Detection of Cytopathic Effects Induced by Influenza, Parainfluenza, and Enterovirus Using Deep Convolution Neural Network
Abstract
:1. Introduction
2. Materials and Methods
2.1. System Equipment
2.2. Construct Dataset and Data Processing
2.2.1. Data Collection
2.2.2. Data Processing
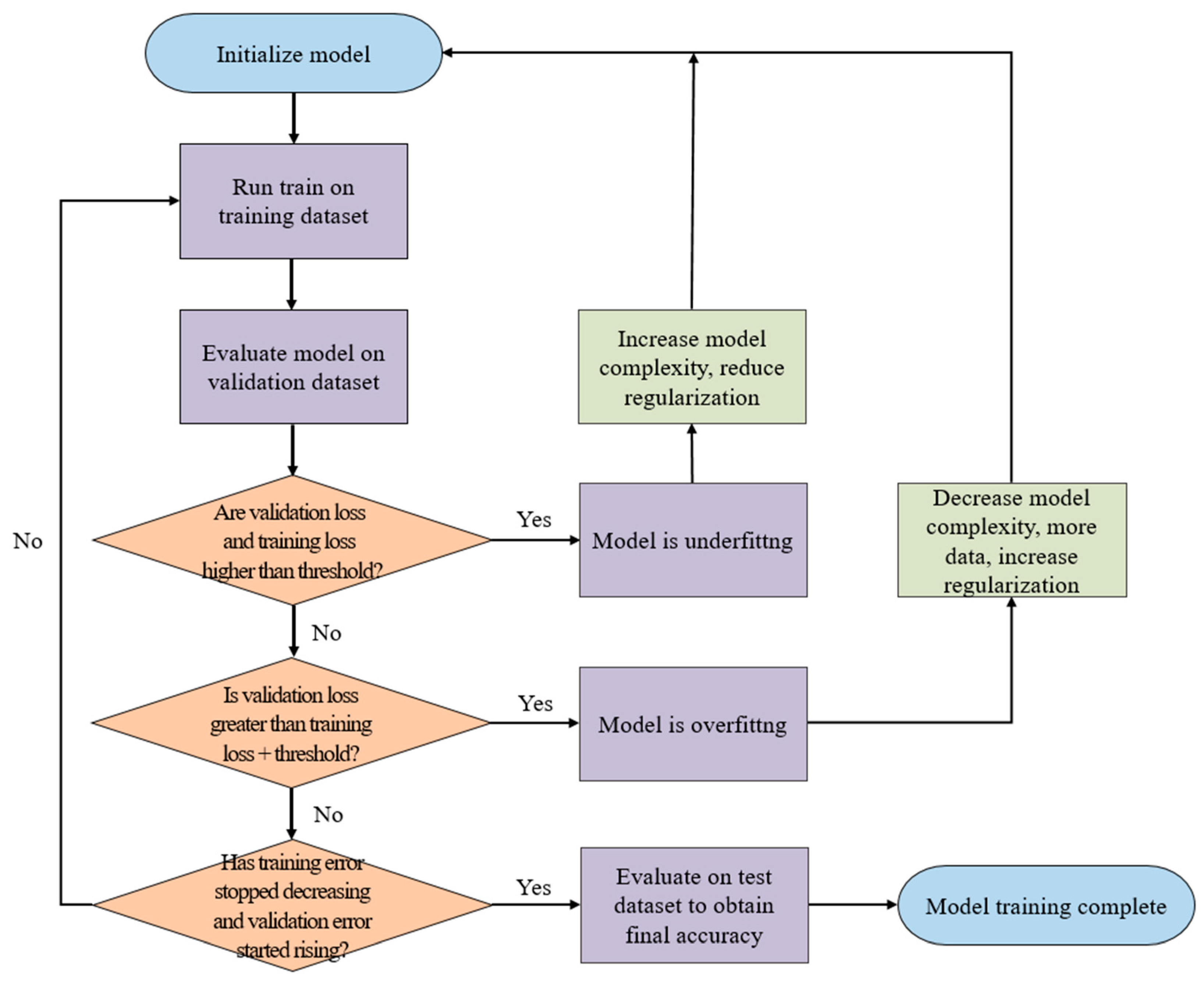
2.3. Convolutional Neural Network (CNN) Model Architecture
2.3.1. VGG16
2.3.2. Inception-V3
2.3.3. MobileNet-V2
2.3.4. ResNet
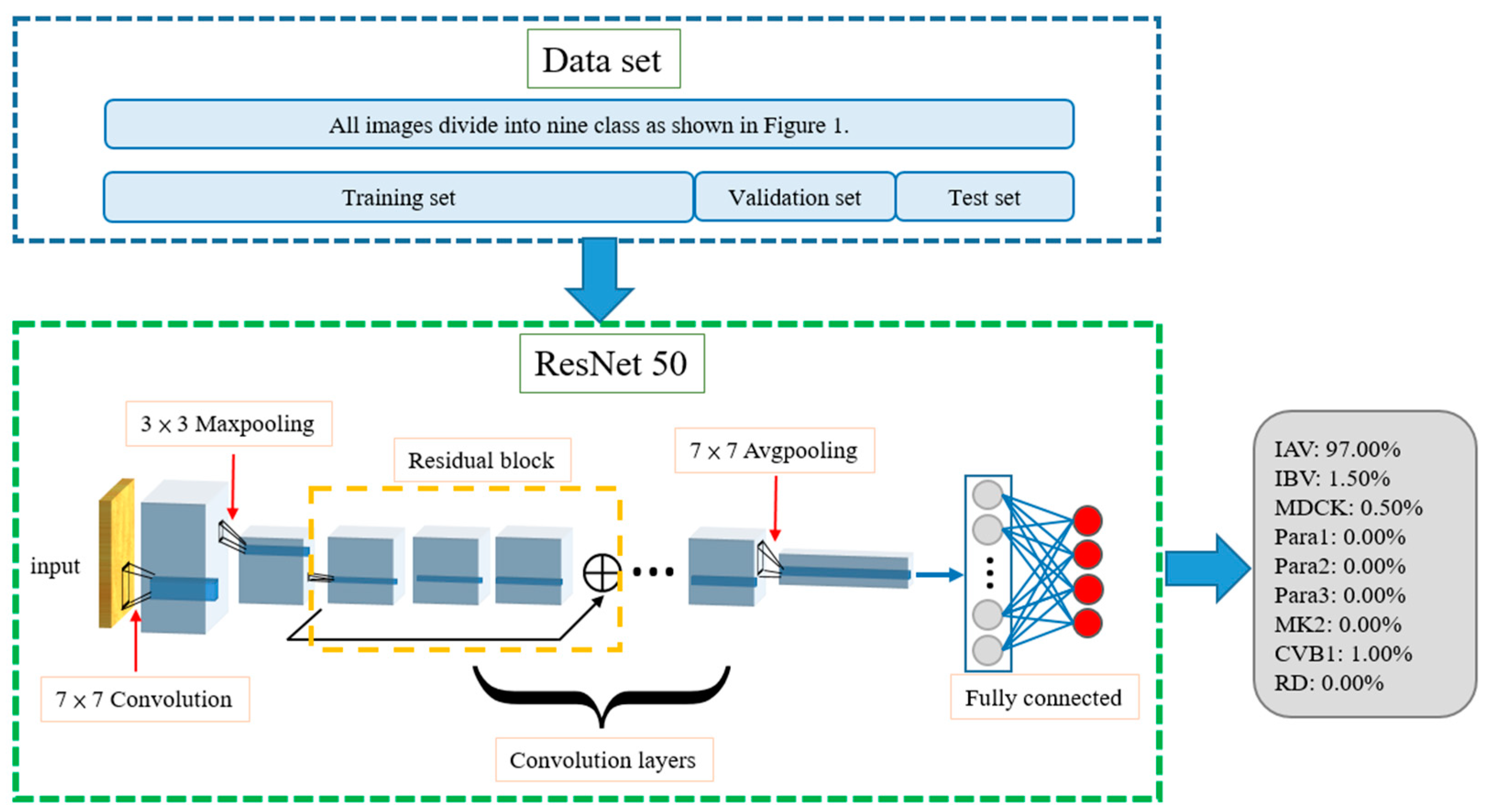
2.4. Single Model
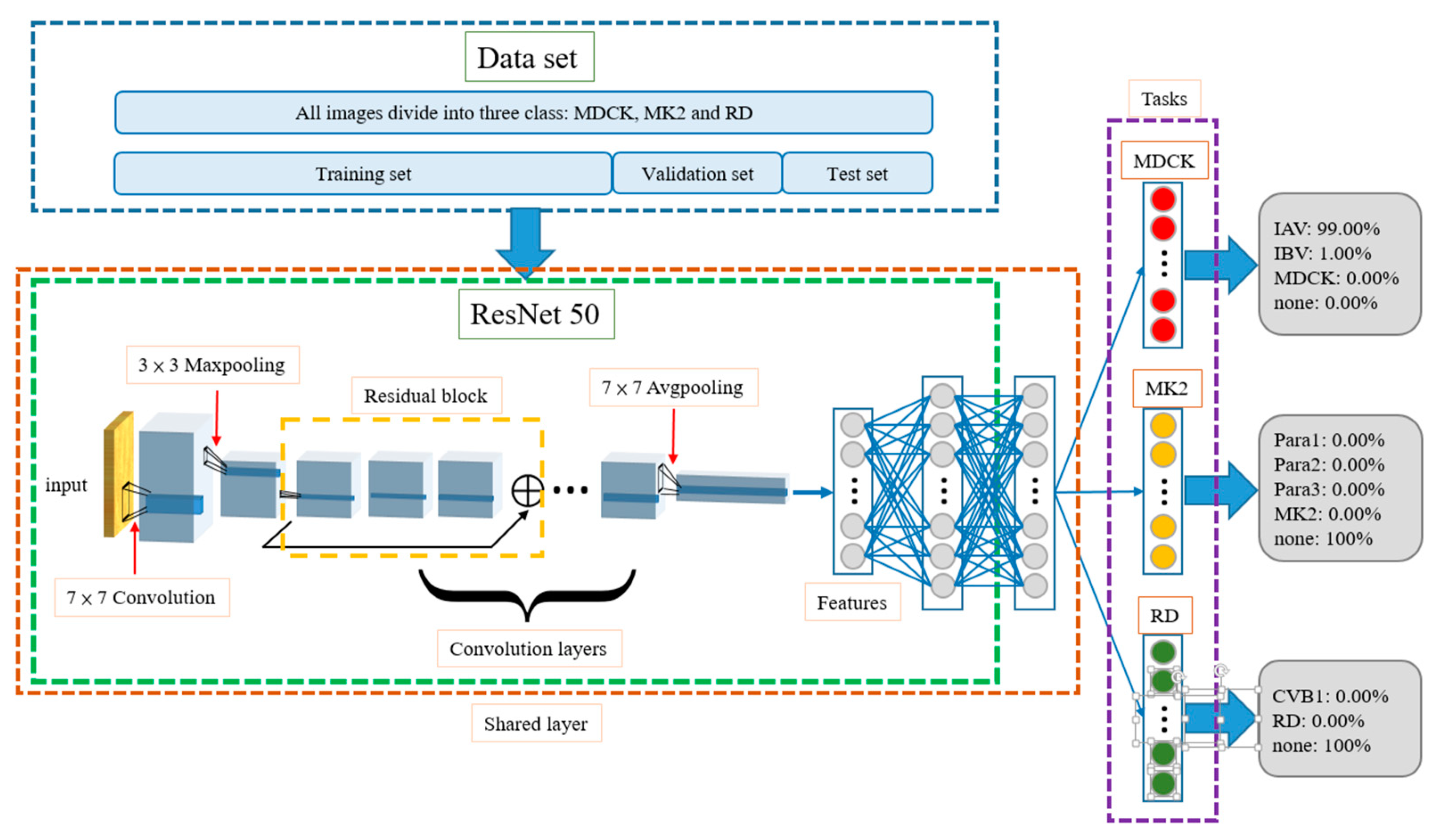
2.5. Multi-Task Learning Model
2.6. Fusion Layer
- Number of None Tasks: 0When none of the three tasks output Pnone as the largest probability in their results, it means that the features of the input image are similar to multiple categories and are difficult to distinguish. In this case, the fusion algorithm will take a total of nine categories from the three tasks into consideration and will select the one with the highest probability as the final result. For example, when IAV is the most likely category in the MDCK task, at 95%; Para2 is the most likely category in the MK2 Task, at 90.1%; and RD is the most likely category in the RD Task, at 65%, the probability of IAV among the three categories is the largest, i.e., argmax {PIAV, PPara2, PRD} = IAV; thus, the model outputs IAV as the result.
