Solution of Mixed-Integer Optimization Problems in Bioinformatics with Differential Evolution Method
Abstract
:1. Introduction
2. Related Work
3. Materials and Methods
3.1. Problem Statement
3.2. Differential Evolution Entirely Parallel Method
- The values are sorted in ascending order.
- The index of the parameter in the floating point array becomes the value of the parameter in the integer array.
- The specification of the problem to be solved:
- -
- The objective function;
- -
- The data with which the solution is to be compared;
- -
- Constraints for the parameters;
- Control parameters of the algorithm.
| Algorithm 1: Differential evolution entirely parallel method |
| INITIALIZATION: |
| The individuals of the population are initialized randomly. |
| Generation |
| repeat |
| RECOMBINATION: |
| if The predefined number of iterations passed. then |
| Make Scatter Search Step |
| else |
| for all individuals in population do |
| Triangulation Recombination of model parameters (2)-(6) |
| Update scaling coefficients and weights (7), (8) |
| end for |
| end if |
| for all offsprings do |
| Make Deduplication of integer-valued parameters |
| end for |
| EVALUATION: |
| for all offsprings do |
| Calculate objective function Q |
| end for |
| SELECTION: |
| for all offsprings do |
| Accept or Reject an offspring to the next generation |
| end for |
| if The predefined number of generations passed then |
| Substitute oldest individuals with the best ones |
| end if |
| until Stopping criterion is met |
3.3. Triangulation Recombination Rule
3.4. Deduplication
- -
- Sample another index h uniformly, different from b, t and r used in recombination.
- -
- Create a helper array: assign 1 if the value firstly occurs in , assign 2 if the value firstly occurs in and 3—if the value is duplicated.
- -
- Sort the helper array in ascending order.
- -
- Fill with the values that corresponds to the first K elements of the helper array.
3.5. Genomic Selection
3.6. Optimization of the Set of Predictors
3.7. Compilation of Training Sample
3.8. Regression Model of Gene Expression
3.9. Experimental Setup
4. Results
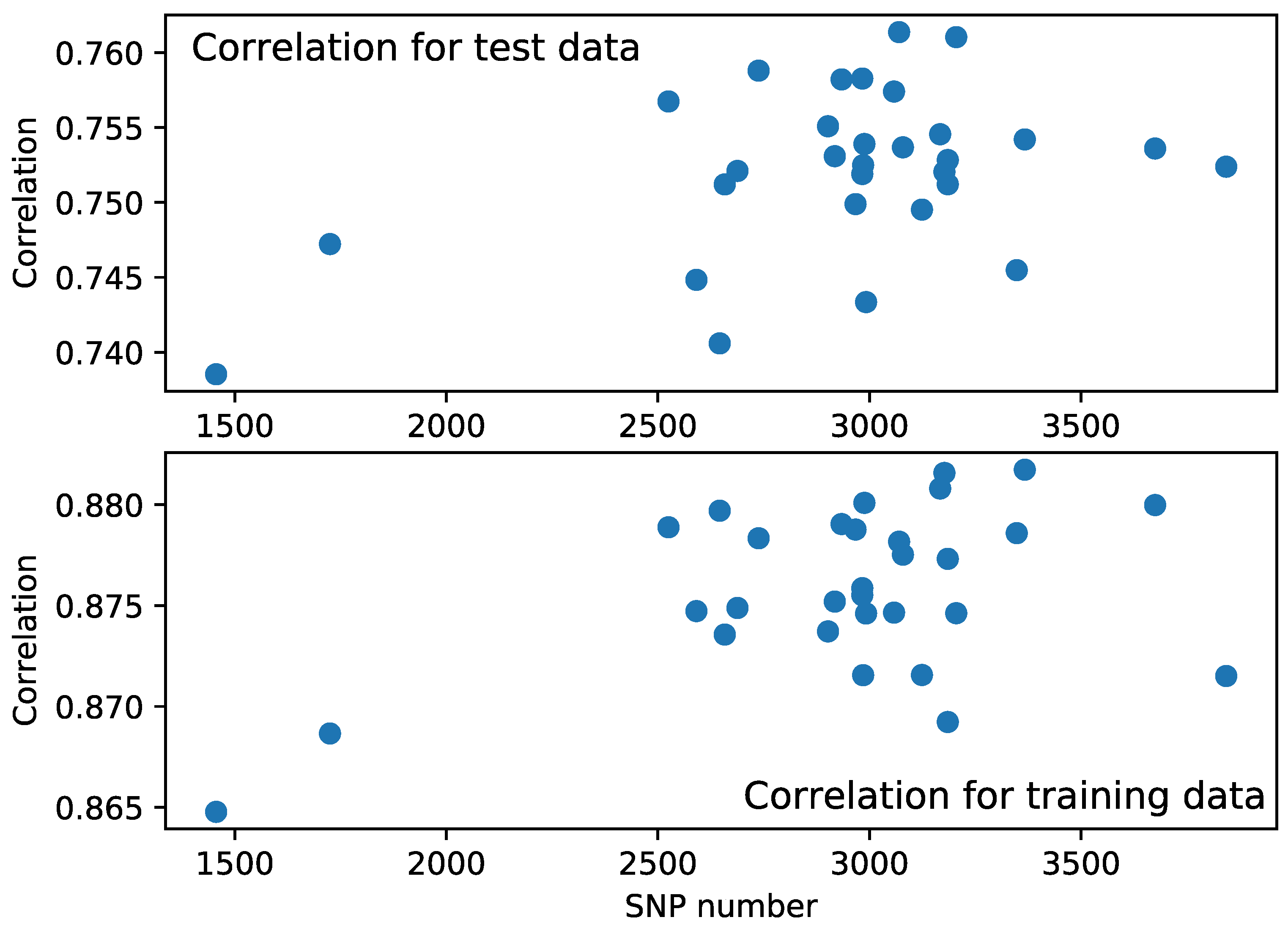
4.1. Optimization of the Set of Predictors
4.2. Compilation of Training Sample
4.3. Regression Model of Gene Expression
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaeger, J.; Surkova, S.; Blagov, M.; Janssens, H.; Kosman, D.; Kozlov, K.N.; Myasnikova, E.; Vanario-Alonso, C.E.; Samsonova, M.; Sharp, D.H.; et al. Dynamic control of positional information in the early Drosophila embryo. Nature 2004, 430, 368–371. [Google Scholar] [CrossRef]
- Storn, R.; Price, K. Differential Evolution—A Simple and Efficient Heuristic for Global Optimization over Continuous Spaces; Technical Report Technical Report TR-95-012; International Computer Science Institute: Berkeley, CA, USA, 1995. [Google Scholar]
- Fomekong-Nanfack, Y.; Kaandorp, J.A.; Blom, J. Efficient parameter estimation for spatio-temporal models of pattern formation: Case study of Drosophila melanogaster. Bioinformatics 2007, 23, 3356–3363. [Google Scholar] [CrossRef] [Green Version]
- Fomekong-Nanfack, Y.; Postma, M.; Kaandorp, J.A. Inferring Drosophila gap gene regulatory network: A parameter sensitivity and perturbation analysis. BMC Syst. Biol. 2009, 3, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suleimenov, Y.; Ay, A.; Samee, M.A.H.; Dresch, J.M.; Sinha, S.; Arnosti, D.N. Global parameter estimation for thermodynamic models of transcriptional regulation. Methods 2013, 62, 99–108. [Google Scholar] [CrossRef]
