Identification of Distinct and Common Subpopulations of Myxoid Liposarcoma and Ewing Sarcoma Cells Using Self-Organizing Maps
Abstract
1. Introduction
2. Materials and Methods
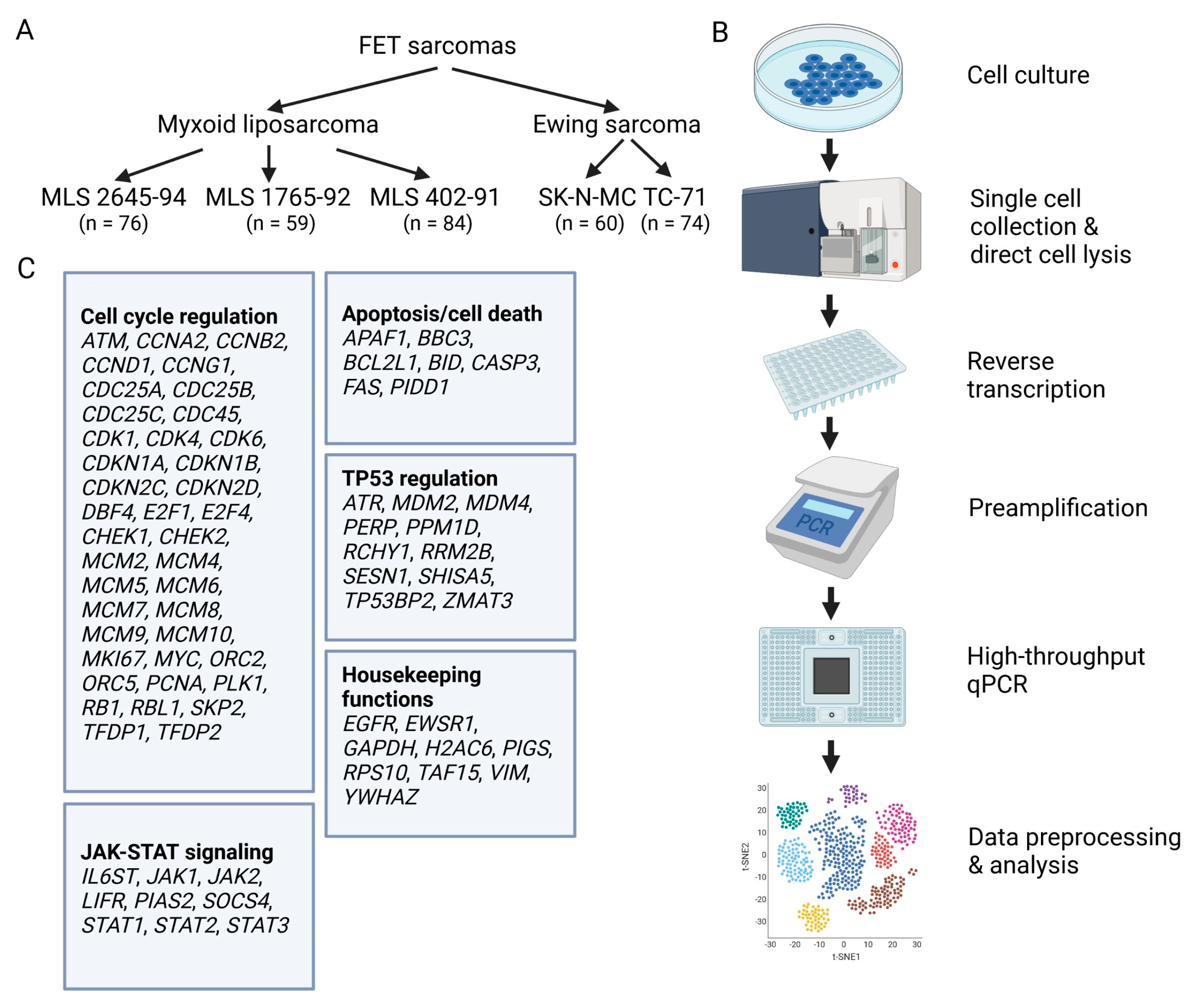
2.1. Cell Culture
2.2. Single-Cell Analysis
2.3. Single-Cell Data Preprocessing
2.4. Single-Cell Data Analysis
3. Results
3.1. Single-Cell Profiling of Myxoid Liposarcoma and Ewing Sarcoma Cells
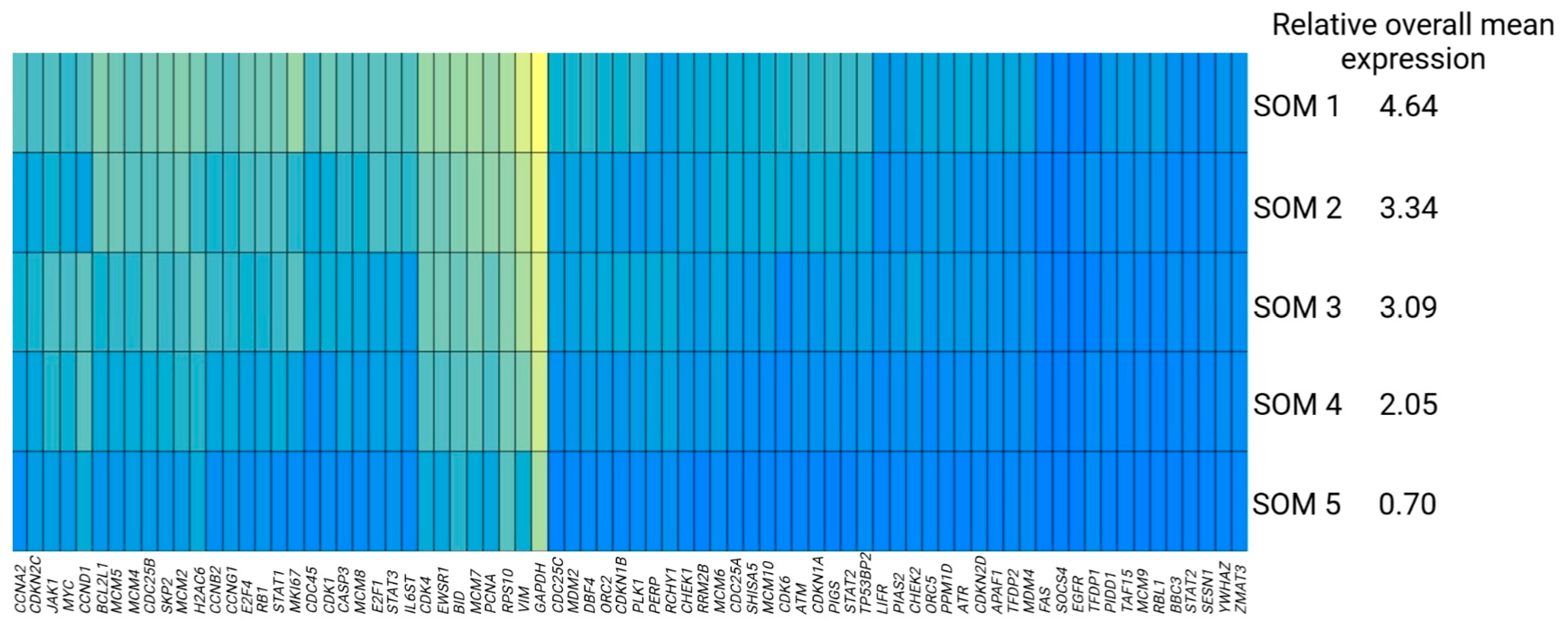
3.2. Identification of Subpopulations of Tumor Cells Using Self-Organizing Maps
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Lindén, M.; Thomsen, C.; Grundevik, P.; Jonasson, E.; Andersson, D.; Runnberg, R.; Dolatabadi, S.; Vannas, C.; Luna Santamarίa, M.; Fagman, H.; et al. FET family fusion oncoproteins target the SWI/SNF chromatin remodeling complex. EMBO Rep. 2019, 20, e45766. [Google Scholar] [CrossRef]
- Åman, P. Fusion oncogenes in tumor development. Semin. Cancer Biol. 2005, 15, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Ståhlberg, A.; Kåbjörn Gustafsson, C.; Engtröm, K.; Thomsen, C.; Dolatabadi, S.; Jonasson, E.; Li, C.Y.; Ruff, D.; Chen, S.M.; Åman, P. Normal and functional TP53 in genetically stable myxoid/round cell liposarcoma. PLoS ONE 2014, 9, e113110. [Google Scholar] [CrossRef]
- Tirode, F.; Surdez, D.; Ma, X.; Parker, M.; Le Deley, M.C.; Bahrami, A.; Zhang, Z.; Lapouble, E.; Grossetête-Lalami, S.; Rusch, M.; et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014, 4, 1342–1353. [Google Scholar] [CrossRef]
- Dolatabadi, S.; Candia, J.; Akrap, N.; Vannas, C.; Tesan Tomic, T.; Losert, W.; Landberg, G.; Åman, P.; Ståhlberg, A. Cell Cycle and Cell Size Dependent Gene Expression Reveals Distinct Subpopulations at Single-Cell Level. Front. Genet. 2017, 8, 1. [Google Scholar] [CrossRef]