- Number of None Tasks: 1When only one task outputs Pnone as the largest probability in its result, the fusion algorithm will only compare the output probabilities of the remaining two tasks. Among them, the largest probability will be regarded as the final output. For example, if the first MDCK task shows that Pnone = 100%, the MK2 task shows that Para3 is the most likely category at 94.9%, and the RD task shows that CVB1 is the most likely category at 83.7%, Para3 has the largest probability, i.e., argmax{PPara3, PCVB1} = Para3, so the result is Para3.
- Number of None Tasks: 2Two tasks outputting Pnone as the largest probability in their result is the most common situation. In this case, the fusion layer directly outputs the category with the largest probability in the remaining one task as the result.
- Number of None Tasks: 3This type of situation rarely occurs. Usually, only when the content of the test image is seriously distorted will the model consider that the image does not belong in any category. Once this situation happens, the fusion algorithm provides two options for output. One is to find the most probable category from the nine categories as the output answer, and the other is to directly output “none”. The default is the former.
2.7. Known Cell Line Classification
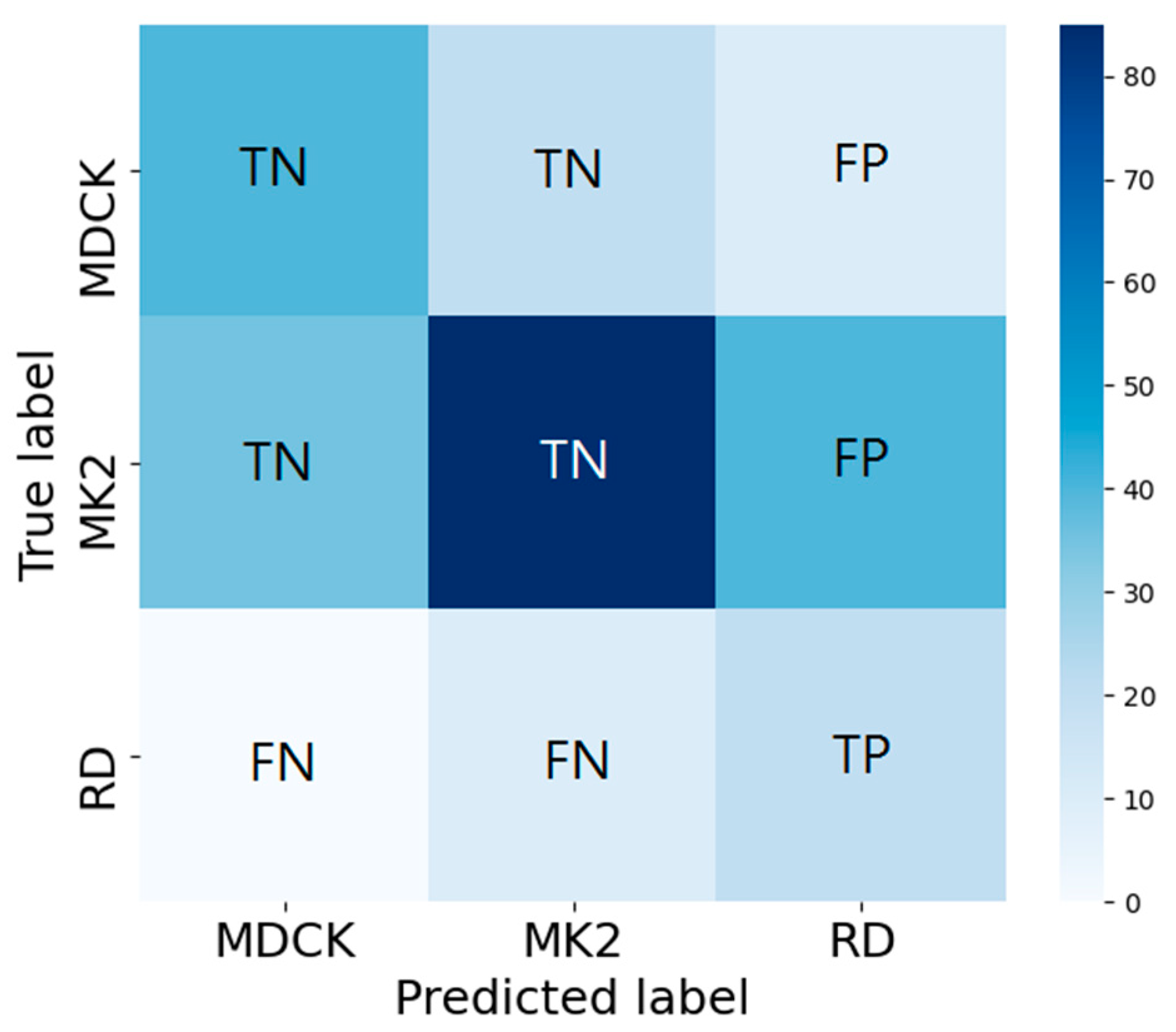
2.8. Multi-Class Model Index Analysis
2.9. Comparison with Human Readers
3. Results
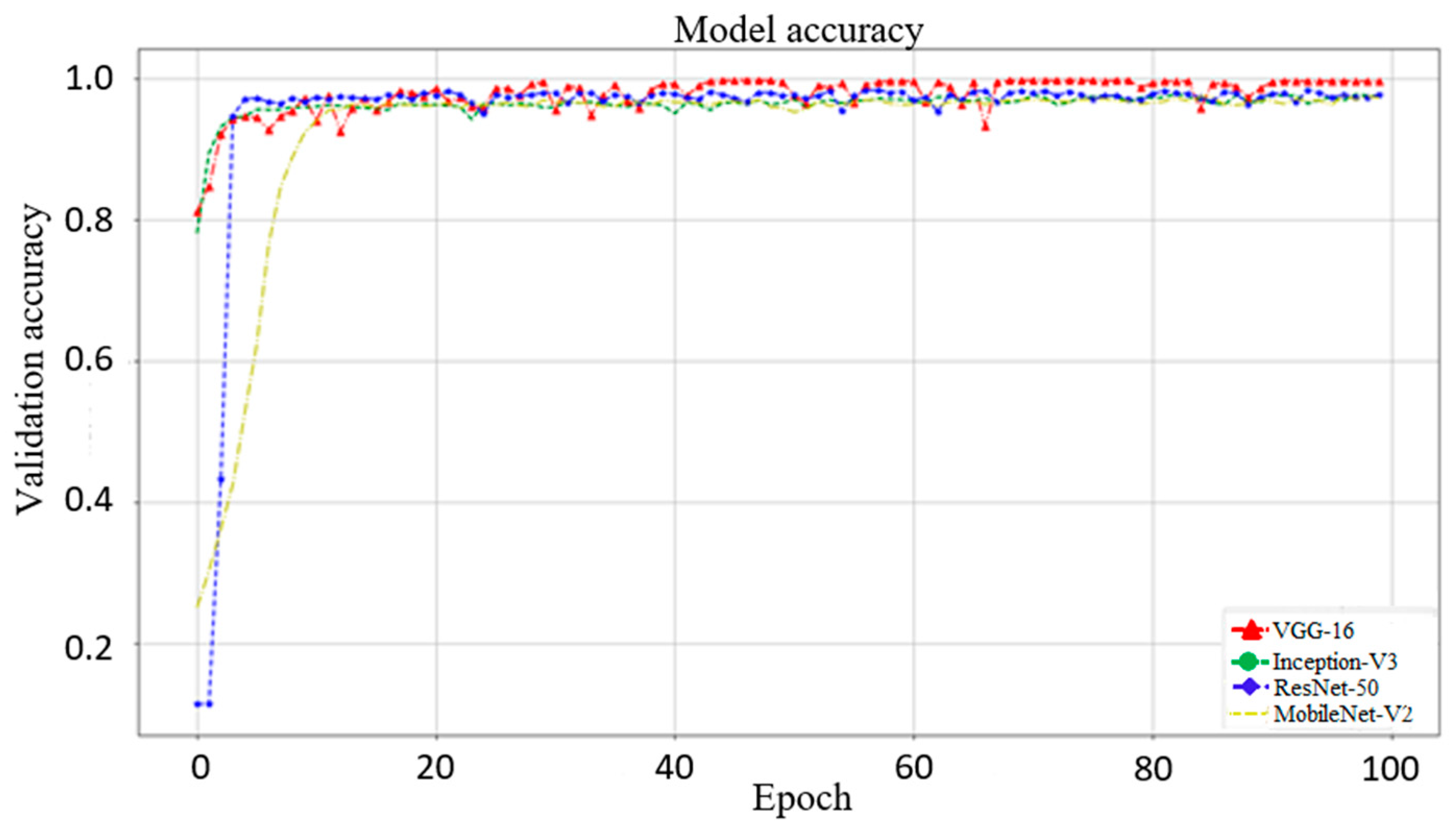
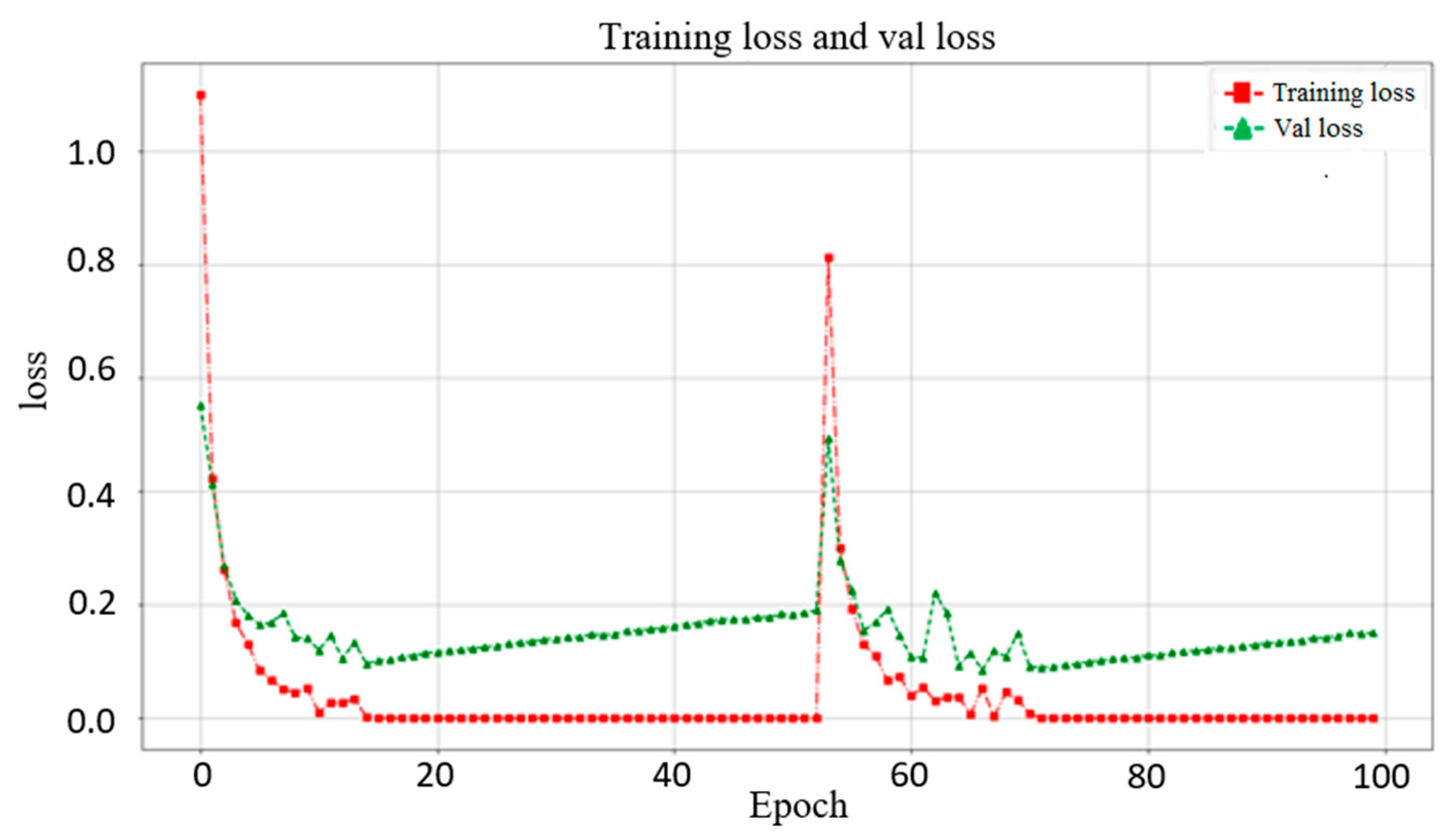
3.1. Effect of Different Networks as Backbone
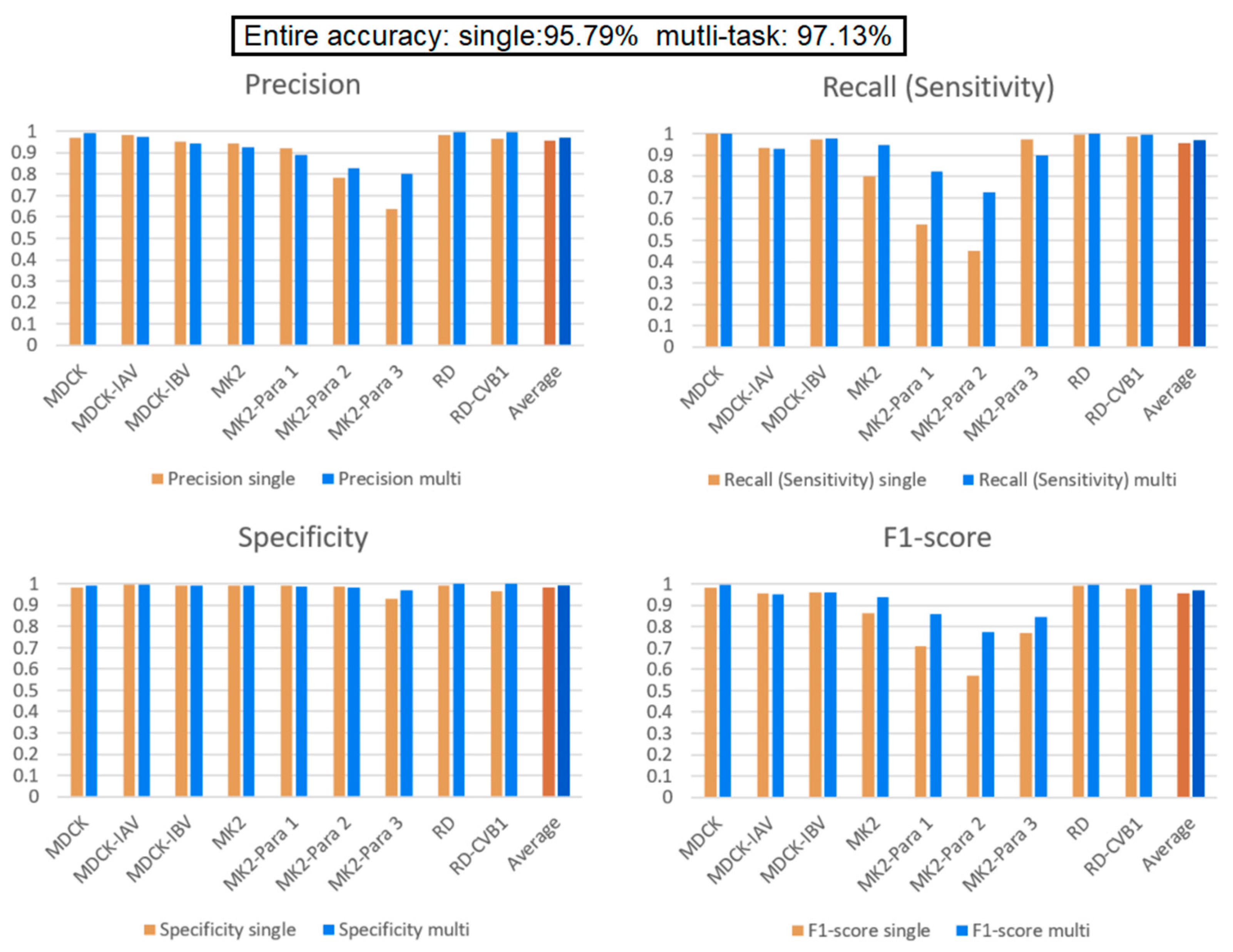
3.2. Comparison of Single Model and Multi-Task Learning Model
3.3. Effect of a Small Data Category
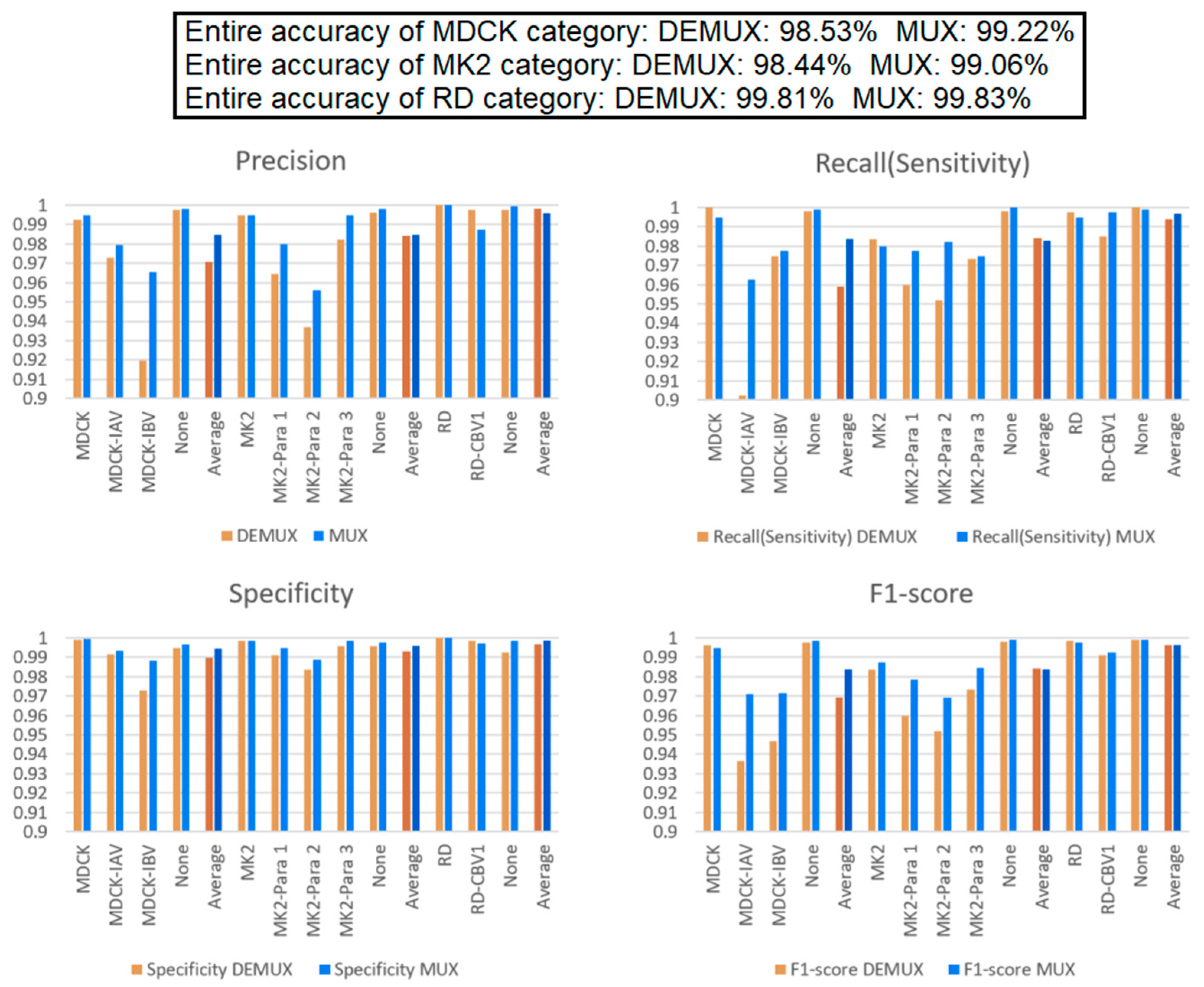
3.4. Comparison of “DEMUX-Single” Model and “MUX-Multi-Task” Learning Model