- Caraffini, F.; Kononova, A.V.; Corne, D. Infeasibility and structural bias in differential evolution. Inf. Sci. 2019, 496, 161–179. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Emmerich, M.T.; Eggermont, J.; Bäck, T.; Schütz, M.; Dijkstra, J.; Reiber, J. Mixed Integer Evolution Strategies for Parameter Optimization. Evol. Comput. 2013, 21, 29–64. [Google Scholar] [CrossRef] [PubMed]
- Che, H.; Wang, J. A Two-Timescale Duplex Neurodynamic Approach to Mixed-Integer Optimization. IEEE Trans. Neural Networks Learn. Syst. 2021, 32, 36–48. [Google Scholar] [CrossRef]
- Sadowski, K.L.; Thierens, D.; Bosman, P.A. GAMBIT: A Parameterless Model-Based Evolutionary Algorithm for Mixed-Integer Problems. Evol. Comput. 2018, 26, 117–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bussieck, M.R.; Meeraus, A. General Algebraic Modeling System (GAMS). In Modeling Languages in Mathematical Optimization; Series Title: Applied, Optimization; Pardalos, P.M., Hearn, D.W., Kallrath, J., Eds.; Springer: Boston, MA, USA, 2004; Volume 88, pp. 137–157. [Google Scholar] [CrossRef]
- Hansen, N. ACMA-ES for Mixed-Integer Nonlinear Optimization; Technical Report 7751; Institut National De Recherche En Informatique Et En Automatique: Le Chesnay-Rocquencourt, France, 2011. [Google Scholar]
- Exler, O.; Schittkowski, K. A trust region SQP algorithm for mixed-integer nonlinear programming. Optim. Lett. 2007, 1, 269–280. [Google Scholar] [CrossRef]
- Exler, O.; Lehmann, T.; Schittkowski, K. A comparative study of SQP-type algorithms for nonlinear and nonconvex mixed-integer optimization. Math. Program. Comput. 2012, 4, 383–412. [Google Scholar] [CrossRef]
- Emmerich, M.T.M.; Li, R.; Zhang, A.; Lucas, P.; Flesch, I. Mixed-Integer Bayesian Optimization Utilizing A-Priori Knowledge on Parameter Dependences. In Proceedings of the 20th Belgium-Netherlands Conference on Artificial Intelligence, BNAIC, Leiden, The Netherlands, 19–20 November 2008; pp. 65–72. [Google Scholar]
- Penas, D.R.; Henriques, D.; González, P.; Doallo, R.; Saez-Rodriguez, J.; Banga, J.R. A parallel metaheuristic for large mixed-integer dynamic optimization problems, with applications in computational biology. PLoS ONE 2017, 12, e0182186. [Google Scholar] [CrossRef] [Green Version]
- Egea, J.A.; Martí, R.; Banga, J.R. An evolutionary method for complex-process optimization. Comput. Oper. Res. 2010, 37, 315–324. [Google Scholar] [CrossRef]
- Egea, J.A.; Henriques, D.; Cokelaer, T.; Villaverde, A.F.; MacNamara, A.; Danciu, D.P.; Banga, J.R.; Saez-Rodriguez, J. MEIGO: An open-source software suite based on metaheuristics for global optimization in systems biology and bioinformatics. BMC Bioinform. 2014, 15, 136. [Google Scholar] [CrossRef] [Green Version]
- Storn, R. Differential Evolution—A Simple and Efficient Heuristic for Global Optimization over Continuous Spaces. J. Glob. Optim. 1997, 11, 341–359. [Google Scholar] [CrossRef]
- Khushaba, R.N.; Al-Jumaily, A.; Al-Ani, A. Evolutionary fuzzy discriminant analysis feature projection technique in myoelectric control. Pattern Recognit. Lett. 2009, 30, 699–707. [Google Scholar] [CrossRef]
- Wu, C.Y.; Tseng, K.Y. Truss structure optimization using adaptive multi-population differential evolution. Struct. Multidiscip. Optim. 2010, 42, 575–590. [Google Scholar] [CrossRef]
- Gong, T.; Tuson, A.L. Differential Evolution for Binary Encoding. In Soft Computing in Industrial Applications; Saad, A., Dahal, K., Sarfraz, M., Roy, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 39, pp. 251–262. [Google Scholar] [CrossRef]
- He, X.; Zhang, Q.; Sun, N.; Dong, Y. Feature Selection with Discrete Binary Differential Evolution. In Proceedings of the 2009 International Conference on Artificial Intelligence and Computational Intelligence, Shanghai, China, 7–8 November 2009; pp. 327–330. [Google Scholar] [CrossRef]
