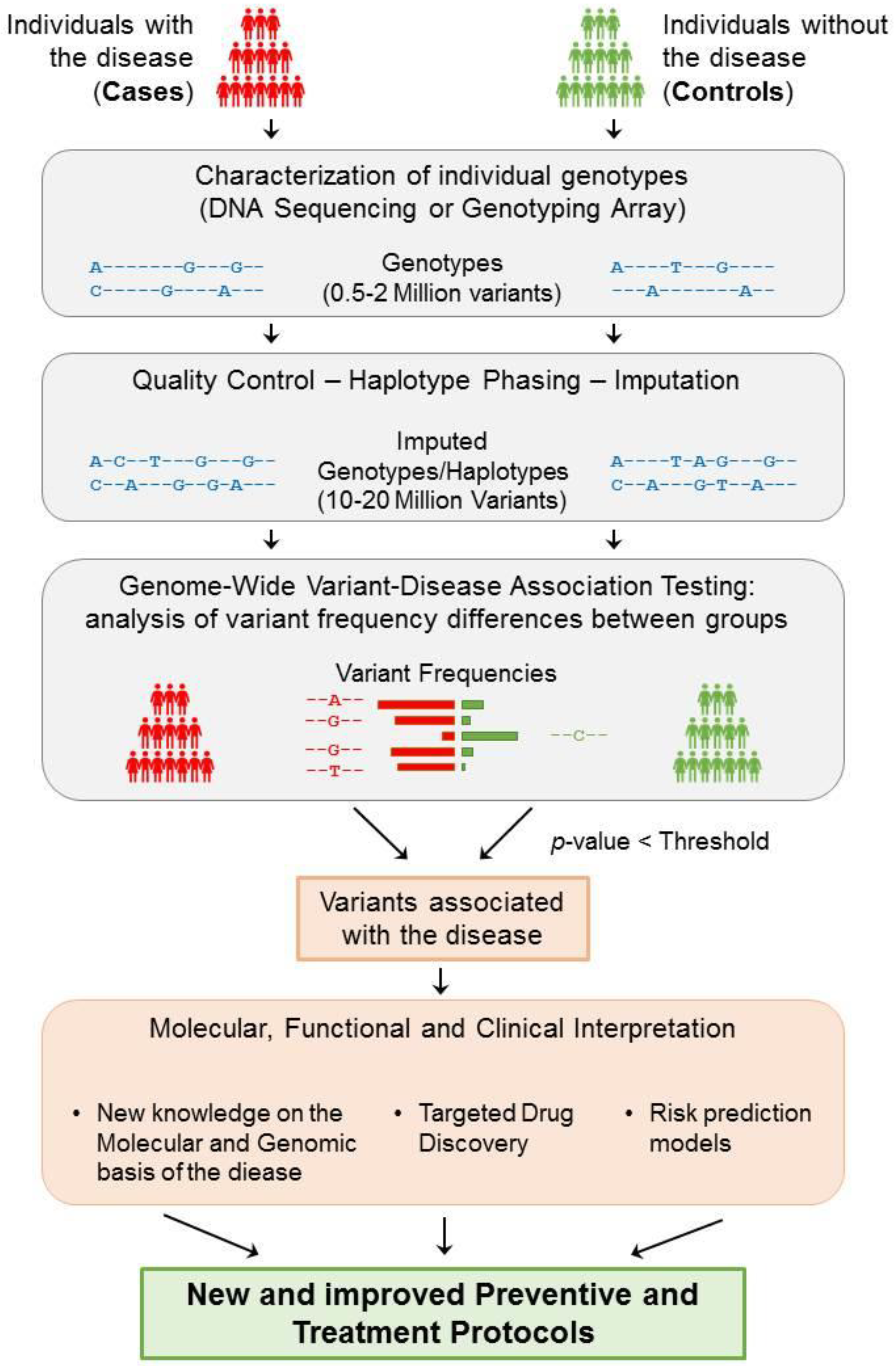
3. Genome Wide Association Studies (GWAS)
3.1. Definition
3.2. Analytical Frameworks for GWAS
- The number of individuals included in the sample of the study . In binary traits, these individuals are divided according to their phenotype, i.e., into cases (diseased) and controls (non-diseased), where .
- A set of genomic variants that are analysed for each individual present in the population.
- The genotype for each variant, which can take a genotype value from . This genotype can be encoded differently depending on the hypothesised inheritance model by defining a function , where encodes for additive , for dominant , for recessive , or for heterodominant . For the purpose of statistical testing, one of the alleles, typically the alternate, is defined as the effect allele.
- Based on the space defined by the genotype, each genomic variant can be considered as a simple random variable so that for which , with as the space of events.
- The phenotype for each individual in the population is given a trait of study, which, in the case of binary traits, is assigned as . The phenotype can be modelled by a Bernoulli distribution , where is the unknown probability of an individual having the disease.
- A measure of the statistical confidence on the association with the phenotype in the form of a p-value.
- A measure of the effect size of having one of the alleles, which is typically expressed by beta () for quantitative traits and an odds ratio () for binary traits.
3.2.1. Contingency Tables
3.2.2. Logistic Regression
3.2.3. Further Extensions and Developments of Regression Models in GWAS
3.2.4. Bayesian Statistics
3.3. Statistical Interpretation of GWAS Results
- Beta: An estimation of the effect size of a variant for a quantitative phenotype: the coefficients obtained from fitting a regression of the genotypes to the phenotype.
- Cohort: A group of individuals.