- Kåbjörn Gustafsson, C.; Ståhlberg, A.; Engtröm, K.; Danielsson, A.; Turesson, I.; Aman, P. Cell senescence in myxoid/round cell liposarcoma. Sarcoma 2014, 2014, 208786. [Google Scholar] [CrossRef]
- Aynaud, M.M.; Mirabeau, O.; Gruel, N.; Grossetête, S.; Boeva, V.; Durand, S.; Surdez, D.; Saulnier, O.; Zaïdi, S.; Gribkova, S.; et al. Transcriptional Programs Define Intratumoral Heterogeneity of Ewing Sarcoma at Single-Cell Resolution. Cell Rep. 2020, 30, 1767–1779.e6. [Google Scholar] [CrossRef] [PubMed]
- Kubista, M.; Dreyer-Lamm, J.; Ståhlberg, A. The secrets of the cell. Mol. Aspects Med. 2018, 59, 1–4. [Google Scholar] [CrossRef]
- Hedlund, E.; Deng, Q. Single-cell RNA sequencing: Technical advancements and biological applications. Mol. Asp. Med. 2018, 59, 36–46. [Google Scholar] [CrossRef]
- Andrews, T.S.; Hemberg, M. Identifying cell populations with scRNASeq. Mol. Asp. Med. 2018, 59, 114–122. [Google Scholar] [CrossRef]
- Peng, L.; Tian, X.; Tian, G.; Xu, J.; Huang, X.; Weng, Y.; Yang, J.; Zhou, L. Single-cell RNA-seq clustering: Datasets, models, and algorithms. RNA Biol. 2020, 17, 765–783. [Google Scholar] [CrossRef]
- Vandeginste, B.G.M.; Massart, D.L.; Buydens, L.M.C.; de Jong, S.; Lewi, P.J.; Smeyers-Verbeke, J. Handbook of Chemometrics and Qualimetrics (Part B); Elsevier: Amsterdam, The Netherlands, 1998; pp. 687–695. [Google Scholar]
- Kohonen, T. MATLAB Implementations and Applications of the Self-Organizing Map; Unigrafia Oy: Helsinki, Finland, 2014; pp. 2, 19–22. [Google Scholar]
- Borges, C.; Gómez-Carracedo, M.P.; Andrade, J.M.; Duarte, M.F.; Biscaya, J.L.; Aires-de-Sousa, J. Geographical classification of weathered crude oil samples with unsupervised self-organizing maps and a consensus criterion. Chemom. Intell. Lab. Syst. 2010, 101, 43–55. [Google Scholar] [CrossRef]
