A Novel Method for COVID-19 Diagnosis Using Artificial Intelligence in Chest X-ray Images
Abstract
:1. Introduction
- The proposed deep-learning model consisted of various efficient modules for COVID-19 classification. The inception residual deep-learning model inspires them. We presented different inception residual blocks that cater to information using different depths feature maps at different scales with other layers. The features are concatenated at each proposed classification block, using average pooling and concatenated features to the fully connected layer. The efficient proposed deep-learning blocks used different regularization techniques to minimize the overfitting due to the small COVID-19 dataset.
- The multiscale features are extracted at different levels of the proposed DL model and embed these features to different ML algorithms to validate the combination of DL and ML models.
- The RISE-based python-based library was used to visualize the activation of feature maps, and also, the SHAP library was used to check the importance of features extracted from the deep-learning model. The visualization results showed the convergence region receiver operating characteristics (ROC), and precision–recall curves showed our proposed technique performance and validation.
2. Method
2.1. Data Collection
2.2. Development of System Architecture: For Discussion
2.2.1. Contribution Using First Approach
2.2.2. Contribution Using the Second Approach
- The objective of the second approach is to combine deep-learning and machine-learning models.
- The features are extracted from the proposed deep-learning model and passed features to the traditional machine-learning model for classification of COVID-19.
- The multiscale features are extracted from various blocks from proposed Inception-ResNet blocks are concatenated, and after the concatenation of these features are used in machine-learning classifiers.
2.2.3. Approach 1: Proposed CoVIRNet Deep Learning
2.2.4. Approach 2: Deep-Feature-Extraction and Machine-Learning Models
Random Forest
Bagging Tree
Gradient Boosting Classifier
Perceptron Model Multilayer
Logistic Regression
3. Results and Discussions
3.1. Data Visualization
3.2. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization, Coronavirus 2020. Available online: https://www.who.int/health-topics/coronavirus#tab=tab_1 (accessed on 10 July 2020).
- Wuhan Municipal Health Commission, Report on Unexplained Viral Pneumonia. Available online: http://wjw.wuhan.gov.cn/front/web/showDetail/2020010509020 (accessed on 25 January 2020).
- Tsang, T.K.; Wu, P.; Lin, Y.; Lau, E.H.; Leung, G.M.; Cowling, B.J. Effect of changing case definitions for COVID-19 on the epidemic curve and transmission parameters in mainland China: A modeling study. Lancet Public Health 2020, 5, e289–e296. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.W. A novel coronavirus from patients with pneumonia in China 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Coronavirus Symptoms and How to Protect Yourself: What We Know 2020. Available online: https://www.wsj.com/articles/what-we-know-about-the-coronavirus-11579716128?mod=theme_coronavirus-ribbon (accessed on 12 July 2020).
- COVID-19 Pandemic in Locations with a Humanitarian Response Plan 2020. Available online: https://data.humdata.org/dataset/coronavirus-covid-19-cases-data-for-china-and-the-rest-of-the-world (accessed on 12 July 2020).
- WHO-Covid 19. Available online: https://covid19.who.int/ (accessed on 12 July 2020).
- Agrawal, R.; Prabakaran, S. Big data in digital healthcare: Lessons learnt and recommendations for general practice. Heredity 2020, 124, 525–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinuesa, R.; Azizpour, H.; Leite, I.; Balaam, M.; Dignum, V.; Domisch, S.; Felländer, A.; Langhans, S.D.; Nerini, F. The role of artificial intelligence in achieving the Sustainable Development Goals. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Desai, A.; Warner, J.; Kuderer, N.; Thompson, M.; Painter, C.; Lyman, G.; Gilberto Lopes, G. Crowdsourcing a crisis response for COVID-19 in oncology. Nat. Cancer 2020, 1, 473–476. [Google Scholar]