- DNA hybridisation array: A technology to identify the genotypes of a specific subset of variants of an individual.
- Effect size: A measure of the contribution of a genomic variant to a specific phenotype.
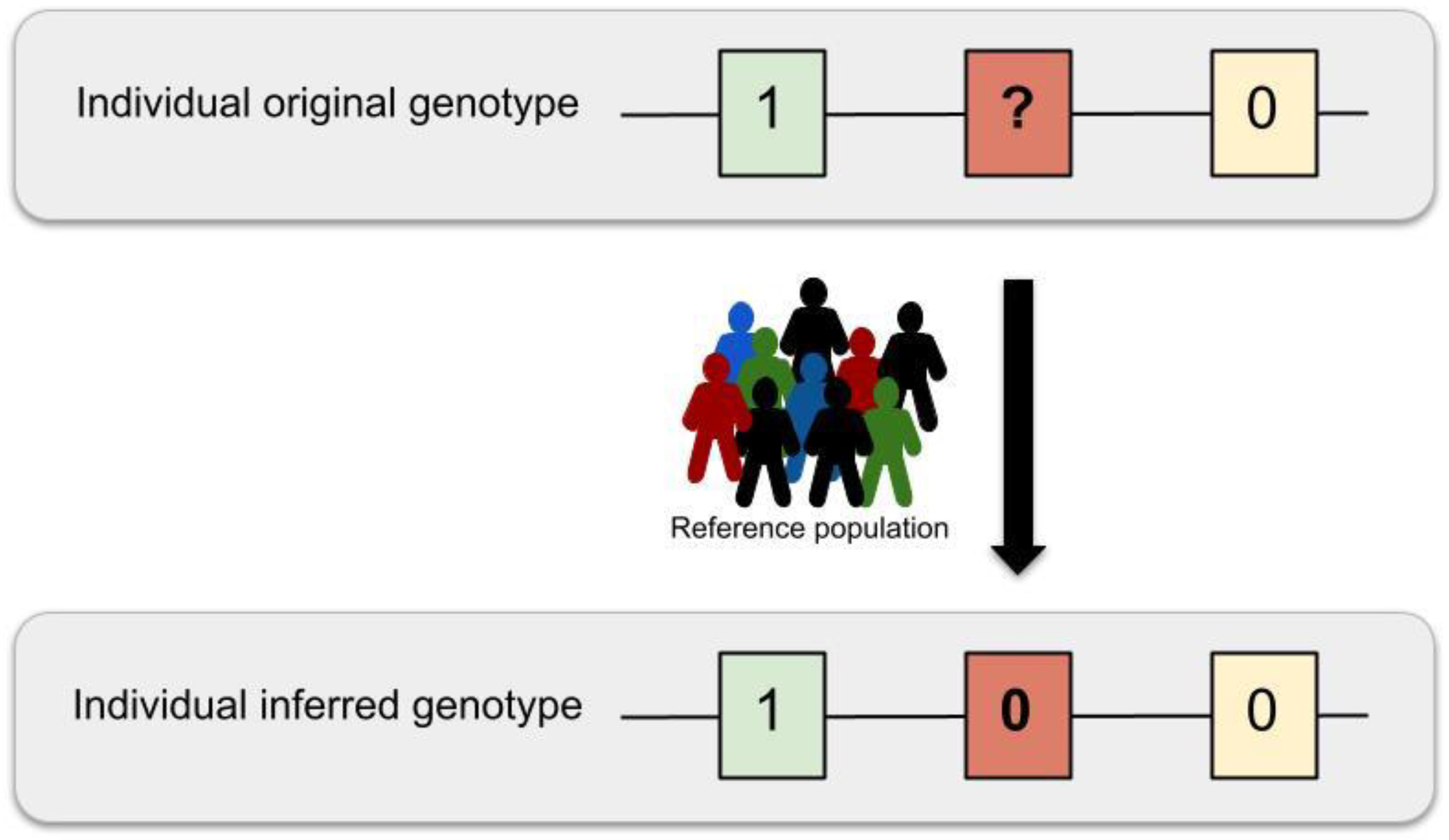
- Imputation: A statistical method to infer missing genotypes given a reduced set of known genotypes and a reference panel.
- Odds Ratio: An estimation of the effect size of a variant for a binary phenotype: the odds of having the disease with a variant divided by the odds of having the disease without it.
- Phasing: A statistical method to infer the haplotypes of an individual to determine which alleles belong to the same chromosomal sequence.
- Reference panel: A set of well characterised haplotypes of a group of individuals, used as a reference to infer non-genotyped variants in other individuals.
- Whole genome sequencing: A technology that provides the complete nucleotide sequence of an individual genome.