- Melssen, W.; Wehrens, R.; Buydens, L. Supervised Kohonen networks for classification problems. Chemom. Intell. Lab. Syst. 2006, 83, 99–113. [Google Scholar] [CrossRef]
- Aires de Sousa, J.M. Data visualization and analysis using Kohonen Self-Organizing Maps (Chapter 7). In Tutorials in Chemoinformatics; Varnek, A., Ed.; John Willey & Sons: Oxford, UK, 2017; pp. 119–126. [Google Scholar]
- Ståhlberg, A.; Bengtsson, M.; Hemberg, M.; Semb, H. Quantitative transcription factor analysis of undifferentiated single human embryonic stem cells. Clin. Chem. 2009, 55, 2162–2170. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.; Akrap, N.; Svec, D.; Godfrey, T.E.; Kubista, M.; Landberg, G.; Ståhlberg, A. Properties of targeted preamplification in DNA and cDNA quantification. Expert Rev. Mol. Diagn. 2015, 15, 1085–1100. [Google Scholar] [CrossRef] [PubMed]
- Ståhlberg, A.; Rusnakova, V.; Forootan, A.; Anderova, M.; Kubista, M. RT-qPCR work-flow for single-cell data analysis. Methods 2013, 59, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, M.; Ståhlberg, A.; Rorsman, P.; Kubista, M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005, 15, 1388–1392. [Google Scholar] [CrossRef]
- Raj, A.; Peskin, C.S.; Tranchina, D.; Vargas, D.Y.; Tyagi, S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006, 4, e309. [Google Scholar] [CrossRef]
- Lindén, M.; Vannas, C.; Österlund, T.; Andersson, L.; Osman, A.; Escobar, M.; Fagman, H.; Ståhlberg, A.; Åman, P. FET fusion oncoproteins interact with BRD4 and SWI/SNF chromatin remodelling complex subtypes in sarcoma. Mol. Oncol. 2022, 16, 2470–2495. [Google Scholar] [CrossRef]
