Rapid Polyp Classification in Colonoscopy Using Textural and Convolutional Features
Abstract
:1. Introduction
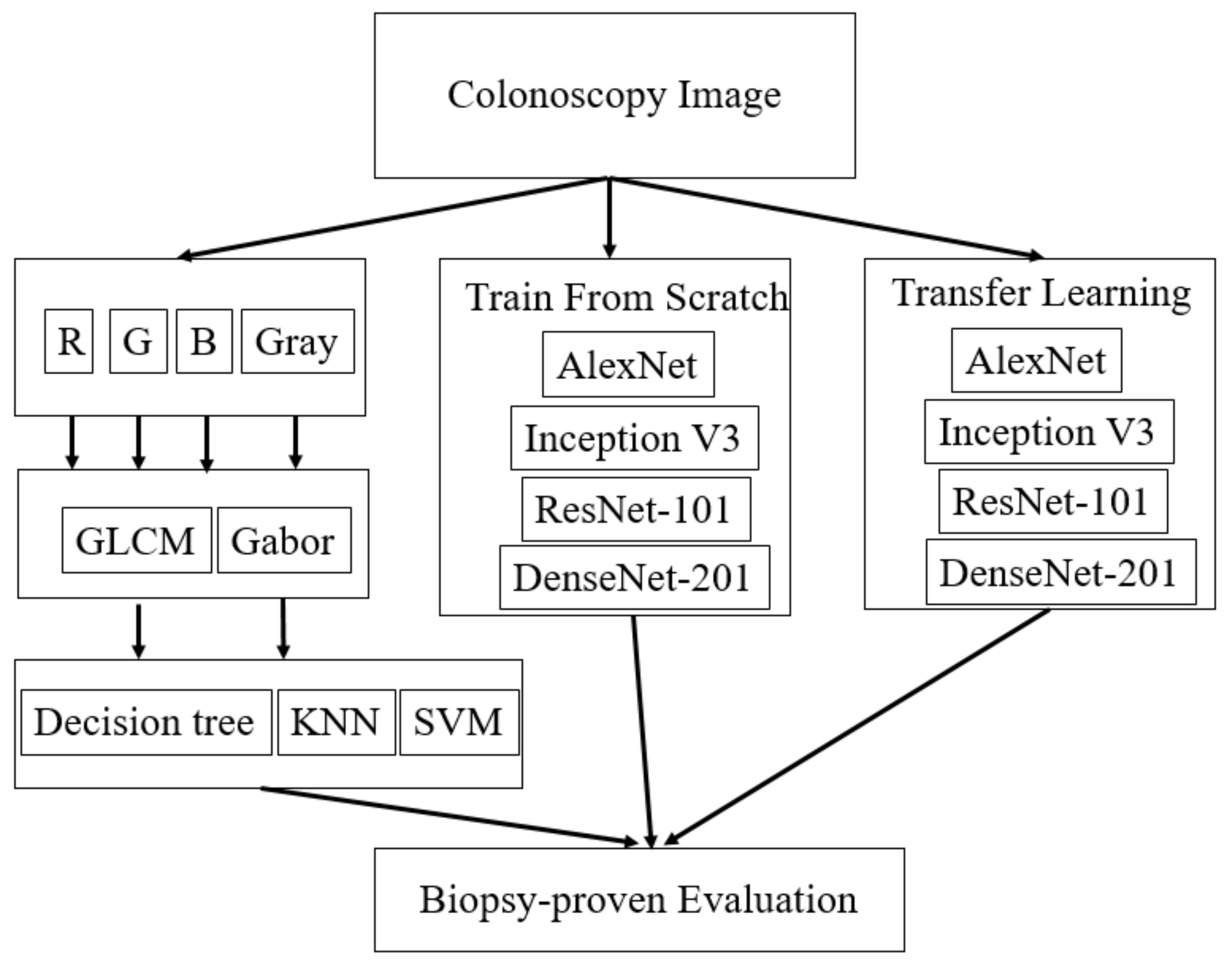
2. Materials and Methods
2.1. Colonoscopy Images
2.2. Textural Features
2.3. DCNN Features
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Siegel, R.L.; Fedewa, S.A.; Anderson, W.F.; Miller, K.D.; Ma, J.; Rosenberg, P.S.; Jemal, A. Colorectal Cancer Incidence Patterns in the United States, 1974–2013. J. Natl. Cancer Inst. 2017, 109, djw322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. World Cancer Report; World Health Organization Press: Geneva, Switzerland, 2014; Chapter 5.5. [Google Scholar]
- Institute, N.C. Colorectal Cancer Prevention (PDQ®)—Health Professional Version; North Carolina Institute: Chapel Hill, NC, USA, 2014. [Google Scholar]
- Hiramatsu, K.; Sakata, H.; Horita, Y.; Orita, N.; Kida, A.; Mizukami, A.; Miyazawa, M.; Hirai, S.; Shimatani, A.; Matsuda, K.; et al. Mesenteric phlebosclerosis associated with long-term oral intake of geniposide, an ingredient of herbal medicine. Aliment. Pharmacol. Ther. 2012, 36, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Carethers, J.M.; Jung, B.H. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology 2015, 149, 1177–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrne, M.F.; Chapados, N.; Soudan, F.; Oertel, C.; Linares Pérez, M.; Kelly, R.; Iqbal, N.; Chandelier, F.; Rex, D.K. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 2019, 68, 94–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corley, D.A.; Jensen, C.D.; Marks, A.R.; Zhao, W.K.; Lee, J.K.; Doubeni, C.A.; Zauber, A.G.; de Boer, J.; Fireman, B.H.; Schottinger, J.E.; et al. Adenoma detection rate and risk of colorectal cancer and death. N. Engl. J. Med. 2014, 370, 1298–1306. [Google Scholar] [CrossRef] [Green Version]
- Mann, R.; Gajendran, M.; Umapathy, C.; Perisetti, A.; Goyal, H.; Saligram, S.; Echavarria, J. Endoscopic Management of Complex Colorectal Polyps: Current Insights and Future Trends. Front. Med. 2022, 8, 728704. [Google Scholar] [CrossRef] [PubMed]
- Zauber, A.G.; Winawer, S.J.; O’Brien, M.J.; Lansdorp-Vogelaar, I.; van Ballegooijen, M.; Hankey, B.F.; Shi, W.; Bond, J.H.; Schapiro, M.; Panish, J.F.; et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N. Engl. J. Med. 2012, 366, 687–696. [Google Scholar] [CrossRef]