- Ghosh, A.; Datta, A.; Ghosh, S. Self-adaptive differential evolution for feature selection in hyperspectral image data. Appl. Soft Comput. 2013, 13, 1969–1977. [Google Scholar] [CrossRef]
- Akbar, A.; Kuanar, A.; Joshi, R.K.; Sandeep, I.S.; Mohanty, S.; Naik, P.K.; Mishra, A.; Nayak, S. Development of Prediction Model and Experimental Validation in Predicting the Curcumin Content of Turmeric (Curcuma longa L.). Front. Plant Sci. 2016, 7, 1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zorarpacı, E.; Özel, S.A. A hybrid approach of differential evolution and artificial bee colony for feature selection. Expert Syst. Appl. 2016, 62, 91–103. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Mishra, S.; Goswami, S. Feature selection using differential evolution with binary mutation scheme. In Proceedings of the 2016 International Conference on Microelectronics, Computing and Communications (MicroCom), Durgapur, India, 23–25 January 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Zhao, W.; Beach, T.H.; Rezgui, Y. A systematic mixed-integer differential evolution approach for water network operational optimization. Proc. R. Soc. A 2018, 474, 20170879. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.S.; Bao, L.L.; Ning, Q.; Ji, J.C.; Zhao, X.W. An Improved Binary Differential Evolution Algorithm for Feature Selection in Molecular Signatures. Mol. Inform. 2018, 37, 1700081. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, K.; Samsonov, A. DEEP—differential evolution entirely parallel method for gene regulatory networks. J. Supercomput. 2011, 57, 172–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozlov, K.; Samsonov, A.M.; Samsonova, M. A software for parameter optimization with Differential Evolution Entirely Parallel method. PeerJ Comput. Sci. 2016, 2, e74. [Google Scholar] [CrossRef] [Green Version]
- Kozlov, K.; Chebotarev, D.; Hassan, M.; Triska, M.; Triska, P.; Flegontov, P.; Tatarinova, T. Differential Evolution approach to detect recent admixture. BMC Genom. 2015, 16, S9. [Google Scholar] [CrossRef] [Green Version]
- Ageev, A.; Aydogan, A.; Bishop-von Wettberg, E.; Nuzhdin, S.V.; Samsonova, M.; Kozlov, K. Simulation Model for Time to Flowering with Climatic and Genetic Inputs for Wild Chickpea. Agronomy 2021, 11, 1389. [Google Scholar] [CrossRef]
- Ageev, A.Y.; Bishop-von Wettberg, E.J.; Nuzhdin, S.V.; Samsonova, M.G.; Kozlov, K.N. Forecasting the Timing of Floral Initiation in Wild Chickpeas under Climate Change. Biophysics 2021, 66, 107–116. [Google Scholar] [CrossRef]
- Kozlov, K.; Singh, A.; Berger, J.; Wettberg, E.B.V.; Kahraman, A.; Aydogan, A.; Cook, D.; Nuzhdin, S.; Samsonova, M. Non-linear regression models for time to flowering in wild chickpea combine genetic and climatic factors. BMC Plant Biol. 2019, 19, 94. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, K.; Sokolkova, A.; Lee, C.R.; Ting, C.T.; Schafleitner, R.; Bishop-von Wettberg, E.; Nuzhdin, S.; Samsonova, M. Dynamical climatic model for time to flowering in Vigna radiata. BMC Plant Biol. 2020, 20, 202. [Google Scholar] [CrossRef]
- Zaharie, D. Parameter Adaptation in Differential Evolution by Controlling the Population Diversity. In Seria Matematica-Informatica, Proceedings of the 4th InternationalWorkshop on Symbolic and Numeric Algorithms for Scientific Computing, Timişoara, Romania, 9–12 October 2002; Petcu, D., Ed.; Analele Universitatii Timisoara: Timisoara, Romania, 2002; Volume XL, pp. 385–397. [Google Scholar]
- Mohamed, A.W.; Almazyad, A.S. Differential Evolution with Novel Mutation and Adaptive Crossover Strategies for Solving Large Scale Global Optimization Problems. Appl. Comput. Intell. Soft Comput. 2017, 2017, 7974218. [Google Scholar] [CrossRef]
