1. Introduction
Cardiovascular disease is responsible for the deaths of approximately 17.9 million people per year, accounting for 31% of all global fatalities [
1]. Cardiovascular disease has become the disease with the highest mortality rate and has seriously threatened human life and health. Electrocardiogram (ECG) is the most widely used noninvasive heart disease diagnosis technology. The ECG signal represents electrical changes during the cardiac cycle and can be recorded with the surface electrodes. Analysis of ECG [
2,
3,
4] signals allows doctors to gather important insights into the health condition of patients and to promptly identify heart abnormalities, thereby prolonging life and improving quality of life through appropriate treatment.
With the rapid advancement of mobile devices for ECG monitoring, automatic interpretation of ECG recordings is becoming more and more important for the early detection and diagnosis of arrhythmias [
5]. In particular, many effective arrhythmia classification methods for single-lead ECG have been proposed [
6,
7,
8], but single-lead ECG alone is not enough to accurately diagnose various heart diseases. 12-lead ECG can comprehensively evaluate cardiac electrical activity (including arrhythmia and myocardial infarction) and each lead reflects heart states through electrical signal changes from different perspectives. Therefore, as a standard clinical ECG examination, 12-lead ECG has attracted more and more interest from researchers [
9,
10,
11,
12,
13].
Over the past few years, various machine learning methods have been employed to analyze ECG signals, such as decision tree [
14], support vector machine [
15] and hidden Markov model [
16]. The most critical part of these methods is to extract discriminant information from original ECG data, i.e., feature extraction. In order to extract features, many methods have been proposed, which can be divided into two categories: manual approaches [
17,
18] and automatic approaches [
19,
20,
21]. The manual approaches mainly rely on professional medical knowledge and the rich expertise of cardiologists. For example, Martis et al. extracted features such as RR interval, R peak amplitude and QRS duration from the original ECG data and used decision trees to diagnose arrhythmia types [
21]. In addition, wavelet transform [
22], short-time Fourier transform and principal component analysis [
23] are often used to extract time-frequency features. It is difficult to obtain satisfying performance for manual approaches because ECG features such as PR interval show a large diversity for different individuals.
Deep learning [
24,
25,
26,
27] is an efficient feature learning method that employs neural networks for mining useful features from a large amount of data. Due to its ability to effectively express unstructured data with non-linear representation, deep learning has been successfully applied in a variety of fields including computer vision, speech recognition and natural language processing [
28,
29,
30]. Due to its powerful ability, many researchers have used deep learning-based methods for ECG classification [
31,
32,
33,
34], which show higher accuracy than traditional methods. For example, Rahhal et al. [
29] proposed to use continuous wavelet transform to convert one-dimensional ECG signals into two-dimensional images and then used convolutional neural network (CNN) to extract useful features of images, yielding a high accuracy for identifying abnormal ventricular beats. Hannun et al. [
34] built a 34-layer CNN network and obtained a cardiologist-level F1 score of 0.837 for classification of twelve arrhythmias. Other common deep learning models that have been used for ECG classification include Long-Short Term Memory (LSTM) network [
35], Recurrent Neural Network (RNN) [
36] and bidirectional LSTM network [
37].
Although deep learning methods have achieved promising performance in ECG classification, they still have some problems. The feature map derived from different leads of the 12-lead ECG signal may exhibit varying levels of contributions for detecting arrhythmias. Specifically, the spatial characteristics of arrhythmias, such as the morphology of waveform, may differ between leads. For example, atrial fibrillation is most obvious in leads II and V1 [
38]. In addition, previous studies mainly focus on one ECG classification task, though there are many kinds of ECG tasks (e.g., anomaly detection, heartbeat classification, ECG delineation and ECG diagnosis) in the real world and there exist correlations between tasks, which indicates that the combination of different tasks is conducive to learning better ECG representations [
39].