- Shalman, D.; Gerson, L.B. Systematic review with meta-analysis: The risk of gastrointestinal haemorrhage post-polypectomy in patients receiving anti-platelet, anti-coagulant and/or thienopyridine medications. Aliment. Pharmacol. Ther. 2015, 42, 949–956. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H.; Nappi, J.; MacEneaney, P.; Rubin, D.T.; Dachman, A.H. Computer-aided diagnosis scheme for detection of polyps at CT colonography. Radiographics 2002, 22, 963–979. [Google Scholar] [CrossRef]
- Tan, J.; Gao, Y.; Cao, W.; Pomeroy, M.; Zhang, S.; Huo, Y.; Li, L.; Liang, Z. GLCM-CNN: Gray Level Co-occurrence Matrix based CNN Model for Polyp Diagnosis. In Proceedings of the 2019 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Chicago, IL, USA, 19–22 May 2019; pp. 1–4. [Google Scholar]
- Lee, J.-G.; Hyo Kim, J.; Hyung Kim, S.; Sun Park, H.; Ihn Choi, B. A straightforward approach to computer-aided polyp detection using a polyp-specific volumetric feature in CT colonography. Comput. Biol. Med. 2011, 41, 790–801. [Google Scholar] [CrossRef]
- Sánchez-González, A.; García-Zapirain, B.; Sierra-Sosa, D.; Elmaghraby, A. Automatized colon polyp segmentation via contour region analysis. Comput. Biol. Med. 2018, 100, 152–164. [Google Scholar] [CrossRef]
- Pacal, I.; Karaman, A.; Karaboga, D.; Akay, B.; Basturk, A.; Nalbantoglu, U.; Coskun, S. An efficient real-time colonic polyp detection with YOLO algorithms trained by using negative samples and large datasets. Comput. Biol. Med. 2022, 141, 105031. [Google Scholar] [CrossRef]
- Pacal, I.; Karaboga, D. A robust real-time deep learning based automatic polyp detection system. Comput. Biol. Med. 2021, 134, 104519. [Google Scholar] [CrossRef]
- Hu, K.; Zhao, L.; Feng, S.; Zhang, S.; Zhou, Q.; Gao, X.; Guo, Y. Colorectal polyp region extraction using saliency detection network with neutrosophic enhancement. Comput. Biol. Med. 2022, 147, 105760. [Google Scholar] [CrossRef]
- Ay, B.; Turker, C.; Emre, E.; Ay, K.; Aydin, G. Automated classification of nasal polyps in endoscopy video-frames using handcrafted and CNN features. Comput. Biol. Med. 2022, 147, 105725. [Google Scholar] [CrossRef]
- Cai, W.-L.; Hong, G.-B. Quantitative image analysis for evaluation of tumor response in clinical oncology. Chronic Dis. Transl. Med. 2018, 4, 18–28. [Google Scholar] [CrossRef]
- Yang, Q.; Li, L.; Zhang, J.; Shao, G.; Zheng, B. A new quantitative image analysis method for improving breast cancer diagnosis using DCE-MRI examinations. Med. Phys. 2015, 42, 103–109. [Google Scholar] [CrossRef]
- Moreno, P.; Bernardino, A.; Santos-Victor, J. Gabor Parameter Selection for Local Feature Detection. In Iberian Conference on Pattern Recognition and Image Analysis; Springer: Berlin/Heidelberg, Germany, 2005; pp. 11–19. [Google Scholar]