- Uher, V.; Gajdoš, P.; Radecký, M.; Snášel, V. Utilization of the Discrete Differential Evolution for Optimization in Multidimensional Point Clouds. Comput. Intell. Neurosci. 2016, 2016, 6329530. [Google Scholar] [CrossRef] [Green Version]
- Kozlov, K. DEEP. Available online: https://gitlab.com/mackoel/deepmethod (accessed on 29 April 2021).
- Noorian, F.; de Silva, A.M.; Leong, P.H.W. gramEvol: Grammatical Evolution in R. J. Stat. Softw. 2016, 71, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Qin, A.; Huang, V.; Suganthan, P. Differential Evolution Algorithm with Strategy Adaptation for Global Numerical Optimization. IEEE Trans. Evol. Comput. 2009, 13, 398–417. [Google Scholar] [CrossRef]
- Wang, S.L.; Morsidi, F.; Ng, T.F.; Budiman, H.; Neoh, S.C. Insights into the effects of control parameters and mutation strategy on self-adaptive ensemble-based differential evolution. Inf. Sci. 2020, 514, 203–233. [Google Scholar] [CrossRef]
- Zhang, J.; Sanderson, A. JADE: Adaptive Differential Evolution with Optional External Archive. IEEE Trans. Evol. Comput. 2009, 13, 945–958. [Google Scholar] [CrossRef]
- Tanabe, R.; Fukunaga, A. Success-history based parameter adaptation for Differential Evolution. In Proceedings of the 2013 IEEE Congress on Evolutionary Computation, Cancun, Mexico, 20–23 June 2013; pp. 71–78. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Li, Y.; Yang, H. Self-adaptive differential evolution algorithm with improved mutation mode. Appl. Intell. 2017, 47, 644–658. [Google Scholar] [CrossRef]
- Li, S.; Gu, Q.; Gong, W.; Ning, B. An enhanced adaptive differential evolution algorithm for parameter extraction of photovoltaic models. Energy Convers. Manag. 2020, 205, 112443. [Google Scholar] [CrossRef]
- Xue, X.; Chen, J. Matching biomedical ontologies through compact differential evolution algorithm. Syst. Sci. Control. Eng. 2019, 7, 85–89. [Google Scholar] [CrossRef] [Green Version]
- Kononova, A.V.; Caraffini, F.; Bäck, T. Differential evolution outside the box. Inf. Sci. 2021, 581, 587–604. [Google Scholar] [CrossRef]
- Qiu, X.; Tan, K.C.; Xu, J.X. Multiple Exponential Recombination for Differential Evolution. IEEE Trans. Cybern. 2017, 47, 995–1006. [Google Scholar] [CrossRef]
- Brest, J.; Zumer, V.; Maucec, M. Self-Adaptive Differential Evolution Algorithm in Constrained Real-Parameter Optimization. In Proceedings of the 2006 IEEE International Conference on Evolutionary Computation, Vancouver, BC, Canada, 16–21 July 2006; pp. 215–222. [Google Scholar] [CrossRef]
- Tian, M.; Gao, X.; Dai, C. Differential evolution with improved individual-based parameter setting and selection strategy. Appl. Soft Comput. 2017, 56, 286–297. [Google Scholar] [CrossRef]
- Meng, Z.; Pan, J.S.; Tseng, K.K. PaDE: An enhanced Differential Evolution algorithm with novel control parameter adaptation schemes for numerical optimization. Knowl.-Based Syst. 2019, 168, 80–99. [Google Scholar] [CrossRef]
- Poláková, R.; Tvrdík, J.; Bujok, P. Differential evolution with adaptive mechanism of population size according to current population diversity. Swarm Evol. Comput. 2019, 50, 100519. [Google Scholar] [CrossRef]
- Zhu, W.; Tang, Y.; Fang, J.A.; Zhang, W. Adaptive population tuning scheme for differential evolution. Inf. Sci. 2013, 223, 164–191. [Google Scholar] [CrossRef]
- Ali, M.Z.; Awad, N.H.; Suganthan, P.N. Multi-population differential evolution with balanced ensemble of mutation strategies for large-scale global optimization. Appl. Soft Comput. 2015, 33, 304–327. [Google Scholar] [CrossRef]
- Ali, M.Z.; Awad, N.H.; Suganthan, P.N.; Reynolds, R.G. An Adaptive Multipopulation Differential Evolution with Dynamic Population Reduction. IEEE Trans. Cybern. 2017, 47, 2768–2779. [Google Scholar] [CrossRef]