Multi-task learning has achieved great success and has performed well in many fields, such as natural language processing [
40], speech recognition [
41], computer vision [
42] and face verification [
43]. In this paper, we propose a deep multi-task learning [
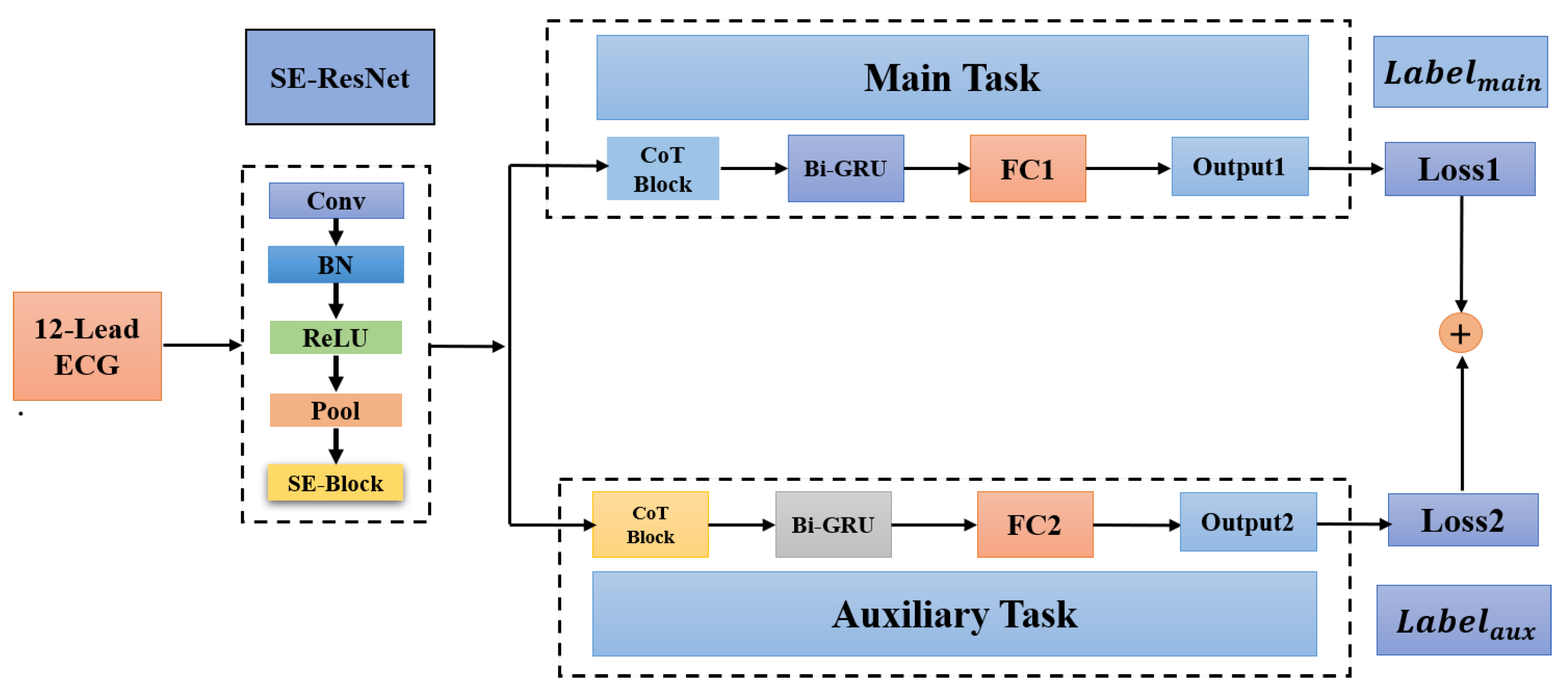
44] method for ECG classification. Specifically, we first construct a related auxiliary classification task with a corresponding dataset by merging similar classes (e.g., left bundle branch block and right bundle branch block) or splitting large classes. We propose a multi-task deep neural network, which includes a shared low-level feature extraction module (i.e., SE-ResNet) and a task-specific classification module. The main contributions of this study are as follows:
A new multi-task network consisting of a shared low-level feature extraction module and a task-specific classification module is proposed for ECG classification.