3.5. Comparison between the Four Models and the Medical Technologist for CPE Reading
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gonzales, R.; Malone, D.C.; Maselli, J.H.; Sande, M.A. Excessive antibiotic use for acute respiratory infections in the United States. Clin. Infect. Dis. 2001, 33, 757–762. [Google Scholar] [CrossRef]
- Grijalva, C.G.; Griffin, M.R.; Edwards, K.M.; Williams, J.V.; Gil, A.I.; Verastegui, H.; Hartinger, S.M.; Vidal, J.E.; Klugman, K.P.; Lanata, C.F. The role of influenza and parainfluenza infections in nasopharyngeal pneumococcal acquisition among young children. Clin. Infect. Dis. 2014, 58, 1369–1376. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.H.S.; Cheng, T.J. Reduction of influenza and enterovirus infection in Taiwan during the COVID-19 pandemic. Aerosol Air Qual. Res. 2020, 20, 2071–2074. [Google Scholar] [CrossRef]
- Storch, G.A. Diagnostic virology. Clin. Infect. Dis. 2000, 31, 739–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Sweter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Hsieh, K.; Wang, Y.; Chen, L.; Zhao, Z.; Savitz, S.; Jiang, X.; Tang, J.; Kim, Y. Drug Repurposing for COVID-19 using Graph Neural Network with Genetic, Mechanistic, and Epidemiological Validation. arXiv 2020, arXiv:2009.10931v1. [Google Scholar]
- Kikkisetti, S.; Zhu, J.; Shen, B.; Li, H.; Duong, T.Q. Deep-learning convolutional neural networks with transfer learning accurately classify COVID-19 lung infection on portable chest radiographs. PeerJ 2020, 8, e10309. [Google Scholar] [CrossRef]
- Jo, Y.; Park, S.; Jung, J.; Yoon, J.; Joo, H.; Kim, M.H.; Kang, S.J.; Choi, M.C.; Lee, S.Y.; Park, Y. Holographic deep learning for rapid optical screening of anthrax spores. Sci. Adv. 2017, 3, e1700606. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Jiang, Z.; Shen, H.; Pan, W.D.; Williams, L.A.; Reddy, V.V.B.; Benjamin, W.H.; Bryan, A.W. Evaluations of deep convolutional neural networks for automatic identification of malaria infected cells. In Proceedings of the 2017 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Orlando, FL, USA, 16–19 February 2017; pp. 101–104. [Google Scholar]