- Epitropakis, M.G.; Tasoulis, D.K.; Pavlidis, N.G.; Plagianakos, V.P.; Vrahatis, M.N. Enhancing Differential Evolution Utilizing Proximity-Based Mutation Operators. IEEE Trans. Evol. Comput. 2011, 15, 99–119. [Google Scholar] [CrossRef]
- Islam, S.M.; Das, S.; Ghosh, S.; Roy, S.; Suganthan, P.N. An Adaptive Differential Evolution Algorithm with Novel Mutation and Crossover Strategies for Global Numerical Optimization. IEEE Trans. Syst. Man Cybern. Part B (Cybern.) 2012, 42, 482–500. [Google Scholar] [CrossRef]
- Zheng, L.M.; Zhang, S.X.; Tang, K.S.; Zheng, S.Y. Differential evolution powered by collective information. Inf. Sci. 2017, 399, 13–29. [Google Scholar] [CrossRef]
- Li, Y.; Wang, S.; Yang, B. An improved differential evolution algorithm with dual mutation strategies collaboration. Expert Syst. Appl. 2020, 153, 113451. [Google Scholar] [CrossRef]
- Cheng, J.; Pan, Z.; Liang, H.; Gao, Z.; Gao, J. Differential evolution algorithm with fitness and diversity ranking-based mutation operator. Swarm Evol. Comput. 2021, 61, 100816. [Google Scholar] [CrossRef]
- Ramadas, M.; Abraham, A.; Kumar, S. ReDE-A Revised mutation strategy for Differential Evolution Algorithm. Int. J. Intell. Eng. Syst. 2016, 9, 51–58. [Google Scholar] [CrossRef]
- Peng, H.; Guo, Z.; Deng, C.; Wu, Z. Enhancing differential evolution with random neighbors based strategy. J. Comput. Sci. 2018, 26, 501–511. [Google Scholar] [CrossRef]
- Guo, S.M.; Yang, C.C. Enhancing Differential Evolution Utilizing Eigenvector-Based Crossover Operator. IEEE Trans. Evol. Comput. 2015, 19, 31–49. [Google Scholar] [CrossRef]
- Wang, Y.; Cai, Z.; Zhang, Q. Differential Evolution with Composite Trial Vector Generation Strategies and Control Parameters. IEEE Trans. Evol. Comput. 2011, 15, 55–66. [Google Scholar] [CrossRef]
- Ghosh, A.; Das, S.; Mullick, S.S.; Mallipeddi, R.; Das, A.K. A switched parameter differential evolution with optional blending crossover for scalable numerical optimization. Appl. Soft Comput. 2017, 57, 329–352. [Google Scholar] [CrossRef]
- Gong, W.; Cai, Z. Differential Evolution with Ranking-Based Mutation Operators. IEEE Trans. Cybern. 2013, 43, 2066–2081. [Google Scholar] [CrossRef]
- Wang, Y.; Cai, Z.; Zhang, Q. Enhancing the search ability of differential evolution through orthogonal crossover. Inf. Sci. 2012, 185, 153–177. [Google Scholar] [CrossRef]
- Mallipeddi, R.; Suganthan, P.N. Differential Evolution Algorithm with Ensemble of Parameters and Mutation and Crossover Strategies. In Swarm, Evolutionary, and Memetic Computing; Lecture Notes in Computer, Science; Panigrahi, B.K., Das, S., Suganthan, P.N., Dash, S.S., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 6466, pp. 71–78. [Google Scholar] [CrossRef]
- Zhang, J.; Naik, H.S.; Assefa, T.; Sarkar, S.; Reddy, R.V.C.; Singh, A.; Ganapathysubramanian, B.; Singh, A.K. Computer vision and machine learning for robust phenotyping in genome-wide studies. Sci. Rep. 2017, 7, 44048. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Chen, Z.; Liu, Z. Improved ensemble of differential evolution variants. PLoS ONE 2021, 16, e0256206. [Google Scholar] [CrossRef]
- Zhou, X.G.; Zhang, G.J. Abstract Convex Underestimation Assisted Multistage Differential Evolution. IEEE Trans. Cybern. 2017, 47, 2730–2741. [Google Scholar] [CrossRef]
- Ghosh, A.; Das, S.; Das, A.K.; Gao, L. Reusing the Past Difference Vectors in Differential Evolution—A Simple But Significant Improvement. IEEE Trans. Cybern. 2020, 50, 4821–4834. [Google Scholar] [CrossRef]