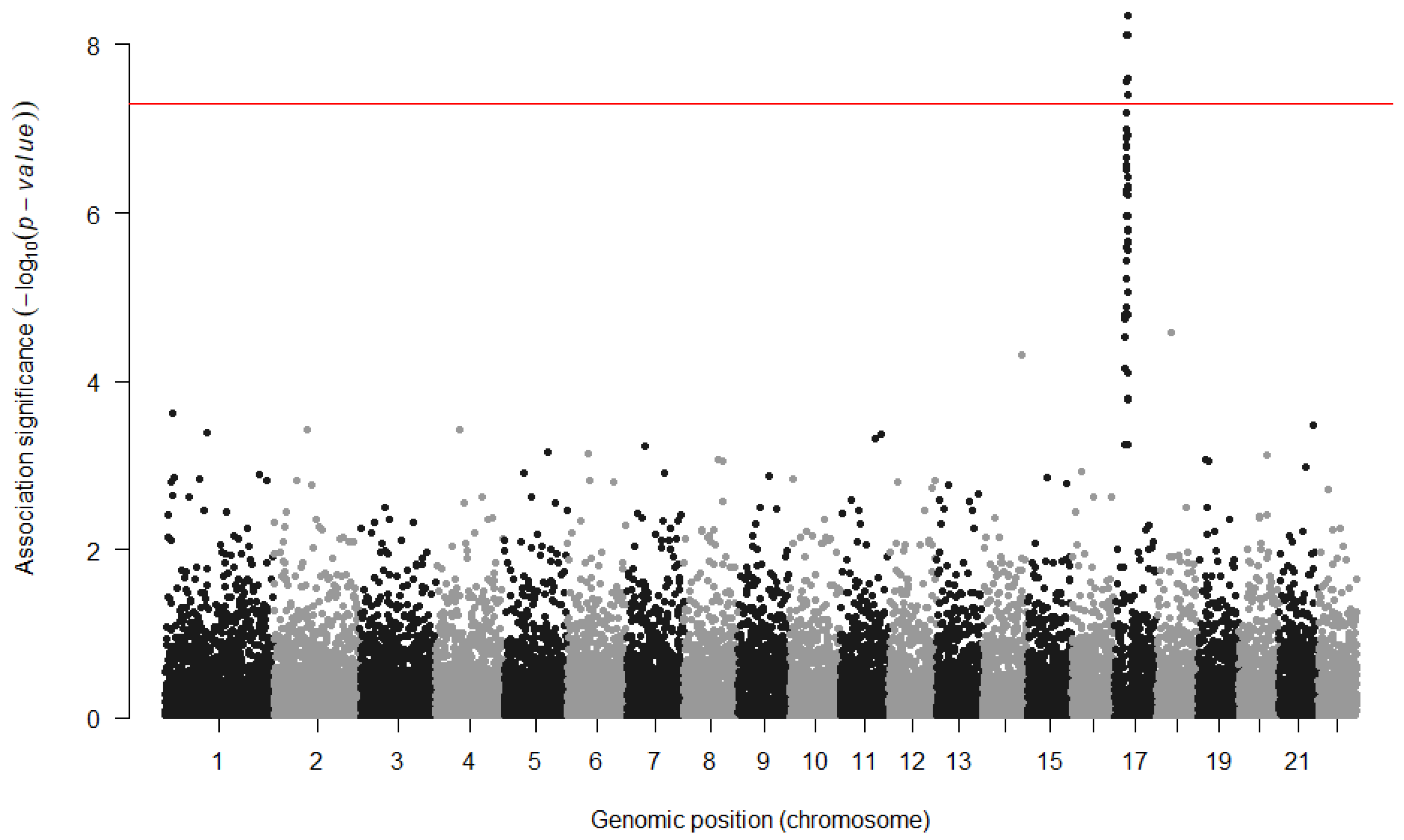
4. Current Practice and GWAS Limitations
4.1. Power and Sample Size
4.2. Increasing the Number of Genomic Variants
4.3. Genetic and Population Heterogeneity
4.4. Complex Interactions
4.4.1. Gene–Gene Interactions (GxG) and Genomic Variant Epistasis
4.4.2. Gene-Environment Interactions (GxE)
4.5. Biological Interpretation and Clinical Implications
4.6. Comprehensive GWAS Strategies for New Discoveries: An Example
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| EWAS | Environment Wide Association Studies |
| GxE | Gene–environment interactions |
| GxG | Gene–gene interactions |
| GLM | Generalized Linear Models |
| GRM | Genetic Relationship Matrix |
| GWAS | Genome Wide Association Studies |
| HMM | Hidden Markov Model |
| LD | Linkage Disequilibrium |
| OR | Odds Ratio |
| PRS | Polygenic Risk Score |
| SNV | Single Nucleotide Variation |
References
- Manolio, T.A.; Brooks, L.D.; Collins, F.S. A HapMap harvest of insights into the genetics of common disease. J. Clin. Investig. 2008, 118, 1590–1605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, K.J. What is complex about complex disorders? Genome Biol. 2012, 13, 237. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.R.; Wray, N.R.; Visscher, P.M. Explaining additional genetic variation in complex traits. Trends Genet. 2014, 30, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodge, S.; Greenberg, D. How Can We Explain Very Low Odds Ratios in GWAS? I. Polygenic Models. Hum. Hered. 2016, 81, 173–180. [Google Scholar] [CrossRef]
- Mahajan, A.; Taliun, D.; Thurner, M.; Robertson, N.R.; Torres, J.M.; Rayner, N.W.; Payne, A.J.; Steinthorsdottir, V.; Scott, R.A.; Grarup, N.; et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018, 50, 1505–1513. [Google Scholar] [CrossRef] [Green Version]
- Génin, E. Missing heritability of complex diseases: Case solved? Hum. Genet. 2020, 139, 103–113. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M.I. Genomics, Type 2 Diabetes, and Obesity. N. Engl. J. Med. 2010, 363, 2339–2350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vercelli, D. Discovering susceptibility genes for asthma and allergy. Nat. Rev. Immunol. 2008, 8, 169–182. [Google Scholar] [CrossRef]
- O’Donnell, C.J.; Nabel, E.G. Genomics of Cardiovascular Disease. N. Engl. J. Med. 2011, 365, 2098–2109. [Google Scholar] [CrossRef]
- Van Cauwenberghe, C.; Van Broeckhoven, C.; Sleegers, K. The genetic landscape of Alzheimer disease: Clinical implications and perspectives. Genet. Med. 2015, 18, 421–430. [Google Scholar] [CrossRef] [Green Version]
- American Diabetes Association. Economic Costs of Diabetes in the U.S. in 2017. Diabetes Care 2018, 41, 917–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vansteelandt, S.; Goetgeluk, S.; Lutz, S.; Waldman, I.; Lyon, H.; Schadt, E.E.; Weiss, S.T.; Lange, C. On the adjustment for covariates in genetic association analysis: A novel, simple principle to infer direct causal effects. Genet. Epidemiol. 2009, 33, 394–405. [Google Scholar] [CrossRef] [Green Version]
- Bonàs-Guarch, S.; Guindo-Martínez, M.; Miguel-Escalada, I.; Grarup, N.; Sebastian, D.; Rodriguez-Fos, E.; Sánchez, F.; Planas-Fèlix, M.; Cortes-Sánchez, P.; González, S.; et al. Re-analysis of public genetic data reveals a rare X-chromosomal variant associated with type 2 diabetes. Nat. Commun. 2018, 9, 321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindo-Martínez, M.; Amela, R.; Bonàs-Guarch, S.; Puiggròs, M.; Salvoro, C.; Miguel-Escalada, I.; Carey, C.E.; Cole, J.B.; Rüeger, S.; Atkinson, E.; et al. The impact of non-additive genetic associations on age-related complex diseases. Nat. Commun. 2021, 12, 2436. [Google Scholar] [CrossRef]