- Kumar, H.; Yadav, P. A Neural Network based Model with Lockdown Condition for Checking the Danger Stage Level of COVID-19 Infection Risk. Int. J. Recent. Technol. 2020, 9. [Google Scholar] [CrossRef]
- Yakimovich, A.; Witte, R.; Andriasyan, V.; Georgi, F.; Grebera, U.F. Label-free digital holo-tomographic microscopy reveals virus-induced cytopathic effects in live cells. mSphere 2018, 3, e00599-18. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Araújo, T.; Aresta, G.; Castro, E.; Rouco, J.; Aguiar, P.; Eloy, C.; Polonia, A.; Campilho, A. Classification of breast cancer histology images using Convolutional Neural Networks. PLoS ONE 2017, 12, e0177544. [Google Scholar] [CrossRef]
- Abubakar, A.; Ajuji, M.; Usman-Yahya, I. Comparison of Deep Transfer Learning Techniques in Human Skin Burns Discrimination. Appl. Syst. Innov. 2020, 3, 20. [Google Scholar] [CrossRef] [Green Version]
- Kiran, G.V.K.; Ganesina, M.R. Automatic Classification of Whole Slide Pap Smear Images Using CNN with PCA Based Feature Interpretation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, Long Beach, CA, USA, 16–20 June 2019; pp. 1074–1079. [Google Scholar]
- Antony, B.J.; Maetschke, S.; Garnavi, R. Automated summarisation of SDOCT volumes using deep learning: Transfer learning vs de novo trained networks. PLoS ONE 2019, 14, e0203726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zieliński, B.; Sroka-Oleksiak, A.; Rymarczyk, D.; Piekarczyk, A.; Brzychczy-Wloch, M. Deep learning approach to describe and classify fungi microscopic images. PLoS ONE 2020, 15, e0234806. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.W. Adapting Mask-RCNN for Automatic Nucleus Segmentation. In Proceedings of the 2019 Computer Vision Conference, Las Vegas, NV, USA, 2–3 May 2019; Volume 2. [Google Scholar]