- Butt, C.; Gill, J.; Chun, D.; Babu, B.A. Deep Learning System to Screen Coronavirus Disease 2019 Pneumonia. Appl. Intell. 2020, 22, 1–7. [Google Scholar]
- Zu, Z.Y.; Jiang, M.D.; Xu, P.-P.; Chen, W. Coronavirus Disease 2019 (COVID-19): A Perspective from China. Radiology 2020, 296, 2. [Google Scholar] [CrossRef] [Green Version]
- Ophir, G.; Maayan, F.-A.; Greenspan, H.; Browning, P.D.; Zhang, H.; Ji, W.; Bernheim, A.; Siegel, E. Rapid AI Development Cycle for the Coronavirus (COVID-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring using Deep Learning CT Image Analysis. arXiv 2020, arXiv:2003.05037. [Google Scholar]
- Jin, C.; Chen, W. Development and Evaluation of an AI System for COVID-19 Diagnosis. MedRxiv 2020. [Google Scholar] [CrossRef]
- Nguyen, T.T. Artificial Intelligence in the Battle against Coronavirus (COVID-19): A Survey and Future Research Directions Thepeninsula.org. Artif. Intell. 2020. [Google Scholar] [CrossRef]
- Allam, Z.; Dey, G.; Jones, D.S. Artificial Intelligence (AI) Provided Early Detection of the Coronavirus (COVID-19) in China and Will Influence Future Urban Health Policy Internationally. Artif. Intell. 2020, 1, 9. [Google Scholar] [CrossRef] [Green Version]
- Khalifa, N.E.M.; Taha, M.H.N.; Hassanien, A.E.; Elghamrawy, S. Detection of Coronavirus (COVID-19) Associated Pneumonia based on Generative Adversarial Networks and a Finetuned Deep Transfer Learning Model using Chest X-ray Dataset. arXiv 2020, arXiv:2004.01184. [Google Scholar]
- Wang, W.; Xu, Y.; Gao, R.; Lu, K.; Han, G.; Wu, W.T. Detection of SARS–CoV-2 in different types of clinical specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [Green Version]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020, 23, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Bernheim, A.; Mei, X.; Huang, M.; Yang, Y.; Fayad, Z.A.; Zhang, N.; Diao, K.; Zhu, B.L.X.; Shan, H.; Chung, M. Chest CT findings in coronavirus disease-19(COVID-19): Relationship to duration of infection. Radiology 2020, 295, 200463. [Google Scholar] [CrossRef] [Green Version]
- Irfan, M.; Iftikhar, M.A.; Yasin, S.; Draz, U.; Ali, T.; Hussain, S.; Bukhari, S.; Alwadie, A.S.; Rahman, S.; Glowacz, A.; et al. Role of Hybrid Deep Neural Networks (HDNNs), Computed Tomography, and Chest X-rays for the Detection of COVID-19. Int. J. Environ. Res. Public Health 2021, 18, 3056. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology 2020. [Google Scholar] [CrossRef]
- Xie, X.; Zhong, Z.; Zhao, W.; Zheng, C.; Wang, F.; Liu, J. Chest CT for typical 2019-nCoV pneumonia: Relationship to negative RT-PCR testing. Radiology 2020. [Google Scholar] [CrossRef] [Green Version]
- Das, A.K.; Ghosh, S.; Thunder, S.; Dutta, R.; Agarwal, S.; Chakrabarti, A. Automatic COVID-19 detection from X-ray images using ensemble learning with convolutional neural network. Pattern Anal. Applic. 2021. [Google Scholar] [CrossRef]
- Arora, N.; Banerjee, A.K.; Narasu, M.L. The role of artificial intelligence in tackling COVID-19. Future Virol. 2020, 15. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. Chex-net: Radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Wang, H.; Xia, Y. Chest-net: A deep neural network for classification of thoracic diseasesonchestradiography. arXiv 2018, arXiv:1807.03058. [Google Scholar]
- Hemdan, E.E.-D.; Shouman, M.A.; Karar, M.E. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in X-ray images. arXiv 2003, arXiv:2003.11055. [Google Scholar]
- Apostolopoulos, I.D.; Bessiana, T. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 6. [Google Scholar] [CrossRef] [Green Version]
- Fenga, L.; Gaspari, M. Predictive Capacity of COVID-19 Test Positivity Rate. Sensors 2021, 21, 2435. [Google Scholar] [CrossRef]
- Saifur, R. The Development of Deep Learning AI based Facial Expression Recognition Technique for Identifying the Patients With Suspected Coronavirus—Public Health Issues in the Context of the COVID-19 Pandemic session. Infect. Dis. Epidemiol. 2021. [Google Scholar] [CrossRef]
- Cohen, J.P.; Morrison, P.; Dao, L. COVID-19 Image Data Collection. 2003. Available online: https://github.com/ieee8023/covid-chestxray-dataset (accessed on 21 August 2020).