- The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, M.I.; Abecasis, G.R.; Cardon, L.R.; Goldstein, D.B.; Little, J.; Ioannidis, J.P.A.; Hirschhorn, J.N. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat. Rev. Genet. 2008, 9, 356–369. [Google Scholar] [CrossRef] [PubMed]
- LaFramboise, T. Single nucleotide polymorphism arrays: A decade of biological, computational and technological advances. Nucleic Acids Res. 2009, 37, 4181–4193. [Google Scholar] [CrossRef] [Green Version]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Prim. 2021, 1, 59. [Google Scholar] [CrossRef]
- Lander, E.S.; Schork, N.J. Genetic dissection of complex traits. Science 1994, 265, 2037–2048. [Google Scholar] [CrossRef] [Green Version]
- Ozaki, K.; Ohnishi, Y.; Iida, A.; Sekine, A.; Yamada, R.; Tsunoda, T.; Sato, H.; Sato, H.; Hori, M.; Nakamura, Y.; et al. Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nat. Genet. 2002, 32, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.J.; Zeiss, C.; Chew, E.Y.; Tsai, J.-Y.; Sackler, R.S.; Haynes, C.; Henning, A.K.; SanGiovanni, J.P.; Mane, S.M.; Mayne, S.T.; et al. Complement Factor H Polymorphism in Age-Related Macular Degeneration. Science 2005, 308, 385. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.; Henry, A.; Roselli, C.; Lin, H.; Sveinbjörnsson, G.; Fatemifar, G.; Hedman, Å.K.; Wilk, J.B.; Morley, M.P.; Chaffin, M.D.; et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat. Commun. 2020, 11, 163. [Google Scholar] [CrossRef]
- van Zuydam, N.R.; Ahlqvist, E.; Sandholm, N.; Deshmukh, H.; Rayner, N.W.; Abdalla, M.; Ladenvall, C.; Ziemek, D.; Fauman, E.; Robertson, N.R.; et al. A Genome-Wide Association Study of Diabetic Kidney Disease in Subjects with Type 2 Diabetes. Diabetes 2018, 67, 1414–1427. [Google Scholar] [CrossRef] [Green Version]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; van Duijn, C.M. GenABEL: An R library for genome-wide association analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutalik, Z.; Johnson, T.; Bochud, M.; Mooser, V.; Vollenweider, P.; Waeber, G.; Waterworth, D.; Beckmann, J.S.; Bergmann, S. Methods for testing association between uncertain genotypes and quantitative traits. Biostatistics 2011, 12, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Marchini, J.; Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010, 11, 499–511. [Google Scholar] [CrossRef]
- Yang, J.J.; Li, J.; Williams, L.K.; Buu, A. An efficient genome-wide association test for multivariate phenotypes based on the Fisher combination function. BMC Bioinform. 2016, 17, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelder, J.A.; Wedderburn, R.W.M. Generalized Linear Models. J. R. Stat. Soc. Ser. A 1972, 135, 370. [Google Scholar] [CrossRef]
- Loh, P.-R.; Kichaev, G.; Gazal, S.; Schoech, A.P.; Price, A.L. Mixed-model association for biobank-scale datasets. Nat. Genet. 2018, 50, 906–908. [Google Scholar] [CrossRef]
- Loh, P.-R.; Tucker, G.; Bulik-Sullivan, B.K.; Vilhjalmsson, B.J.; Finucane, H.K.; Salem, R.M.; Chasman, D.I.; Ridker, P.M.; Neale, B.M.; Berger, B.; et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015, 47, 284–290. [Google Scholar] [CrossRef]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mbatchou, J.; Barnard, L.; Backman, J.; Marcketta, A.; Kosmicki, J.A.; Ziyatdinov, A.; Benner, C.; O’Dushlaine, C.; Barber, M.; Boutkov, B.; et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 2021, 53, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Nielsen, J.B.; Fritsche, L.G.; Dey, R.; Gabrielsen, M.E.; Wolford, B.N.; LeFaive, J.; VandeHaar, P.; Gagliano, S.A.; Gifford, A.; et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018, 50, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Rohan, L.F.; Dorian, G. Bayesian Methods Applied to GWAS. Methods Mol. Biol. 2013, 1019, 237–274. [Google Scholar] [CrossRef]