- Li, W.; Gong, W. An Improved Multioperator-Based Constrained Differential Evolution for Optimal Power Allocation in WSNs. Sensors 2021, 21, 6271. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Duan, M.; Zhang, X.; Cheng, P. A hybrid differential evolution based on gaining-sharing knowledge algorithm and harris hawks optimization. PLoS ONE 2021, 16, e0250951. [Google Scholar] [CrossRef]
- Jadon, S.S.; Tiwari, R.; Sharma, H.; Bansal, J.C. Hybrid Artificial Bee Colony algorithm with Differential Evolution. Appl. Soft Comput. 2017, 58, 11–24. [Google Scholar] [CrossRef]
- Mohamed, A.W.; Hadi, A.A.; Fattouh, A.M.; Jambi, K.M. LSHADE with semi-parameter adaptation hybrid with CMA-ES for solving CEC 2017 benchmark problems. In Proceedings of the 2017 IEEE Congress on Evolutionary Computation (CEC), Donostia-San Sebastián, Spain, 5–8 June 2017; pp. 145–152. [Google Scholar] [CrossRef]
- Zhao, F.; Xue, F.; Zhang, Y.; Ma, W.; Zhang, C.; Song, H. A hybrid algorithm based on self-adaptive gravitational search algorithm and differential evolution. Expert Syst. Appl. 2018, 113, 515–530. [Google Scholar] [CrossRef]
- Wang, S.; Li, Y.; Yang, H. Self-adaptive mutation differential evolution algorithm based on particle swarm optimization. Appl. Soft Comput. 2019, 81, 105496. [Google Scholar] [CrossRef]
- Luo, J.; Shi, B. A hybrid whale optimization algorithm based on modified differential evolution for global optimization problems. Appl. Intell. 2019, 49, 1982–2000. [Google Scholar] [CrossRef]
- Li, S.; Gong, W.; Wang, L.; Yan, X.; Hu, C. A hybrid adaptive teaching-learning-based optimization and differential evolution for parameter identification of photovoltaic models. Energy Convers. Manag. 2020, 225, 113474. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, R.; Li, C.; Zhang, L.; Cui, Z. Hybrid Symbiotic Differential Evolution Moth-Flame Optimization Algorithm for Estimating Parameters of Photovoltaic Models. IEEE Access 2020, 8, 156328–156346. [Google Scholar] [CrossRef]
- Aarts, E.H.L.; Korst, J.H.M. Boltzmann machines as a model for parallel annealing. Algorithmica 1991, 6, 437–465. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Wang, L.; Zhou, X. A modified Boltzmann Annealing Differential Evolution algorithm for inversion of directional resistivity logging-while-drilling measurements. J. Pet. Sci. Eng. 2020, 188, 106916. [Google Scholar] [CrossRef]
- Ahmadianfar, I.; Kheyrandish, A.; Jamei, M.; Gharabaghi, B. Optimizing operating rules for multi-reservoir hydropower generation systems: An adaptive hybrid differential evolution algorithm. Renew. Energy 2021, 167, 774–790. [Google Scholar] [CrossRef]
- Huo, L.; Zhu, J.; Li, Z.; Ma, M. A Hybrid Differential Symbiotic Organisms Search Algorithm for UAV Path Planning. Sensors 2021, 21, 3037. [Google Scholar] [CrossRef]
- Deng, C.; Dong, X.; Tan, Y.; Peng, H. Enhanced Differential Evolution Algorithm with Local Search Based on Hadamard Matrix. Comput. Intell. Neurosci. 2021, 2021, 8930980. [Google Scholar] [CrossRef]
- Rahnamayan, S.; Tizhoosh, H.; Salama, M. Opposition-Based Differential Evolution. IEEE Trans. Evol. Comput. 2008, 12, 64–79. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Zhang, Q.; Tsang, E. DE/EDA: A new evolutionary algorithm for global optimization. Inf. Sci. 2005, 169, 249–262. [Google Scholar] [CrossRef]
- Peng, H.; Wu, Z.; Deng, C. Enhancing Differential Evolution with Commensal Learning and Uniform Local Search. Chin. J. Electron. 2017, 26, 725–733. [Google Scholar] [CrossRef]
- Guo, H.; Li, Y.; Li, J.; Sun, H.; Wang, D.; Chen, X. Differential evolution improved with self-adaptive control parameters based on simulated annealing. Swarm Evol. Comput. 2014, 19, 52–67. [Google Scholar] [CrossRef]