- Chest X-ray Images (Pneumonia). Available online: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia (accessed on 1 April 2020).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. IEEE conference on computer vision and pattern recognition, Research on Reservoir Lithology Prediction Based on Residual Neural Network and Squeeze-and- Excitation Neural Network. Int. J. Comput. Inf. Eng. 2020, 14, 770–778. [Google Scholar]
- Szegedy, C.; Sergey, I.; Vincent, V.; Alex, A. Inception-v4, inception-resnet and the impact of residual connections on learning. arXiv 2016, arXiv:1602.07261. [Google Scholar]
- Feng, S.; Xia, L.; Shan, F.; Wu, D.; Wei, Y.; Yuan, H.; Jiang, H.; Gao, Y.; Sui, H.; Shen, D. Large-scale screening of covid-19 from community-acquired pneumonia using infection size-aware classification. arXiv 2020, arXiv:2003.09860. [Google Scholar]
- Sheetal, R.; Kumar, N.; Rajpal, A. COV-ELM classifier: An Extreme Learning Machine based identification of COVID-19 using Chest-ray Images. arXiv 2020, arXiv:2007.08637. [Google Scholar]
- Delong, C.; Liu, F.; Li, Z. A Review of Automatically Diagnosing COVID-19 based on Scanning Image. arXiv 2006, arXiv:2006.05245. [Google Scholar]
- Abdelkader, B.; Ouhbi, S.; Lakas, A.; Benkhelifa, E.; Chen, C. End-to-End AI-Based Point-of-Care Diagnosis System for Classifying Respiratory Illnesses and Early Detection of COVID-19. Front. Med. 2021. [Google Scholar] [CrossRef]
- Liang, S.; Mo, Z.; Yan, F.; Xia, L.; Shan, F.; Ding, Z.; Song, B. Adaptive feature selection guided deep forest for covid-19 classification with chest ct. IEEE J. Biomed. Health Inform. 2020. [Google Scholar] [CrossRef]
- Lundberg, S.; Lee, S.I. An unexpected unity among methods for interpreting model predictions. arXiv 2016, arXiv:1611.07478. [Google Scholar]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv 2003, arXiv:2003.10849. [Google Scholar]
- Panwar, H.; Gupta, P.K.; Siddiqui, M.K.; Morales-Menendez, R.; Singh, V. Application of Deep Learning for Fast Detection of COVID-19 in X-Rays using nCOVnet. Chaos Solitons Fractals 2020, 138, 109944. [Google Scholar] [CrossRef]
- Altan, A.; Karasu, S. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm, and deep learning technique. Chaos Solitons Fractals 2020, 140, 110071. [Google Scholar] [CrossRef]
- Chowdhury, M.E.H.; Rahman, T.; Khandakar, A.; Mazhar, R.; Kadir, M.A.; Mahbub, Z. Can AI help in screening viral and COVID-19 pneumonia? arXiv 2003, arXiv:2003.13145. [Google Scholar] [CrossRef]