- Buciu, I.; Gacsadi, A. Gabor wavelet based features for medical image analysis and classification. In Proceedings of the 2009 2nd International Symposium on Applied Sciences in Biomedical and Communication Technologies, Bratislava, Slovakia, 24–27 November 2009; pp. 1–4. [Google Scholar]
- Chaddad, A.; Desrosiers, C.; Bouridane, A.; Toews, M.; Hassan, L.; Tanougast, C. Multi Texture Analysis of Colorectal Cancer Continuum Using Multispectral Imagery. PLoS ONE 2016, 11, e0149893. [Google Scholar] [CrossRef]
- Haghighat, M.; Zonouz, S.; Abdel-Mottaleb, M. CloudID: Trustworthy cloud-based and cross-enterprise biometric identification. Expert Syst. Appl. 2015, 42, 7905–7916. [Google Scholar] [CrossRef]
- Jolliffe, I. Principal Component Analysis. In Encyclopedia of Statistics in Behavioral Science; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2005. [Google Scholar] [CrossRef]
- Gales, M.a.S.Y. The application of hidden Markov models in speech recognition. Found. Trends Signal Process. 2008, 1, 195–304. [Google Scholar] [CrossRef]
- Bengio, Y.; Courville, A.; Vincent, P. Representation learning: A review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef] [PubMed]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [Green Version]
- Boyd, S.; Parikh, N.; Chu, E.; Peleato, B.; Eckstein, J. Distributed optimization and statistical learning via the alternating direction method of multipliers. Found. Trends Mach. Learn. 2011, 3, 1–122. [Google Scholar] [CrossRef]
- Han, S.; Pool, J.; Tran, J.; Dally, W. Learning both weights and connections for efficient neural network. In Advances in Neural Information Processing Systems; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 2015. [Google Scholar]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Yuan, Z.-W.; Zhang, J. Feature Extraction and Image Retrieval Based on AlexNet; SPIE: Bellingham, WA, USA, 2016; Volume 10033. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Zhai, Y.; Fu, J.; Lu, Y.; Li, H. Feature Selective Networks for Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Huang, G.; Liu, S.; Van der Maaten, L.; Weinberger, K.Q. Condensenet: An efficient densenet using learned group convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 2752–2761. [Google Scholar]
- Pogorelov, K.; Ostroukhova, O.; Jeppsson, M.; Espeland, H.; Griwodz, C.; Lange, T.d.; Johansen, D.; Riegler, M.; Halvorsen, P. Deep Learning and Hand-Crafted Feature Based Approaches for Polyp Detection in Medical Videos. In Proceedings of the 2018 IEEE 31st International Symposium on Computer-Based Medical Systems (CBMS), Karlstad, Sweden, 18–21 June 2018; pp. 381–386. [Google Scholar]