- Kozlov, K.; Ivanisenko, N.; Ivanisenko, V.; Kolchanov, N.; Samsonova, M.; Samsonov, A.M. Enhanced Differential Evolution Entirely Parallel Method for Biomedical Applications. In Parallel Computing Technologies; Hutchison, D., Kanade, T., Kittler, J., Kleinberg, J.M., Mattern, F., Mitchell, J.C., Naor, M., Nierstrasz, O., Pandu Rangan, C., Steffen, B., et al., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 7979, pp. 409–416. [Google Scholar] [CrossRef]
- Kozlov, K.N.; Baumann, P.; Waldmann, J.; Samsonova, M.G. TeraPro, a system for processing large biomedical images. Pattern Recognit. Image Anal. 2013, 23, 488–497. [Google Scholar] [CrossRef]
- Hammer, G.L.; Vaderlip, R.; Gibson, G.; Wade, L.; Henzell, R.; Younger, D.; Warren, J.; Dale, A. Genotype-by-environment interaction in grain sorghum. II. Effects of temperature and photoperiod on ontogeny. Crop Sci. 1989, 29, 376–384. [Google Scholar] [CrossRef]
- Van Oosterom, E.; Bidinger, F.; Weltzien, E. A yield architecture framework to explain adaptation of pearl millet to environmental stress. Field Crop. Res. 2003, 80, 33–56. [Google Scholar] [CrossRef] [Green Version]
- Whish, J.; Butler, G.; Castor, M.; Cawthray, S.; Broad, I.; Carberry, P.; Hammer, G.; McLean, G.; Routley, R.; Yeates, S. Modelling the effects of row configuration on sorghum yield reliability in north-eastern Australia. Aust. J. Agric. Res. 2005, 56, 11. [Google Scholar] [CrossRef]
- Jordan, D.R.; Tao, Y.Z.; Godwin, I.D.; Henzell, R.G.; Cooper, M.; McIntyre, C.L. Comparison of identity by descent and identity by state for detecting genetic regions under selection in a sorghum pedigree breeding program. Mol. Breed. 2005, 14, 441–454. [Google Scholar] [CrossRef]
- Cooper, M.; van Eeuwijk, F.A.; Hammer, G.L.; Podlich, D.W.; Messina, C. Modeling QTL for complex traits: Detection and context for plant breeding. Curr. Opin. Plant Biol. 2009, 12, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, T.R.; Messina, C.D.; Beatty, A.; Samples, M. Assessment across the United States of the Benefits of Altered Soybean Drought Traits. Agron. J. 2010, 102, 475–482. [Google Scholar] [CrossRef]
- Technow, F.; Messina, C.D.; Totir, L.R.; Cooper, M. Integrating Crop Growth Models with Whole Genome Prediction through Approximate Bayesian Computation. PLoS ONE 2015, 10, e0130855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boote, K.J.; Jones, J.W.; Hoogenboom, G. Incorporating realistic trait physiology into crop growth models to support genetic improvement. Silico Plants 2021, 3, diab002. [Google Scholar] [CrossRef]
- Asseng, S.; van Herwaarden, A.F. Analysis of the benefits to wheat yield from assimilates stored prior to grain filling in a range of environments. Plant Soil 2003, 256, 217–229. [Google Scholar] [CrossRef]
- Hammer, G.L.; Chapman, S.; van Oosterom, E.; Podlich, D.W. Trait physiology and crop modelling as a framework to link phenotypic complexity to underlying genetic systems. Aust. J. Agric. Res. 2005, 56, 947. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Solberg, T.R.; Shepherd, R.; Woolliams, J.A. A fast algorithm for BayesB type of prediction of genome-wide estimates of genetic value. Genet. Sel. Evol. 2009, 41, 2. [Google Scholar] [CrossRef] [Green Version]
- De los Campos, G.; Hickey, J.M.; Pong-Wong, R.; Daetwyler, H.D.; Calus, M.P.L. Whole-Genome Regression and Prediction Methods Applied to Plant and Animal Breeding. Genetics 2013, 193, 327–345. [Google Scholar] [CrossRef] [Green Version]