- Wang, L.; Wong, A. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-Ray Images. arXiv 2003, arXiv:2003.09871. [Google Scholar] [CrossRef]
- Kumar, R.; Arora, R.; Bansal, V.; Sahayasheela, V.J.; Buckchash, H.; Imran, J. Accurate Prediction of COVID-19 using Chest X-Ray Images through Deep Feature Learning model with SMOTE and Machine Learning Classifiers. MedRxiv 2020. [Google Scholar] [CrossRef]
- Sethy, P.K.; Behera, S.K. Detection of coronavirus disease (covid-19) based on deep features. Preprints 2020. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Khan, A.I.; Shah, J.L.; Bhat, M.M. Coronet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef]
- Mahmud, T.; Awsafur, R.; Fattah, A.S. CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020, 122, 103869. [Google Scholar] [CrossRef]
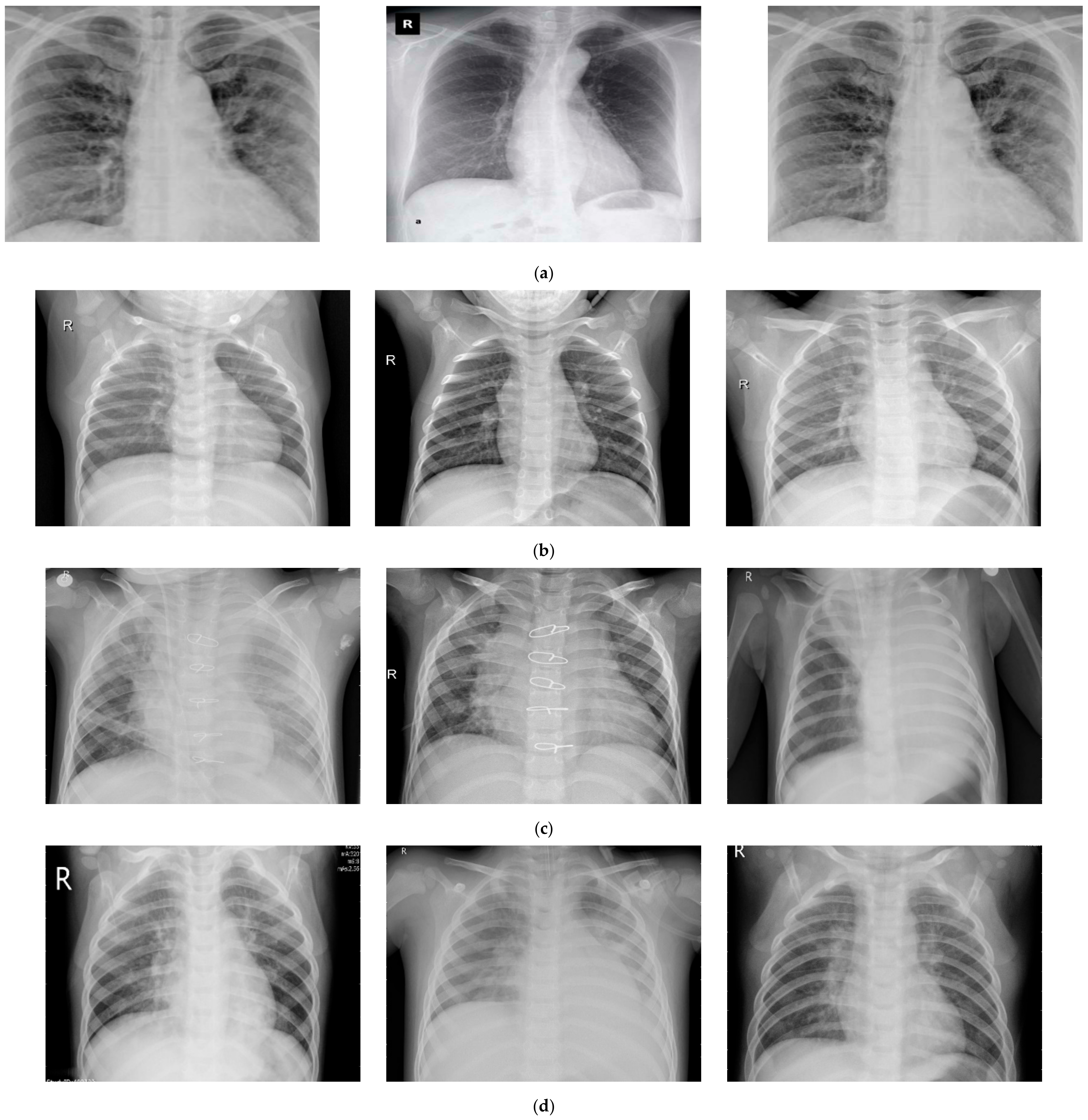












| Sr. No. | Disease | Total Samples | Training Samples | Testing Samples |
|---|---|---|---|---|
| 1 | Normal images | 310 | 248 | 62 |
| 2 | Pneumonia-bacterial-infection images | 330 | 264 | 66 |
| 3 | Viral-pneumonia images | 327 | 261 | 66 |
| 4 | Corona-infection images | 284 | 227 | 57 |
| Model | Accuracy | Precision | Recall | F1score |
|---|---|---|---|---|
| Xception—Fine-Tuned | 0.8822 | 0.88041 | 0.8796 | 0.8787 |
| CoVIRNet Model | 0.9578 | 0.9491 | 0.9544 | 0.9509 |
| ResNet101—Fine-Tuned | 0.8880 | 0.8954 | 0.9033 | 0.8923 |
| MobielNetV2—Fine-Tuned | 0.90347 | 0.9028 | 0.9011 | 0.9005 |
| DenseNet201—Fine-Tuned | 0.9419 | 0.9462 | 0.9514 | 0.9474 |
| CoVIRNet with RF | 0.9729 | 0.9774 | 0.9702 | 0.9732 |
| Model | Accuracy | Precision | Recall | F1score |
|---|---|---|---|---|
| CoVIRNet Model with LR | 0.9279 | 0.9283 | 0.9264 | 0.9263 |
| CoVIRNet Model with MLP | 0.9446 | 0.9439 | 0.9437 | 0.9435 |
| CoVIRNet Model with GB | 0.9523 | 0.95141 | 0.9515 | 0.9512 |
| CoVIRNet Model with BT | 0.9613 | 0.9607 | 0.9607 | 0.9605 |
| CoVIRNet Model with RF | 0.9729 | 0.9774 | 0.9702 | 0.9732 |
| Study | Dataset | Model Used | Classification Accuracy |
|---|---|---|---|
| Narin et al. [43] | 2-class: 50 COVID-19/50 normal | Transfer learning with ResNet50 and Inception-v3 | 98% |
| Panwar et al. [44] | 2-class: 142 COVID-19/ 142 normal | nCOVnet CNN | 88% |
| Altan et al. [45] | 3-class: 219 COVID-19 1341 norma l1345 pneumonia viral | 2D curvelet transform, chaotic salp swarm algorithm (CSSA), EfficientNet-B0 | 99% |
| Chowdhury et al. [46] | 3-class: 423 COVID-19 1579 normal 1485 pneumonia viral | Transfer learning with CheXNet | 97.7% |