- Liu, X.; Faes, L.; Kale, A.U.; Wagner, S.K.; Fu, D.J.; Bruynseels, A.; Mahendiran, T.; Moraes, G.; Shamdas, M.; Kern, C.; et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit. Health 2019, 1, e271–e297. [Google Scholar] [CrossRef]
- Hsu, Y.-C.; Wu, T.-C.; Lo, Y.-C.; Wang, L.-S. Gastrointestinal complications and extraintestinal manifestations of inflammatory bowel disease in Taiwan: A population-based study. J. Chin. Med. Assoc. 2017, 80, 56–62. [Google Scholar] [CrossRef] [Green Version]
- Torres, J.; Mehandru, S.; Colombel, J.-F.; Peyrin-Biroulet, L. Crohn’s disease. Lancet 2017, 389, 1741–1755. [Google Scholar] [CrossRef]
- Beaugerie, L.; Itzkowitz, S.H. Cancers Complicating Inflammatory Bowel Disease. N. Engl. J. Med. 2015, 372, 1441–1452. [Google Scholar] [CrossRef]




| Model Type | Accuracy | Feature |
|---|---|---|
| Ensemble Bagged Trees | 75.6% | GLCM_B |
| Coarse KNN | 75.0% | GLCM_B |
| Ensemble Booted Trees | 73.9% | GLCM_G |
| Ensemble RUSBooted Trees | 73.5% | Gabor_B |
| Quadratic SVM | 72.8% | GLCM_B |
| Train from Scratch | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| Alex | 96.4% | 95.7% | 97.2% |
| Inception-V3 | 82.4% | 78.7% | 85.9% |
| ResNet-101 | 80.6% | 87.2% | 74.5% |
| DenseNet-201 | 87.4% | 86.2% | 87.7% |
| Transfer Learning | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| Alex | 81.3% | 90.4% | 72.6% |
| Inception-V3 | 78.2% | 67.0% | 87.7% |
| ResNet-101 | 85.3% | 81.9% | 87.7% |
| DenseNet-201 | 87.7% | 83.0% | 91.5% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo, C.-M.; Yeh, Y.-H.; Tang, J.-H.; Chang, C.-C.; Yeh, H.-J. Rapid Polyp Classification in Colonoscopy Using Textural and Convolutional Features. Healthcare 2022, 10, 1494. https://doi.org/10.3390/healthcare10081494
Lo C-M, Yeh Y-H, Tang J-H, Chang C-C, Yeh H-J. Rapid Polyp Classification in Colonoscopy Using Textural and Convolutional Features. Healthcare. 2022; 10(8):1494. https://doi.org/10.3390/healthcare10081494
Chicago/Turabian StyleLo, Chung-Ming, Yu-Hsuan Yeh, Jui-Hsiang Tang, Chun-Chao Chang, and Hsing-Jung Yeh. 2022. "Rapid Polyp Classification in Colonoscopy Using Textural and Convolutional Features" Healthcare 10, no. 8: 1494. https://doi.org/10.3390/healthcare10081494
APA StyleLo, C.-M., Yeh, Y.-H., Tang, J.-H., Chang, C.-C., & Yeh, H.-J. (2022). Rapid Polyp Classification in Colonoscopy Using Textural and Convolutional Features. Healthcare, 10(8), 1494. https://doi.org/10.3390/healthcare10081494