- Liu, D.; Zhang, D.; Song, Y.; Zhang, C.; Zhang, F.; O’Donenell, L.; Cai, L. Nuclei Segmentation via a Deep Panoptic Model with Semantic Feature Fusion. In Proceedings of the Twenty-Eighth International Joint Conference on Artificial Intelligence, Macao, China, 10–16 August 2019; pp. 861–868. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.E.; Chao, T.L.; Tsai, H.T.; Lin, P.H.; Tsai, Y.L.; Chang, S.Y. Differentiation of Cytopathic Effects (CPE) induced by influenza virus infection using deep Convolutional Neural Networks (CNN). PLoS Compt. Biol. 2020, 16, e1007883. [Google Scholar] [CrossRef]
- Yan, R.; Ren, F.; Wang, Z.; Wang, L.; Zhang, T.; Liu, Y.; Rao, X.; Zheng, C.; Zhang, F. Breast cancer histopathological image classification using a hybrid deep neural network. Methods 2020, 173, 52–60. [Google Scholar] [CrossRef]
- Wang, Q.; Bi, S.; Sun, M.; Wang, Y.; Wang, D.; Yang, S. Deep learning approach to peripheral leukocyte recognition. PLoS ONE 2019, 14, e0218808. [Google Scholar] [CrossRef] [PubMed]
- Andriasyan, V.; Yakimovich, A.; Georgi, F.; Petkidis, A.; Witte, R.; Puntener, D.; Greber, U.F. Deep learning of virus infections reveals mechanics of lytic cells. bioRxiv Preprint 2019, 798074. [Google Scholar]
- Jiang, Y.; Chen, L.; Zhang, H.; Xiao, X. Breast cancer histopathological image classification using convolutional neural networks with small SE-ResNet module. PLoS ONE 2019, 14, e0214587. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Dong, W.; Socher, R.; Fei-Fei, L.; Li, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Hartwig, A. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A survey of transfer learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A Survey on Deep Transfer Learning. In Artificial Neural Networks and Machine Learning–ICANN 2018, Proceedings of the 27th International Conference on Artificial Neural Networks, Rhodes, Greece, 4–7 October 2018; Lecture Notes in Computer Science; Kůrková, V., Manolopoulos, Y., Hammer, B., Iliadis, L., Maglogiannis, I., Eds.; Springer: Cham, Switzerland, 2018; Volume 11141. [Google Scholar]
- Pathak, Y.; Shukla, P.K.; Tiwari, A.; Stalin, S.; Singh, S. Deep Transfer Learning Based Classification Model for COVID-19 Disease. Ing. Rec. Biomed. 2020, in press. [Google Scholar] [CrossRef]
- Thung, K.H.; Wee, C.Y. A brief review on multi-task learning. Multimed. Tools Appl. 2018, 77, 29705–29725. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Q. A Survey on Multi-Task Learning. IEEE Trans. Knowl. Data Eng. 2021. [Google Scholar] [CrossRef]
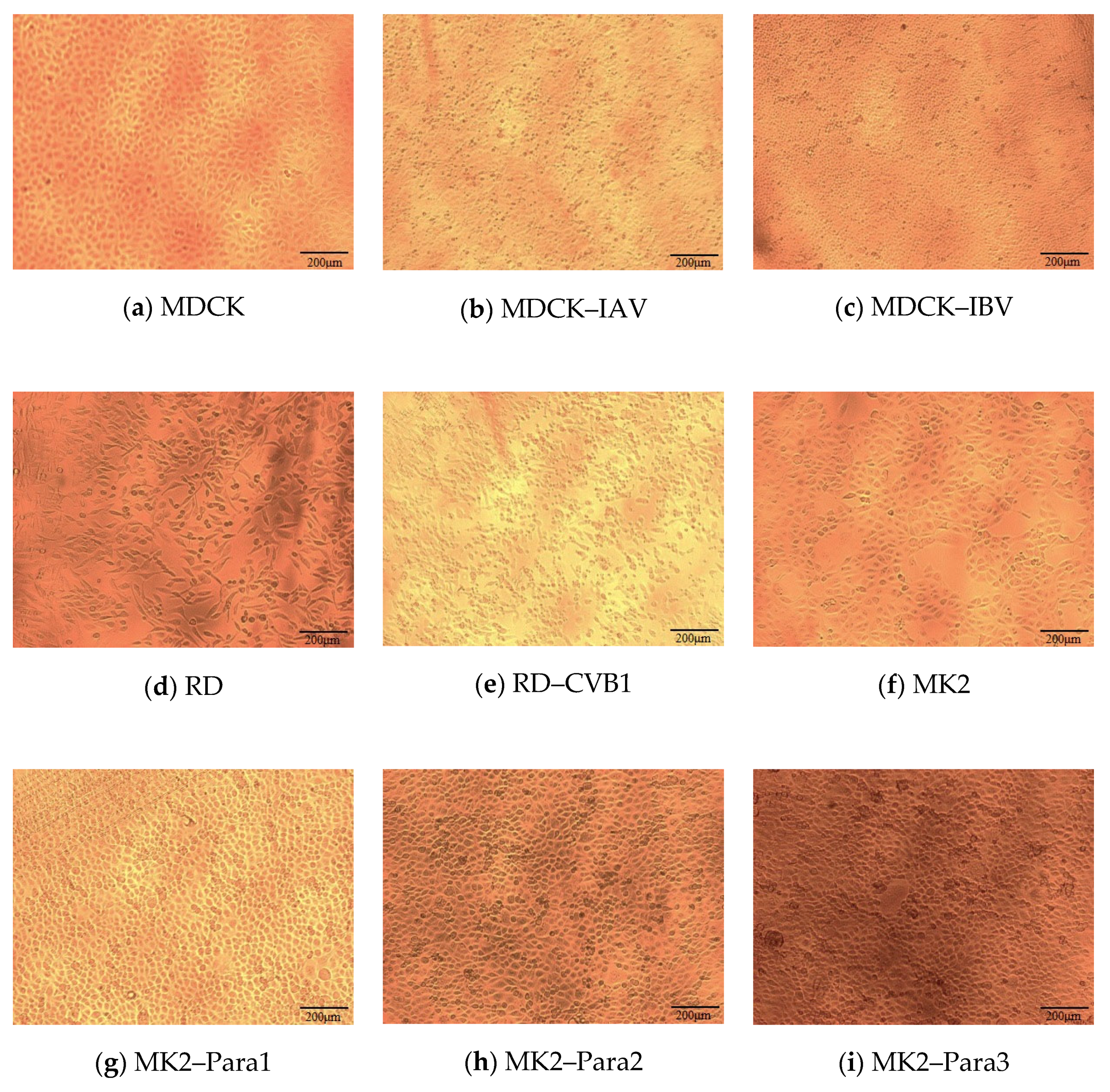


| Cell Line (Quantity) | Inoculation Cell Lines for Various Specimens (Specimen Cells) (Quantity) |
|---|---|
| MDCK (548) | IAV (929), IBV (625), HSV-1 (20), HSV-2 (20) |
| RD (672) | CVB1 (609), HSV-1 (165), HSV-2 (110) |
| HEp-2 (748) | ADV (183), RSV (405), HSV-1 (30), HSV-2 (35) |
| A549 (551) | ADV (300), HSV-1 (180), HSV-2 (50) |
| MK2 (780) | RSV (110), CVB1 (120), Para1 (645), Para2(622), Para3 (552) |
| MRC-5 (520) | CVB1 (60), HSV-1 (90), HSV-2 (60) |
| Cell Line (Quantity) | Inoculation Cell Lines for Various Specimens (Specimen Cells) (Quantity) |
|---|---|
| MDCK (2000) | IAV (2000), IBB (2000) |
| RD (2000) | CVB1 (2000) |
| MK2 (2000) | Para1 (2000), Para2 (2000), Para3 (2000) |
| No. of Samples of Each Category | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Technologist | MDCK | MDCK–IA | MDCK–IB | MK2 | MK2–Para1 | MK2–Para2 | MK2–Para3 | RD | RD–CVB1 | Total No. |