| Wang and Wong [47] | 3-class: 358 COVID-19/5538 normal/8066 pneumonia | COVID-Net | 93.3% |
| Kumar et al. [48] | 3-class: 62 COVID-19/1341 normal/1345 pneumonia | ResNet1523 features and XGBoost classifier | 90% |
| Sethy and Behera [49] | 3-class: 127 COVID-19/127 normal/127 pneumonia | ResNet50 features and SVM | 95.33% |
| Ozturk et al. [50] | 3-class: 125 COVID-19/500 normal 500 pneumonia | DarkCovidNet CNN | 87.2% |
| Khan et al. [51] | 4-class: 284 COVID-19/310 normal/330 pneumonia bacterial/327 pneumonia viral | CoroNet CNN | 89.6% |
| Tanvir Mahmud et al. [52] | 4-class: 305 COVID-19 + 305 Normal + 305 Viral Pneumonia + 305 Bacterial Pneumonia | StackedMulti-resolutionCovXNet | 90.3% |
| Proposed CoVIRNet DL model | 4-class: 284 COVID-19/310 normal/330 pneumonia bacterial/327 pneumonia viral | Multiscale features CoVIRNet | 95.78% |
| Proposed CoVIRNet DL model with RF | 4-class: 284 COVID-19/310 normal/330 pneumonia bacterial/327 pneumonia viral | Multiscale features CoVIRNet+ RF | 97.29% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almalki, Y.E.; Qayyum, A.; Irfan, M.; Haider, N.; Glowacz, A.; Alshehri, F.M.; Alduraibi, S.K.; Alshamrani, K.; Alkhalik Basha, M.A.; Alduraibi, A.; et al. A Novel Method for COVID-19 Diagnosis Using Artificial Intelligence in Chest X-ray Images. Healthcare 2021, 9, 522. https://doi.org/10.3390/healthcare9050522
Almalki YE, Qayyum A, Irfan M, Haider N, Glowacz A, Alshehri FM, Alduraibi SK, Alshamrani K, Alkhalik Basha MA, Alduraibi A, et al. A Novel Method for COVID-19 Diagnosis Using Artificial Intelligence in Chest X-ray Images. Healthcare. 2021; 9(5):522. https://doi.org/10.3390/healthcare9050522
Chicago/Turabian StyleAlmalki, Yassir Edrees, Abdul Qayyum, Muhammad Irfan, Noman Haider, Adam Glowacz, Fahad Mohammed Alshehri, Sharifa K. Alduraibi, Khalaf Alshamrani, Mohammad Abd Alkhalik Basha, Alaa Alduraibi, and et al. 2021. "A Novel Method for COVID-19 Diagnosis Using Artificial Intelligence in Chest X-ray Images" Healthcare 9, no. 5: 522. https://doi.org/10.3390/healthcare9050522
APA StyleAlmalki, Y. E., Qayyum, A., Irfan, M., Haider, N., Glowacz, A., Alshehri, F. M., Alduraibi, S. K., Alshamrani, K., Alkhalik Basha, M. A., Alduraibi, A., Saeed, M. K., & Rahman, S. (2021). A Novel Method for COVID-19 Diagnosis Using Artificial Intelligence in Chest X-ray Images. Healthcare, 9(5), 522. https://doi.org/10.3390/healthcare9050522