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Genome J. 2011, 4, 250. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Lado, B.; Vázquez, D.; Quincke, M.; Silva, P.; Aguilar, I.; Gutiérrez, L. Resource allocation optimization with multi-trait genomic prediction for bread wheat (Triticum aestivum L.) baking quality. Theor. Appl. Genet. 2018, 131, 2719–2731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangin, B.; Rincent, R.; Rabier, C.E.; Moreau, L.; Goudemand-Dugue, E. Training set optimization of genomic prediction by means of EthAcc. PLoS ONE 2019, 14, e0205629. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, K.; Gursky, V.; Kulakovskiy, I.; Samsonova, M. Sequence-based model of gap gene regulatory network. BMC Genom. 2014, 15, S6. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.; Signor, S.A.; Kozlov, K.; Nuzhdin, S.V. Novel approach to quantitative spatial gene expression uncovers genetic stochasticity in the developing Drosophila eye. Evol. Dev. 2019, 21, 157–171. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, M. Grammatical Evolution. IEEE Trans. Evol. Comput. 2001, 5, 349–358. [Google Scholar] [CrossRef] [Green Version]
- Scrucca, L. On Some Extensions to GA Package: Hybrid Optimisation, Parallelisation and Islands Evolution. R J. 2017, 9, 20. [Google Scholar] [CrossRef] [Green Version]
- Guyon, I.; Elisseeff, A. An Introduction to Variable and Feature Selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Kohavi, R.; John, G.H. Wrappers for feature subset selection. Artif. Intell. 1997, 97, 273–324. [Google Scholar] [CrossRef] [Green Version]
- Furey, T.S.; Cristianini, N.; Duffy, N.; Bednarski, D.W.; Schummer, M.; Haussler, D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics 2000, 16, 906–914. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. A Comparative Study on Feature Selection Methods for Drug Discovery. J. Chem. Inf. Comput. Sci. 2004, 44, 1823–1828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meinshausen, N.; Bühlmann, P. Stability selection: Stability Selection. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2010, 72, 417–473. [Google Scholar] [CrossRef]
- Li, X.J.; Hayward, C.; Fong, P.Y.; Dominguez, M.; Hunsucker, S.W.; Lee, L.W.; McLean, M.; Law, S.; Butler, H.; Schirm, M.; et al. A Blood-Based Proteomic Classifier for the Molecular Characterization of Pulmonary Nodules. Sci. Transl. Med. 2013, 5, 207ra142. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| 8 | 22 | 8 | 15 | 8 | 84 | 47 | 105 | ||
|---|---|---|---|---|---|---|---|---|---|
| helper | 1 | 1 | 3 | 1 | … | 3 | 2 | 2 | 2 |
| sorted | 1 | 1 | 1 | 2 | … | 2 | 2 | 3 | 3 |
| 8 | 22 | 15 | 84 | … | 47 | 105 | 8 | 8 |
| Method | Mean EthAcc | p-Value for Comparison with New Method |
|---|---|---|
| new | 0.75 | |
| DE/rand/1/bin | 0.54 | <2.2 |
| DE/rand/1/exp | 0.56 | < |
| DE/best/1/bin | 0.57 | 8.15 |
| DE/best/1/exp | 0.57 | 4.20 |
| GA/rvd | 0.63 | 4.31 |
| GA/bin | 0.62 | 9.44 |
| GA/rvd/loc | 0.57 | 2.36 |
| GA/bin/loc | 0.52 | 4.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salihov, S.; Maltsov, D.; Samsonova, M.; Kozlov, K. Solution of Mixed-Integer Optimization Problems in Bioinformatics with Differential Evolution Method. Mathematics 2021, 9, 3329. https://doi.org/10.3390/math9243329
Salihov S, Maltsov D, Samsonova M, Kozlov K. Solution of Mixed-Integer Optimization Problems in Bioinformatics with Differential Evolution Method. Mathematics. 2021; 9(24):3329. https://doi.org/10.3390/math9243329
Chicago/Turabian StyleSalihov, Sergey, Dmitriy Maltsov, Maria Samsonova, and Konstantin Kozlov. 2021. "Solution of Mixed-Integer Optimization Problems in Bioinformatics with Differential Evolution Method" Mathematics 9, no. 24: 3329. https://doi.org/10.3390/math9243329
APA StyleSalihov, S., Maltsov, D., Samsonova, M., & Kozlov, K. (2021). Solution of Mixed-Integer Optimization Problems in Bioinformatics with Differential Evolution Method. Mathematics, 9(24), 3329. https://doi.org/10.3390/math9243329