We propose two strategies to create auxiliary tasks, which exploit the hierarchical class information to achieve feature sharing between the main task and the auxiliary task.
Contextual Transformer (CoT) block is first introduced in ECG classification to solve the problem that the self-attention mechanism only focuses on the local information of the ECG sequence.
5. Discussion
Experimental results demonstrate the effectiveness of our proposed method. As shown in
Table 5 and
Figure 5, our method produced an overall F1 score of 0.833 and 0.827 on the PTB-XL and CPSC2018 datasets, respectively, which are higher than the optimal F1 scores produced by other methods by 0.01 and 0.014. Among different classes of the CPSC2018 dataset, as shown in
Table 5, our method shows advantages for SNR, LBBB, STD, STE and is inferior to other methods for AF, PVC, etc. In addition, as shown in
Table 3, the F1 score of MI is lower than the other four superclasses. Therefore, we need to try different preprocessing techniques (including different sampling rates or noise reduction methods, etc.) to improve the classification performance of MI in the next work.
We find that the auxiliary task has an impact on the main task. For example, LBBB and RBBB belong to the same superclass in the auxiliary task and our method yields similar scores (0.937 and 0.939) for them, while there is a large lap between the two scores for other methods. When the correlation between the main task and auxiliary task is high, the shared SE-ResNet can be well trained in parallel by two tasks. As shown in
Table 6 and
Table 7, by contrast, the correlation between the main task and random auxiliary task is relatively small, the advantage of multi-task learning cannot be exhibited, indicating the effectiveness of the proposed strategy for creating auxiliary tasks.
Table 8,
Table 9,
Table 10 and
Table 11 demonstrate the effectiveness of the CoT attention mechanism and the Bi-GRU layer. CoT block can simultaneously extract key features of static local information and global dynamic information from ECG sequences. Although the Bi-GRU module also plays an important role in processing sequence information, the accuracy drop caused by removing the Bi-GRU layer is not as much as that caused by removing the CoT block.
In addition, to make the results more distinct, we use three decimals consistently for all results including F1 scores in this study. Two decimals may lead to indistinguishable comparison, e.g., three methods would have the same F1 score of 0.81 in
Table 5. Although a value at the third decimal may be insignificant in the statistical sense, many studies still presented F1 scores with three decimals for better comparison of different methods [
11,
30,
47].
6. Conclusions
In this work, a new multi-task deep neural network, which includes a shared low-level feature extraction module (i.e., SE-ResNet) and a task-specific classification module, is proposed. Contextual Transformer (CoT) block is introduced in the classification module to solve the problem that the self-attention mechanism only focuses on the local information of the ECG feature sequence. Thus, the proposed method can dynamically model the local and global information of the ECG feature sequence. In addition, we propose to create auxiliary tasks by merging similar classes or splitting large classes, which exploit the hierarchical class information. The proposed method produced an overall F1 score of 0.833 and 0.827 on the PTB-XL and CPSC2018 datasets, respectively, which are higher than the optimal F1 scores produced by other methods by 0.01 and 0.014, suggesting the effectiveness of our method. However, our multi-task method still has some limitations. Specifically, our model shows bad performance in detecting some abnormal classes, such as detecting the superclass MI on the PTB-XL dataset. In addition, we only use hard parameter sharing to achieve multi-tasking without considering the soft parameter implementation. Finally, we hope that the arrhythmia detection method proposed in this paper can play a role in the process of early diagnosis of cardiovascular diseases. In the future, we will consider integrating more auxiliary tasks for improving the performance of the main task and investigate other approaches to creating auxiliary tasks. With more tasks, we also need to improve the network structure for computational efficiency.