| 1 | 86 | 49 | 23 | 157 | 11 | 36 | 32 | 76 | 30 | 500 |
| 2 | 64 | 140 | 51 | 58 | 94 | 139 | 69 | 98 | 167 | 880 |
| No. of Accurate Predictions | 141 | 99 | 57 | 183 | 63 | 114 | 83 | 124 | 146 | 1010 (73.19%) |
| Our AI Model | MDCK | MDCK–IA | MDCK–IB | MK2 | MK2–Para1 | MK2–Para2 | MK2–Para3 | RD | RD–CVB1 | Total No. |
| Single | 500 | 500 | 500 | 500 | 500 | 500 | 500 | 500 | 500 | 4500 |
| Multi-Task Learning | 500 | 500 | 500 | 500 | 500 | 500 | 500 | 500 | 500 | 4500 |
| No. of Accurate Predictions | 988 | 975 | 971 | 997 | 960 | 959 | 956 | 998 | 988 | 8792 (97.69%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.-J.; Lin, P.-H.; Lin, Y.-Y.; Pu, K.-Y.; Wang, C.-F.; Lin, S.-Y.; Chen, T.-S. Detection of Cytopathic Effects Induced by Influenza, Parainfluenza, and Enterovirus Using Deep Convolution Neural Network. Biomedicines 2022, 10, 70. https://doi.org/10.3390/biomedicines10010070
Chen J-J, Lin P-H, Lin Y-Y, Pu K-Y, Wang C-F, Lin S-Y, Chen T-S. Detection of Cytopathic Effects Induced by Influenza, Parainfluenza, and Enterovirus Using Deep Convolution Neural Network. Biomedicines. 2022; 10(1):70. https://doi.org/10.3390/biomedicines10010070
Chicago/Turabian StyleChen, Jen-Jee, Po-Han Lin, Yi-Ying Lin, Kun-Yi Pu, Chu-Feng Wang, Shang-Yi Lin, and Tzung-Shi Chen. 2022. "Detection of Cytopathic Effects Induced by Influenza, Parainfluenza, and Enterovirus Using Deep Convolution Neural Network" Biomedicines 10, no. 1: 70. https://doi.org/10.3390/biomedicines10010070
APA StyleChen, J.-J., Lin, P.-H., Lin, Y.-Y., Pu, K.-Y., Wang, C.-F., Lin, S.-Y., & Chen, T.-S. (2022). Detection of Cytopathic Effects Induced by Influenza, Parainfluenza, and Enterovirus Using Deep Convolution Neural Network. Biomedicines, 10(1), 70. https://doi.org/10.3390/biomedicines10010070