- van Erp, N.; Gelder, P. van Bayesian logistic regression analysis. AIP Conf. Proc. 2013, 1553, 147. [Google Scholar] [CrossRef] [Green Version]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Benner, C.; Spencer, C.C.A.; Havulinna, A.S.; Salomaa, V.; Ripatti, S.; Pirinen, M. FINEMAP: Efficient variable selection using summary data from genome-wide association studies. Bioinformatics 2016, 32, 1493. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Zeng, L.; Schunkert, H.; Söding, J. Bayesian multiple logistic regression for case-control GWAS. PLoS Genet. 2018, 14, e1007856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lloyd-Jones, L.R.; Zeng, J.; Sidorenko, J.; Yengo, L.; Moser, G.; Kemper, K.E.; Wang, H.; Zheng, Z.; Magi, R.; Esko, T.; et al. Improved polygenic prediction by Bayesian multiple regression on summary statistics. Nat. Commun. 2019, 10, 5086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Basu, S.; Mirabello, L.; Spector, L.G.; Zhang, L. A Bayesian Gene-Based Genome-Wide Association Study Analysis of Osteosarcoma Trio Data Using a Hierarchically Structured Prior. Cancer Inform. 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Turchin, M.C.; Stephens, M. Bayesian multivariate reanalysis of large genetic studies identifies many new associations. PLoS Genet. 2019, 15, e1008431. [Google Scholar] [CrossRef] [Green Version]
- Pe’er, I.; Yelensky, R.; Altshuler, D.; Daly, M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008, 32, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Risch, N.; Merikangas, K. The future of genetic studies of complex human diseases. Science 1996, 273, 1516–1517. [Google Scholar] [CrossRef] [Green Version]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goddard, M.E.; Wray, N.R.; Verbyla, K.; Visscher, P.M. Estimating Effects and Making Predictions from Genome-Wide Marker Data. Stat. Sci. 2009, 24, 517–529. [Google Scholar] [CrossRef] [Green Version]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, K.; Stringer, S.; Frei, O.; Umićević Mirkov, M.; de Leeuw, C.; Polderman, T.J.C.; van der Sluis, S.; Andreassen, O.A.; Neale, B.M.; Posthuma, D. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019, 51, 1339–1348. [Google Scholar] [CrossRef]
- Beck, T.; Hastings, R.K.; Gollapudi, S.; Free, R.C.; Brookes, A.J. GWAS Central: A comprehensive resource for the comparison and interrogation of genome-wide association studies. Eur. J. Hum. Genet. 2014, 22, 949–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Ripke, S.; Neale, B.M.; Corvin, A.; Walters, J.T.R.; Farh, K.H.; Holmans, P.A.; Lee, P.; Bulik-Sullivan, B.; Collier, D.A.; Huang, H.; et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [CrossRef] [Green Version]
- Steinthorsdottir, V.; Thorleifsson, G.; Sulem, P.; Helgason, H.; Grarup, N.; Sigurdsson, A.; Helgadottir, H.T.; Johannsdottir, H.; Magnusson, O.T.; Gudjonsson, S.A.; et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat. Genet. 2014, 46, 294–298. [Google Scholar] [CrossRef]
- Sakaue, S.; Kanai, M.; Tanigawa, Y.; Karjalainen, J.; Kurki, M.; Koshiba, S.; Narita, A.; Konuma, T.; Yamamoto, K.; Akiyama, M.; et al. A global atlas of genetic associations of 220 deep phenotypes. MedRxiv 2020, 46, 20213652. [Google Scholar] [CrossRef]
- Alonso, L.; Piron, A.; Morán, I.; Guindo-Martinez, M.; Bonas-Guarch, S.; Atla, G.; Miguel-Escalada, I.; Royo, R.; Puiggros, M.; Garcia-Hurtado, X.; et al. TIGER: The gene expression regulatory variation landscape of human pancreatic islets. Cell Rep. 2021, 37, 109807. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015, 12, 1001779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, A.; Hirata, M.; Kamatani, Y.; Muto, K.; Matsuda, K.; Kiyohara, Y.; Ninomiya, T.; Tamakoshi, A.; Yamagata, Z.; Mushiroda, T.; et al. Overview of the BioBank Japan Project: Study design and profile. J. Epidemiol. 2017, 27, S2–S8. [Google Scholar] [CrossRef]