- Dolatabadi, S.; Jonasson, E.; Lindén, M.; Fereydouni, B.; Bäcksten, K.; Nilsson, M.; Martner, A.; Forootan, A.; Fagman, H.; Landberg, G.; et al. JAK-STAT signalling controls cancer stem cell properties including chemotherapy resistance in myxoid liposarcoma. Int. J. Cancer 2019, 145, 435–449. [Google Scholar] [CrossRef]
- Karlsson, J.; Kroneis, T.; Jonasson, E.; Larsson, E.; Ståhlberg, A. Transcriptomic Characterization of the Human Cell Cycle in Individual Unsynchronized Cells. J. Mol. Biol. 2017, 429, 3909–3924. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Lovén, J.; Rahl, P.B.; Paranal, R.M.; Burge, C.B.; Bradner, J.E.; Lee, T.I.; Young, R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012, 151, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Hu, G.; Wei, G.; Cui, K.; Yamane, A.; Resch, W.; Wang, R.; Green, D.R.; Tessarollo, L.; Casellas, R.; et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012, 151, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Marguerat, S.; Bahler, J. Coordinating genome expression with cell size. Trends Genet. 2012, 28, 560–565. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, K.; Xia, Z.; Chavez, M.; Pal, S.; Seol, J.-H.; Chen, C.-C.; Li, W.; Tyler, J.K. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014, 28, 396–408. [Google Scholar] [CrossRef]
- Burn, K.; Home, G. Environmental classification using Kohonen self-organizing maps. Expert Sys. 2008, 25, 98–114. [Google Scholar] [CrossRef]
- Ballabio, D.; Consoni, V.; Todeschini, R. The Kohonen and CPANN toolbox: A collection of MATLAB modules for Self-Organizing Maps and counterpropagation artificial neural networks. Chemom. Intell. Lab. Syst. 2009, 98, 115–122. [Google Scholar] [CrossRef]

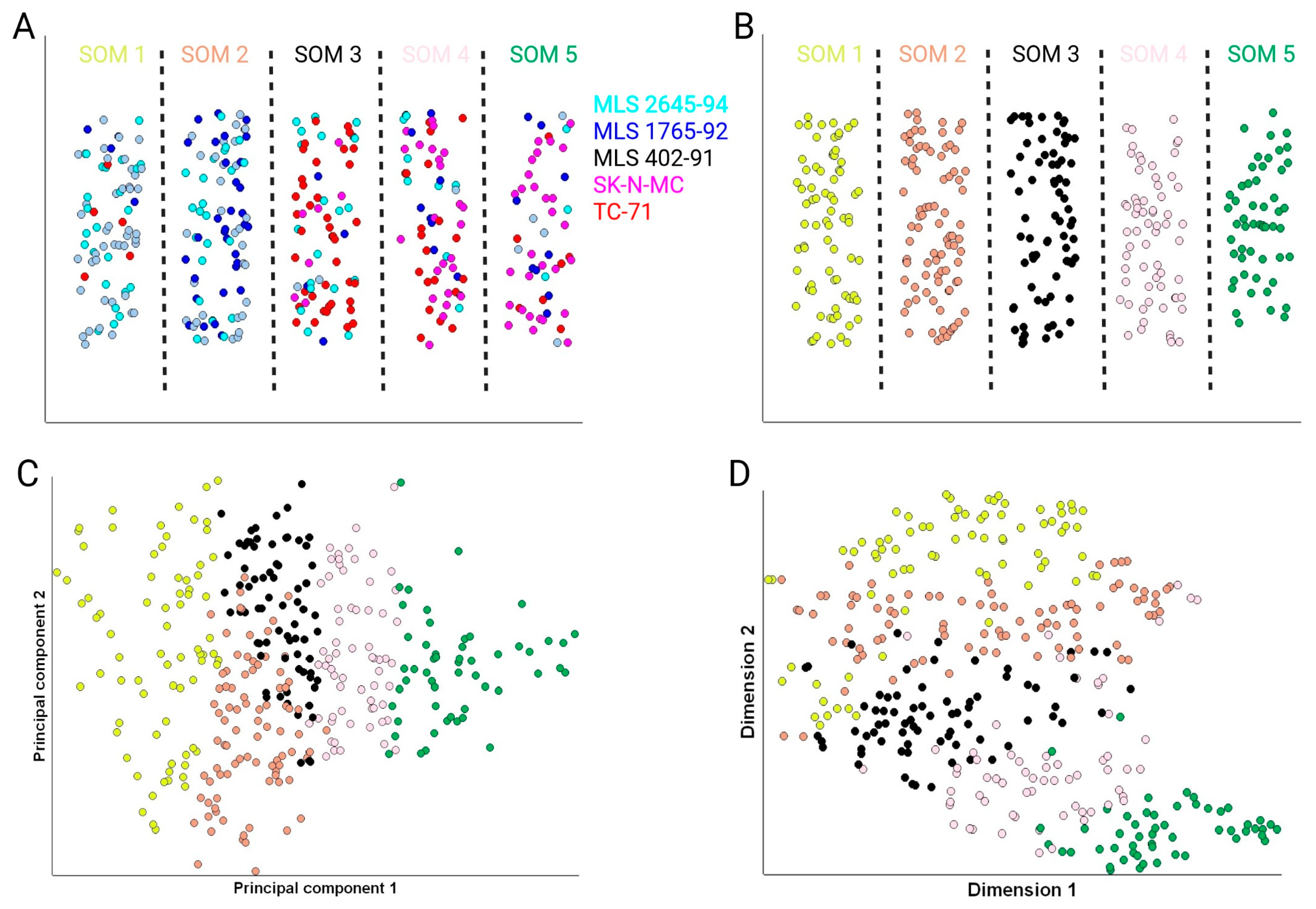
| SOM 1 | SOM 2 | SOM 3 | SOM 4 | SOM 5 | |
|---|---|---|---|---|---|
| MLS 2645-94 | 23 | 22 | 19 | 9 | 3 |
| MLS 1765-92 | 4 | 33 | 4 | 9 | 9 |
| MLS 402-91 | 44 | 32 | 2 | 1 | 5 |