- Borodulin, K.; Tolonen, H.; Jousilahti, P.; Jula, A.; Juolevi, A.; Koskinen, S.; Kuulasmaa, K.; Laatikainen, T.; Männistö, S.; Peltonen, M.; et al. Cohort Profile: The National FINRISK Study. Int. J. Epidemiol. 2018, 47, 696–696i. [Google Scholar] [CrossRef]
- Panagiotou, O.A.; Willer, C.J.; Hirschhorn, J.N.; Ioannidis, J.P.A. The Power of Meta-Analysis in Genome-Wide Association Studies. Annu. Rev. Genom. Hum. Genet. 2013, 14, 441–465. [Google Scholar] [CrossRef] [Green Version]
- Evangelou, E.; Ioannidis, J.P.A. Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet. 2013, 14, 379–389. [Google Scholar] [CrossRef]
- Hivert, V.; Sidorenko, J.; Rohart, F.; Goddard, M.E.; Yang, J.; Wray, N.R.; Yengo, L.; Visscher, P.M. Estimation of non-additive genetic variance in human complex traits from a large sample of unrelated individuals. Am. J. Hum. Genet. 2021, 108, 786–798. [Google Scholar] [CrossRef]
- Lamy, P.; Grove, J.; Wiuf, C. A review of software for microarray genotyping. Hum. Genom. 2011, 5, 304–309. [Google Scholar] [CrossRef] [Green Version]
- Marchini, J.; Howie, B.; Myers, S.; McVean, G.; Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007, 39, 906–913. [Google Scholar] [CrossRef]
- Das, S.; Abecasis, G.R.; Browning, B.L. Genotype Imputation from Large Reference Panels. Annu. Rev. Genom. Hum. Genet. 2018, 19, 73–96. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Willer, C.; Sanna, S.; Abecasis, G. Genotype Imputation. Annu. Rev. Genom. Hum. Genet. 2009, 10, 387. [Google Scholar] [CrossRef]
- Boomsma, D.I.; Wijmenga, C.; Slagboom, E.P.; Swertz, M.A.; Karssen, L.C.; Abdellaoui, A.; Ye, K.; Guryev, V.; Vermaat, M.; Van Dijk, F.; et al. The Genome of the Netherlands: Design, and project goals. Eur. J. Hum. Genet. 2014, 22, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The UK10K Consortium The UK10K project identifies rare variants in health and disease. Nature 2015, 526, 82–90. [CrossRef] [Green Version]
- McCarthy, S.; Das, S.; Kretzschmar, W.; Delaneau, O.; Wood, A.R.; Teumer, A.; Kang, H.M.; Fuchsberger, C.; Danecek, P.; Sharp, K.; et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016, 48, 1279–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taliun, D.; Harris, D.N.; Kessler, M.D.; Carlson, J.; Szpiech, Z.A.; Torres, R.; Taliun, S.A.G.; Corvelo, A.; Gogarten, S.M.; Kang, H.M.; et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 2021, 590, 290–299. [Google Scholar] [CrossRef]
- Valls-Margarit, J.; Galván-Femenía, I.; Matias, D.; Blay, N.; Puiggròs, M.; Carreras, A.; Salvoro, C.; Cortés, B.; Amela, R.; Farre, X.; et al. GCAT|Panel, a comprehensive structural variant haplotype map of the Iberian population from high-coverage whole-genome sequencing. bioRxiv 2021, 21, 453041. [Google Scholar] [CrossRef]
- Marchini, J. Haplotype Estimation and Genotype Imputation. In Handbook of Statistical Genomics; Wiley: Hoboken, NJ, USA, 2019; Volume 1, pp. 87–114. [Google Scholar]
- Scheet, P.; Stephens, M. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006, 78, 629–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burton, P.R.; Clayton, D.G.; Cardon, L.R.; Craddock, N.; Deloukas, P.; Duncanson, A.; Kwiatkowski, D.P.; McCarthy, M.I.; Ouwehand, W.H.; Samani, N.J.; et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007, 447, 661–678. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Willer, C.J.; Ding, J.; Scheet, P.; Abecasis, G.R. MaCH: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010, 34, 816–834. [Google Scholar] [CrossRef] [Green Version]
- Naj, A.C. Genotype Imputation in Genome-Wide Association Studies. Curr. Protoc. Hum. Genet. 2019, 102, e84. [Google Scholar] [CrossRef] [PubMed]
- Lo, C. Algorithms for Haplotype Phasing. Available online: https://cseweb.ucsd.edu//~chl107/pubs/re.pdf (accessed on 30 April 2021).