| SK-N-MC | 0 | 0 | 7 | 24 | 29 |
| TC-71 | 5 | 0 | 41 | 21 | 7 |
| All MLS cells | 71 | 87 | 25 | 19 | 17 |
| All EWS cells | 5 | 0 | 48 | 45 | 36 |
| Total number of cells | 76 | 87 | 73 | 64 | 53 |
| Cell Cycle Regulation | JAK-STAT Signaling | Apoptosis/Cell Death | TP53 Regulation | Housekeeping Functions | |
|---|---|---|---|---|---|
| SOM 1 | 5.08 1 | 3.71 | 3.84 | 3.26 | 6.11 |
| SOM 2 | 3.66 | 2.56 | 2.69 | 2.03 | 5.01 |
| SOM 3 | 3.37 | 2.17 | 2.24 | 2.13 | 4.74 |
| SOM 4 | 2.11 | 1.37 | 1.58 | 1.23 | 3.88 |
| SOM 5 | 0.67 | 0.13 | 0.47 | 0.00 | 2.46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forootan, A.; Andersson, D.; Dolatabadi, S.; Svec, D.; Andrade, J.; Ståhlberg, A. Identification of Distinct and Common Subpopulations of Myxoid Liposarcoma and Ewing Sarcoma Cells Using Self-Organizing Maps. Chemosensors 2023, 11, 67. https://doi.org/10.3390/chemosensors11010067
Forootan A, Andersson D, Dolatabadi S, Svec D, Andrade J, Ståhlberg A. Identification of Distinct and Common Subpopulations of Myxoid Liposarcoma and Ewing Sarcoma Cells Using Self-Organizing Maps. Chemosensors. 2023; 11(1):67. https://doi.org/10.3390/chemosensors11010067
Chicago/Turabian StyleForootan, Amin, Daniel Andersson, Soheila Dolatabadi, David Svec, José Andrade, and Anders Ståhlberg. 2023. "Identification of Distinct and Common Subpopulations of Myxoid Liposarcoma and Ewing Sarcoma Cells Using Self-Organizing Maps" Chemosensors 11, no. 1: 67. https://doi.org/10.3390/chemosensors11010067
APA StyleForootan, A., Andersson, D., Dolatabadi, S., Svec, D., Andrade, J., & Ståhlberg, A. (2023). Identification of Distinct and Common Subpopulations of Myxoid Liposarcoma and Ewing Sarcoma Cells Using Self-Organizing Maps. Chemosensors, 11(1), 67. https://doi.org/10.3390/chemosensors11010067