- Ahlqvist, E.; Storm, P.; Käräjämäki, A.; Martinell, M.; Dorkhan, M.; Carlsson, A.; Vikman, P.; Prasad, R.B.; Aly, D.M.; Almgren, P.; et al. Novel subgroups of adult-onset diabetes and their association with outcomes: A data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018, 6, 361–369. [Google Scholar] [CrossRef] [Green Version]
- Ahlqvist, E.; Prasad, R.B.; Groop, L. Subtypes of Type 2 Diabetes Determined From Clinical Parameters. Diabetes 2020, 69, 2086–2093. [Google Scholar] [CrossRef]
- Waters, K.; Stram, D.; Hassanein, M.; Le Marchand, L.; Wilkens, L.; Maskarinec, G.; Monroe, K.; Kolonel, L.; Altshuler, D.; Henderson, B.; et al. Consistent association of type 2 diabetes risk variants found in europeans in diverse racial and ethnic groups. PLoS Genet. 2010, 6, e1001078. [Google Scholar] [CrossRef]
- Imamura, M.; Takahashi, A.; Yamauchi, T.; Hara, K.; Yasuda, K.; Grarup, N.; Zhao, W.; Wang, X.; Huerta-Chagoya, A.; Hu, C.; et al. Genome-wide association studies in the Japanese population identify seven novel loci for type 2 diabetes. Nat. Commun. 2016, 7, 10531. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Spracklen, C.N.; Marenne, G.; Varshney, A.; Corbin, L.J.; Luan, J.; Willems, S.M.; Wu, Y.; Zhang, X.; Horikoshi, M.; et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 2021, 53, 840–860. [Google Scholar] [CrossRef]
- Chen, M.-H.; Raffield, L.M.; Mousas, A.; Sakaue, S.; Huffman, J.E.; Moscati, A.; Trivedi, B.; Jiang, T.; Akbari, P.; Vuckovic, D.; et al. Trans-ethnic and Ancestry-Specific Blood-Cell Genetics in 746,667 Individuals from 5 Global Populations. Cell 2020, 182, 1198–1213.e14. [Google Scholar] [CrossRef]
- Marchini, J.; Donnelly, P.; Cardon, L.R. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat. Genet. 2005, 37, 413–417. [Google Scholar] [CrossRef]
- Álvarez-Castro, J.M. Gene–Environment Interaction in the Era of Precision Medicine—Filling the Potholes Rather Than Starting to Build a New Road. Front. Genet. 2020, 11, 6. [Google Scholar] [CrossRef]
- Manolio, T.A. Genomewide Association Studies and Assessment of the Risk of Disease. N. Engl. J. Med. 2010, 363, 166–176. [Google Scholar] [CrossRef] [Green Version]
- White, D.; Rabago-Smith, M. Genotype-phenotype associations and human eye color. J. Hum. Genet. 2011, 56, 5–7. [Google Scholar] [CrossRef] [Green Version]
- Cordell, H.J. Detecting gene–gene interactions that underlie human diseases. Nat. Rev. Genet. 2009, 10, 392–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirino, Y.; Bertsias, G.; Ishigatsubo, Y.; Mizuki, N.; Tugal-Tutkun, I.; Seyahi, E.; Ozyazgan, Y.; Sacli, F.S.; Erer, B.; Inoko, H.; et al. Genome-wide association analysis identifies new susceptibility loci for Behçet’s disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet. 2013, 45, 202–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monir, M.M.; Zhu, J. Comparing GWAS Results of Complex Traits Using Full Genetic Model and Additive Models for Revealing Genetic Architecture. Sci. Rep. 2017, 7, 38600. [Google Scholar] [CrossRef] [Green Version]
- Wan, X.; Yang, C.; Yang, Q.; Xue, H.; Fan, X.; Tang, N.L.S.; Yu, W. BOOST: A fast approach to detecting gene-gene interactions in genome-wide case-control studies. Am. J. Hum. Genet. 2010, 87, 325–340. [Google Scholar] [CrossRef] [Green Version]
- Behravan, H.; Hartikainen, J.M.; Tengström, M.; Pylkäs, K.; Winqvist, R.; Kosma, V.; Mannermaa, A. Machine learning identifies interacting genetic variants contributing to breast cancer risk: A case study in Finnish cases and controls. Sci. Rep. 2018, 8, 13149. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.D.; Hahn, L.W.; Roodi, N.; Bailey, L.R.; Dupont, W.D.; Parl, F.F.; Moore, J.H. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 2001, 69, 138–147. [Google Scholar] [CrossRef] [Green Version]
- Hahn, L.W.; Ritchie, M.D.; Moore, J.H. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 2003, 19, 376–382. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.H. Computational analysis of gene-gene interactions using multifactor dimensionality reduction. Expert Rev. Mol. Diagn. 2004, 4, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, J.S. Bayesian inference of epistatic interactions in case-control studies. Nat. Genet. 2007, 39, 1167–1173. [Google Scholar] [CrossRef]
- Kerin, M.; Marchini, J. Gene-environment interactions using a Bayesian whole genome regression model. bioRxiv 2019, 19, 797829. [Google Scholar] [CrossRef] [Green Version]
- Gayán, J.; González-Pérez, A.; Bermudo, F.; Sáez, M.E.; Royo, J.L.; Quintas, A.; Galan, J.J.; Morón, F.J.; Ramirez-Lorca, R.; Real, L.M.; et al. A method for detecting epistasis in genome-wide studies using case-control multi-locus association analysis. BMC Genom. 2008, 9, 360. [Google Scholar] [CrossRef] [Green Version]
- Dempfle, A.; Scherag, A.; Hein, R.; Beckmann, L.; Chang-Claude, J.; Schäfer, H. Gene-environment interactions for complex traits: Definitions, methodological requirements and challenges. Eur. J. Hum. Genet. 2008, 16, 1164–1172. [Google Scholar] [CrossRef] [PubMed]
- Bookman, E.B.; McAllister, K.; Gillanders, E.; Wanke, K.; Balshaw, D.; Rutter, J.; Reedy, J.; Shaughnessy, D.; Agurs-Collins, T.; Paltoo, D.; et al. Gene-environment interplay in common complex diseases: Forging an integrative model-Recommendations from an NIH workshop. Genet. Epidemiol. 2011, 35, 217–225. [Google Scholar] [CrossRef] [Green Version]
- Patel, C.J.; Bhattacharya, J.; Butte, A.J. An environment-wide association study (EWAS) on type 2 diabetes mellitus. PLoS ONE 2010, 5, e10746. [Google Scholar] [CrossRef]
- Thomas, D. Methods for investigating gene-environment interactions in candidate pathway and genome-wide association studies. Annu. Rev. Public Health 2010, 31, 21–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, P.H.G.; Sylvestre, M.P.; Tremblay, J.; Hamet, P. Key Considerations and Methods in the Study of Gene-Environment Interactions. Am. J. Hypertens. 2016, 29, 891–899. [Google Scholar] [CrossRef] [Green Version]
- Han, S.S.; Chatterjee, N. Review of Statistical Methods for Gene-Environment Interaction Analysis. Curr. Epidemiol. Rep. 2018, 5, 39–45. [Google Scholar] [CrossRef]
- McAllister, K.; Mechanic, L.E.; Amos, C.; Aschard, H.; Blair, I.A.; Chatterjee, N.; Conti, D.; Gauderman, W.J.; Hsu, L.; Hutter, C.M.; et al. Current Challenges and New Opportunities for Gene-Environment Interaction Studies of Complex Diseases. Am. J. Epidemiol. 2017, 186, 753–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, D. Gene-Environment-Wide Association Studies: Emerging Approaches. Nat. Rev. Genet. 2010, 11, 259. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Chen, Z.; Pearson, T.; Zhao, J.; Hu, H.; Prosperi, M. Design and methodology challenges of environment-wide association studies: A systematic review. Environ. Res. 2020, 183, 109275. [Google Scholar] [CrossRef]
- Cano-Gamez, E.; Trynka, G. From GWAS to Function: Using Functional Genomics to Identify the Mechanisms Underlying Complex Diseases. Front. Genet. 2020, 11, 424. [Google Scholar] [CrossRef] [PubMed]
- Lichou, F.; Trynka, G. Functional studies of GWAS variants are gaining momentum. Nat. Commun. 2020, 11, 6283. [Google Scholar] [CrossRef]
- Lambert, S.A.; Abraham, G.; Inouye, M. Towards clinical utility of polygenic risk scores. Hum. Mol. Genet. 2019, 28, R133–R142. [Google Scholar] [CrossRef]
- Choi, S.W.; Mak, T.S.H.; O’Reilly, P.F. Tutorial: A guide to performing polygenic risk score analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef] [PubMed]
- The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [Green Version]
- Taylor, D.L.; Jackson, A.U.; Narisu, N.; Hemani, G.; Erdos, M.R.; Chines, P.S.; Swift, A.; Idol, J.; Didion, J.P.; Welch, R.P.; et al. Integrative analysis of gene expression, DNA methylation, physiological traits, and genetic variation in human skeletal muscle. Proc. Natl. Acad. Sci. USA 2019, 116, 10883–10888. [Google Scholar] [CrossRef] [Green Version]
- Beesley, J.; Sivakumaran, H.; Moradi Marjaneh, M.; Shi, W.; Hillman, K.M.; Kaufmann, S.; Hussein, N.; Kar, S.; Lima, L.G.; Ham, S.; et al. eQTL Colocalization Analyses Identify NTN4 as a Candidate Breast Cancer Risk Gene. Am. J. Hum. Genet. 2020, 107, 778–787. [Google Scholar] [CrossRef]
- Martin, A.R.; Kanai, M.; Kamatani, Y.; Okada, Y.; Neale, B.M.; Daly, M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019, 51, 584–591. [Google Scholar] [CrossRef]
- McGuire, A.L.; Gabriel, S.; Tishkoff, S.A.; Wonkam, A.; Chakravarti, A.; Furlong, E.E.M.; Treutlein, B.; Meissner, A.; Chang, H.Y.; López-Bigas, N.; et al. The road ahead in genetics and genomics. Nat. Rev. Genet. 2020, 21, 581–596. [Google Scholar] [CrossRef]
- Mulder, N.; Abimiku, A.; Adebamowo, S.N.; de Vries, J.; Matimba, A.; Olowoyo, P.; Ramsay, M.; Skelton, M.; Stein, D.J. H3Africa: Current perspectives. Pharmgenomics Pers. Med. 2018, 11, 59–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miga, K.H.; Wang, T. The Need for a Human Pangenome Reference Sequence. Rev. Genom. Hum. Genet. 2021, 22, 81–102. [Google Scholar] [CrossRef] [PubMed]
| AA | AB | BB | Total | |
|---|---|---|---|---|
| Cases | ||||
| Controls | ||||
| Total |
| A | B | Total | |
|---|---|---|---|
| Cases | |||
| Controls | |||
| Total |
| A | B | |
|---|---|---|
| Cases | ||
| Controls |
| A | B | |
|---|---|---|
| Cases | ||
| Controls |
| Dominant Model {0,1,1} | Recessive Model {1,0,0} | Heterodominant Model {0,1,0} | |||||
|---|---|---|---|---|---|---|---|
| AA | AB + BB | AA + AB | BB | AA + BB | AB | Total | |
| Cases | |||||||
| Controls | |||||||
| Total | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alonso, L.; Morán, I.; Salvoro, C.; Torrents, D. In Search of Complex Disease Risk through Genome Wide Association Studies. Mathematics 2021, 9, 3083. https://doi.org/10.3390/math9233083
Alonso L, Morán I, Salvoro C, Torrents D. In Search of Complex Disease Risk through Genome Wide Association Studies. Mathematics. 2021; 9(23):3083. https://doi.org/10.3390/math9233083
Chicago/Turabian StyleAlonso, Lorena, Ignasi Morán, Cecilia Salvoro, and David Torrents. 2021. "In Search of Complex Disease Risk through Genome Wide Association Studies" Mathematics 9, no. 23: 3083. https://doi.org/10.3390/math9233083
APA StyleAlonso, L., Morán, I., Salvoro, C., & Torrents, D. (2021). In Search of Complex Disease Risk through Genome Wide Association Studies. Mathematics, 9(23), 3083. https://doi.org/10.3390/math9233083






